Molecular Aspects of the Emergence of Monkeypox Virus Clades
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of Genome Sequences and Alignment
2.2. Phylogenetic Tree Construction and Evolutionary Analyses
2.3. Detection of Evolutionary Pressure
3. Results
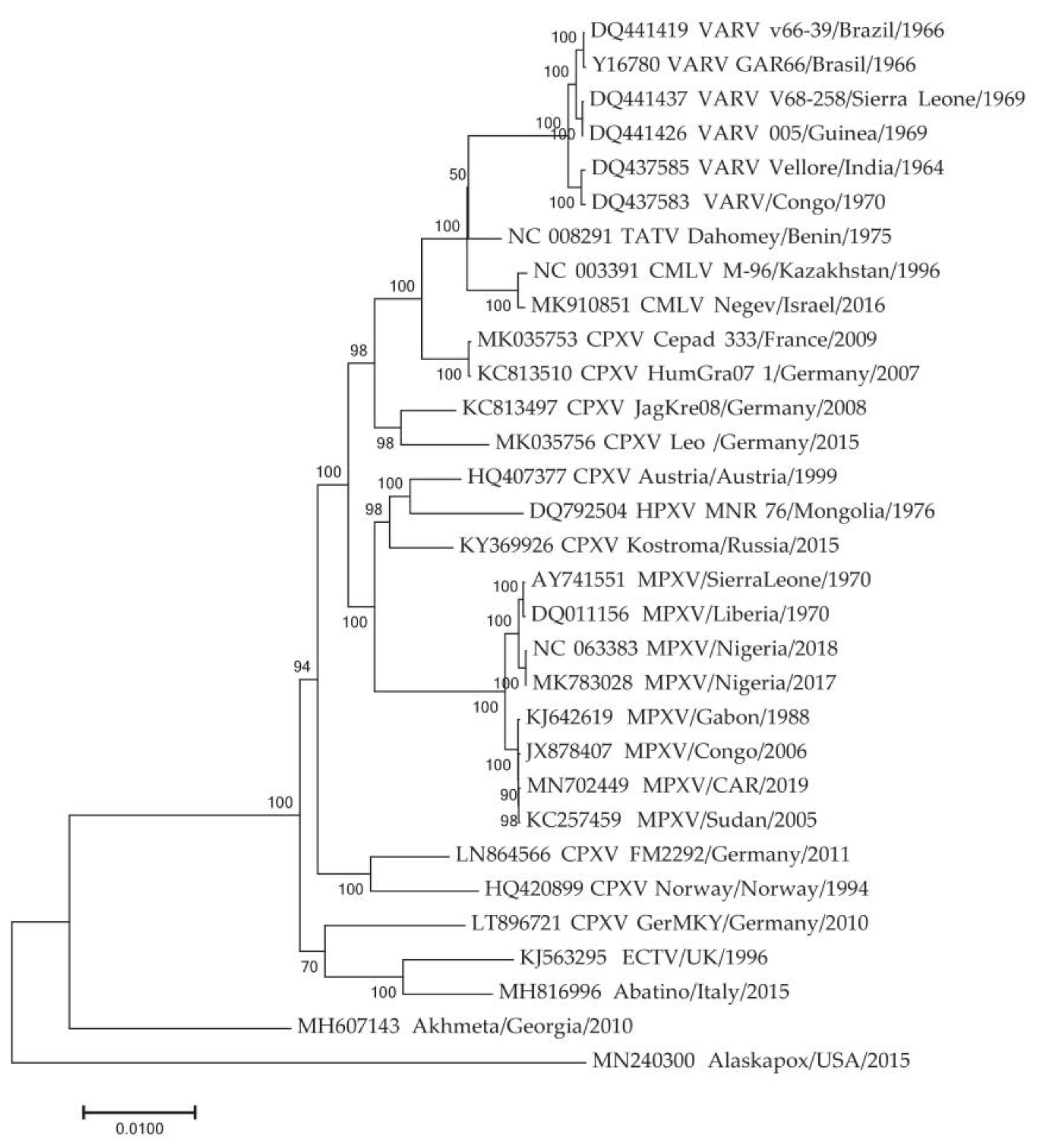
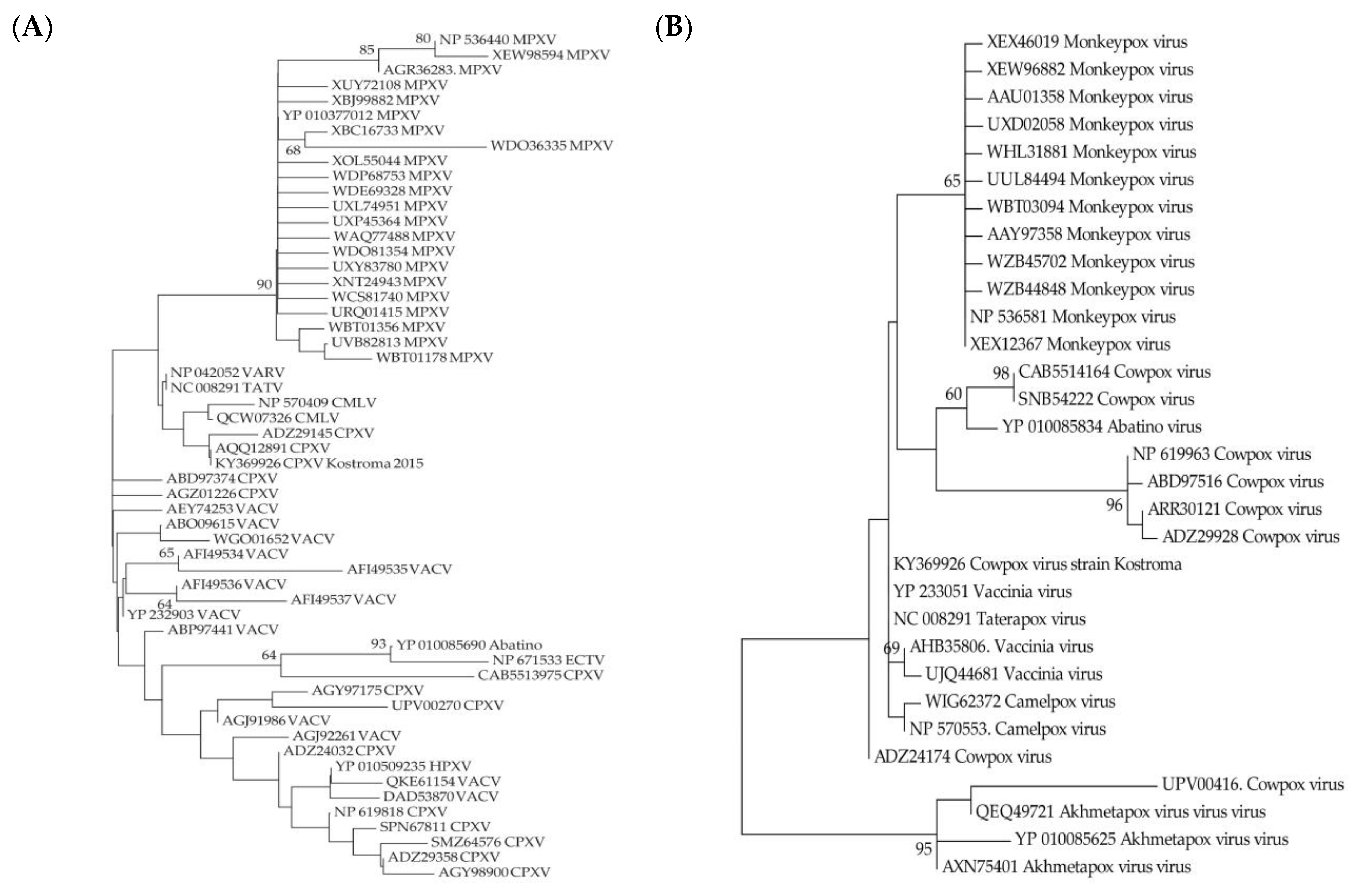
3.1. Selection of Genome Sequences
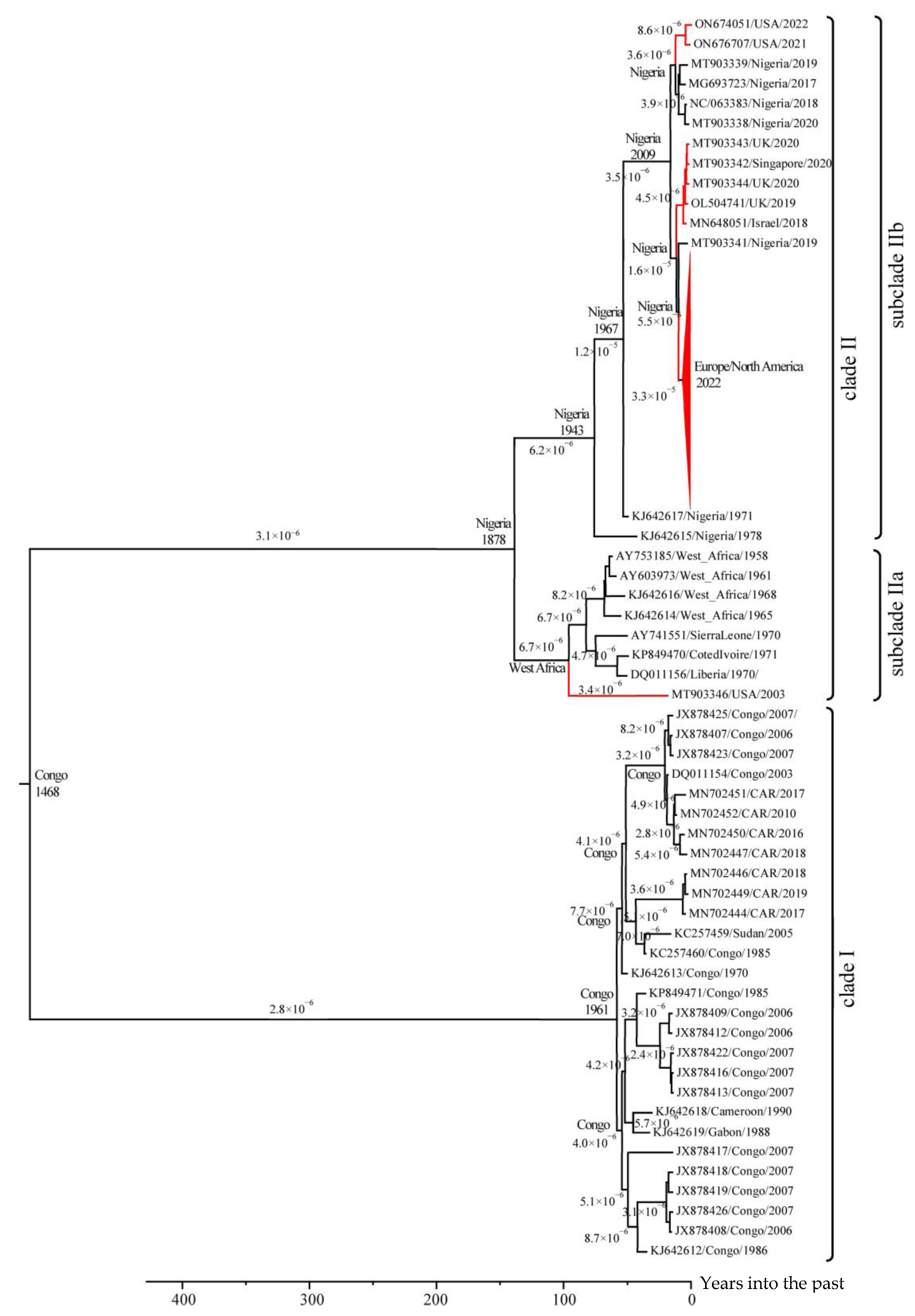
3.2. Molecular Dating in the Evolution of MPXV
3.3. Genome Content and Adaptive Selection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moss, B. Poxviridae. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 2129–2159. [Google Scholar]
- Mitjà, O.; Ogoina, D.; Titanji, B.K.; Galvan, C.; Muyembe, J.J.; Marks, M.; Orkin, C.M. Monkeypox. Lancet 2022, 400, 1603–1617. [Google Scholar] [CrossRef]
- Happi, C.; Adetifa, I.; Mbala, P.; Njouom, R.; Nakoune, E.; Happi, A.; Ndodo, N.; Ayansola, O.; Mboowa, G.; Bedford, T.; et al. Urgent Need for a Non-Discriminatory and Non-Stigmatizing Nomenclature for Monkeypox Virus. PLoS Biol. 2022, 20, e3001769. [Google Scholar] [CrossRef]
- Cho, C.T.; Wenner, H.A. Monkeypox Virus. Bacteriol. Rev. 1973, 37, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N.; Marennikova, S.S.; Moyer, R.W. Orthopoxviruses Pathogenic for Humans; Springer: Berlin, Germany, 2005. [Google Scholar]
- Von Magnus, P.; Andersen, E.K.; Petersen, K.B.; Birch-Andersen, A. A Pox-like Disease in Cynomolgus Monkeys. Acta Pathol. Microbiol. Scand. 1959, 46, 156–176. [Google Scholar] [CrossRef]
- Marennikova, S.S.; Seluhina, E.M.; Mal’ceva, N.N.; Cimiskjan, K.L.; Macevic, G.R. Isolation and Properties of the Causal Agent of a New Variola-like Disease (Monkeypox) in Man. Bull. World Health Organ. 1972, 46, 599–611. [Google Scholar]
- Ladnyj, I.D.; Ziegler, P.; Kima, E. A Human Infection Caused by Monkeypox Virus in Basankusu Territory, Democratic Republic of the Congo. Bull. World Health Organ. 1972, 46, 593–597. [Google Scholar]
- Douglass, N.; Dumbell, K. Independent Evolution of Monkeypox and Variola Viruses. J. Virol. 1992, 66, 7565–7567. [Google Scholar] [CrossRef] [PubMed]
- Esposito, J.J.; Knight, J.C. Orthopoxvirus DNA: A Comparison of Restriction Profiles and Maps. Virology 1985, 143, 230–251. [Google Scholar] [CrossRef]
- Shchelkunov, S.N.; Totmenin, A.V.; Safronov, P.F.; Mikheev, M.V.; Gutorov, V.V.; Ryazankina, O.I.; Petrov, N.A.; Babkin, I.V.; Uvarova, E.A.; Sandakhchiev, L.S.; et al. Analysis of the Monkeypox Virus Genome. Virology 2002, 297, 172–194. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N.; Totmenin, A.V.; Babkin, I.V.; Safronov, P.F.; Ryazankina, O.I.; Petrov, N.A.; Gutorov, V.V.; Uvarova, E.A.; Mikheev, M.V.; Sisler, J.R.; et al. Human Monkeypox and Smallpox Viruses: Genomic Comparison. FEBS Lett. 2001, 509, 66–70. [Google Scholar] [CrossRef]
- Esteban, D.J.; Hutchinson, A.P. Genes in the Terminal Regions of Orthopoxvirus Genomes Experience Adaptive Molecular Evolution. BMC Genomics. 2011, 12, 261. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Wang, C.; Upton, C. Poxviruses: Past, Present and Future. Virus Res. 2006, 117, 105–118. [Google Scholar] [CrossRef]
- Douglass, N.J.; Richardson, M.; Dumbell, K.R. Evidence for Recent Genetic Variation in Monkeypox Viruses. J. Gen. Virol. 1994, 75, 1303–1309. [Google Scholar] [CrossRef]
- Likos, A.M.; Sammons, S.A.; Olson, V.A.; Frace, A.M.; Li, Y.; Olsen-Rasmussen, M.; Davidson, W.; Galloway, R.; Khristova, M.L.; Reynolds, M.G.; et al. A Tale of Two Clades: Monkeypox Viruses. J. Gen. Virol. 2005, 86, 2661–2672. [Google Scholar] [CrossRef] [PubMed]
- Vakaniaki, E.H.; Kacita, C.; Kinganda-Lusamaki, E.; O’Toole, Á.; Wawina-Bokalanga, T.; Mukadi-Bamuleka, D.; Amuri-Aziza, A.; Malyamungu-Bubala, N.; Mweshi-Kumbana, F.; Mutimbwa-Mambo, L.; et al. Sustained Human Outbreak of a New MPXV Clade I Lineage in Eastern Democratic Republic of the Congo. Nat. Med. 2024, 30, 2791–2795. [Google Scholar] [CrossRef] [PubMed]
- Wawina-Bokalanga, T.; Akil-Bandali, P.; Kinganda-Lusamaki, E.; Lokilo, E.; Jansen, D.; Amuri-Aziza, A.; Makangara-Cigolo, J.C.; Pukuta-Simbu, E.; Ola-Mpumbe, R.; Muyembe, M.; et al. Co-circulation of Monkeypox Virus Subclades Ia and Ib in Kinshasa Province, Democratic Republic of the Congo, July to August 2024. Euro Surveill. 2024, 29, 2400592. [Google Scholar] [CrossRef]
- Bunge, E.M.; Hoet, B.; Chen, L.; Lienert, F.; Weidenthaler, H.; Baer, L.R.; Steffen, R. The Changing Epidemiology of Human Monkeypox—A Potential Threat? A Systematic Review. PLoS Neglected Trop. Dis. 2022, 16, e0010141. [Google Scholar] [CrossRef] [PubMed]
- Americo, J.L.; Earl, P.L.; Moss, B. Virulence Differences of Mpox (Monkeypox) Virus Clades I, IIa, and IIb.1 in a Small Animal Model. Proc. Natl. Acad. Sci. USA 2023, 120, e2220415120. [Google Scholar] [CrossRef]
- Alakunle, E.; Kolawole, D.; Diaz-Canova, D.; Alele, F.; Adegboye, O.; Moens, U.; Okeke, M.I. A Comprehensive Review of Monkeypox Virus and Mpox Characteristics. Front. Cell. Infect. Microbiol. 2024, 14, 1360586. [Google Scholar] [CrossRef]
- Mandja, B.M.; Brembilla, A.; Handschumacher, P.; Bompangue, D.; Gonzalez, J.P.; Muyembe, J.J.; Mauny, F.; Mombouli, J.V.; Gessain, A.; Deharo, E.; et al. Temporal and Spatial Dynamics of Monkeypox in Democratic Republic of Congo, 2000–2015. EcoHealth 2019, 16, 476–487. [Google Scholar] [CrossRef]
- Jezek, Z.; Szczeniowski, M.; Paluku, K.M.; Mutombo, M. Human Monkeypox: Clinical Features of 282 Patients. J. Infect. Dis. 1987, 156, 293–298. [Google Scholar] [CrossRef]
- Yinka-Ogunleye, A.; Aruna, O.; Dalhat, M.; Ogoina, D.; McCollum, A.; Disu, Y.; Mamadu, I.; Akinpelu, A.; Ahmad, A.; Burga, J.; et al. Outbreak of Human Monkeypox in Nigeria in 2017–18: A Clinical and Epidemiological Report. Lancet Infect. Dis. 2019, 19, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.D.; Melski, J.W.; Graham, M.B.; Regnery, R.L.; Sotir, M.J.; Wegner, M.V.; Kazmierczak, J.J.; Stratman, E.J.; Li, Y.; Fairley, J.A.; et al. The Detection of Monkeypox in Humans in the Western Hemisphere. N. Engl. J. Med. 2004, 350, 342–350. [Google Scholar] [CrossRef]
- Huhn, G.D.; Bauer, A.M.; Yorita, K.; Graham, M.B.; Sejvar, J.; Likos, A.; Damon, I.K.; Reynolds, M.G.; Kuehnert, M.J. Clinical Characteristics of Human Monkeypox, and Risk Factors for Severe Disease. Clin. Infect. Dis. 2005, 41, 1742–1751. [Google Scholar] [CrossRef] [PubMed]
- Erez, N.; Achdout, H.; Milrot, E.; Schwartz, Y.; Wiener-Well, Y.; Paran, N.; Politi, B.; Tamir, H.; Israely, T.; Weiss, S.; et al. Diagnosis of Imported Monkeypox, Israel, 2018. Emerg. Infect. Dis. 2019, 25, 980–983. [Google Scholar] [CrossRef] [PubMed]
- Enserink, M. Infectious diseases. U.S. monkeypox outbreak traced to Wisconsin pet dealer. Science 2003, 300, 1639. [Google Scholar] [CrossRef]
- Song, Y.; Yan, Y.; Xu, J.; Lv, S.; Ren, G.; Zhou, Y.; Song, W.; Ge, R.; Xu, P.; Zhu, G.; et al. Complete Genome Sequence Analysis of the First Imported Mpox Virus Clade Ib Variant in China. Pathogens 2025, 14, 102. [Google Scholar] [CrossRef]
- Marennikova, S.S.; Seluhina, E.M. Susceptibility of Some Rodent Species to Monkeypox Virus, and Course of the Infection. Bull. World Health Organ. 1976, 53, 13–20. [Google Scholar]
- Doty, J.B.; Malekani, J.M.; Kalemba, L.N.; Stanley, W.T.; Monroe, B.P.; Nakazawa, Y.U.; Mauldin, M.R.; Bakambana, T.L.; Liyandja Dja Liyandja, T.; Braden, Z.H.; et al. Assessing Monkeypox Virus Prevalence in Small Mammals at the Human–Animal Interface in the Democratic Republic of the Congo. Viruses 2017, 9, 283. [Google Scholar] [CrossRef]
- Parker, S.; Nuara, A.; Buller, R.M.L.; Schultz, D.A. Human Monkeypox: An Emerging Zoonotic Disease. Future Microbiol. 2007, 2, 17–34. [Google Scholar] [CrossRef]
- Grant, R.; Nguyen, L.L.; Breban, R. Modelling Human-to-Human Transmission of Monkeypox. Bull. World Health Organ. 2020, 98, 638–640. [Google Scholar] [CrossRef]
- Zahmatyar, M.; Fazlollahi, A.; Motamedi, A.; Zolfi, M.; Seyedi, F.; Nejadghaderi, S.A.; Sullman, M.J.M.; Mohammadinasab, R.; Kolahi, A.A.; Arshi, S.; et al. Human Monkeypox: History, Presentations, Transmission, Epidemiology, Diagnosis, Treatment, and Prevention. Front. Med. 2023, 10, 1157670. [Google Scholar] [CrossRef]
- Jezek, Z.; Gromyko, A.I.; Szczeniowski, M.V. Human Monkeypox. J. Hyg. Epidemiol. Microbiol. Immunol. 1983, 27, 13–28. [Google Scholar]
- Branda, F.; Pierini, M.; Mazzoli, S. Monkeypox: Early Estimation of Basic Reproduction Number R0 in Europe. J. Med. Virol. 2023, 95, e28270. [Google Scholar] [CrossRef]
- Seang, S.; Burrel, S.; Todesco, E.; Leducq, V.; Monsel, G.; Le Pluart, D.; Cordevant, C.; Pourcher, V.; Palich, R. Evidence of Human-to-Dog Transmission of Monkeypox Virus. Lancet 2022, 400, 658–659. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Shapiro, B.; Rambaut, A.; Drummond, A.J. Choosing Appropriate Substitution Models for the Phylogenetic Analysis of Protein-Coding Sequences. Mol. Biol. Evol. 2006, 23, 7–9. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple Methods for Estimating the Numbers of Synonymous and Nonsynonymous Nucleotide Substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Yutin, N.; Wolf, Y.I.; Koonin, E.V.; Moss, B. Ancient Gene Capture and Recent Gene Loss Shape the Evolution of Orthopoxvirus-Host Interaction Genes. mBio 2021, 12, e0149521. [Google Scholar] [CrossRef]
- Posada, D. How does recombination affect phylogeny estimation? Trends Ecol. Evol. 2000, 15, 489–490. [Google Scholar] [CrossRef]
- Esposito, J.J.; Sammons, S.A.; Frace, A.M.; Osborne, J.D.; Olsen-Rasmussen, M.; Zhang, M.; Govil, D.; Damon, I.K.; Kline, R.; Laker, M.; et al. Genome Sequence Diversity and Clues to the Evolution of Variola (Smallpox) Virus. Science 2006, 313, 807–812. [Google Scholar] [CrossRef]
- Gubser, C.; Hue, S.; Kellam, P.; Smith, G.L. Poxvirus Genomes: A Phylogenetic Analysis. J. Gen. Virol. 2004, 85, 105–117. [Google Scholar] [CrossRef]
- Franke, A.; Pfaff, F.; Jenckel, M.; Hoffmann, B.; Höper, D.; Antwerpen, M.; Meyer, H.; Beer, M.; Hoffmann, D. Classification of Cowpox Viruses into Several Distinct Clades and Identification of a Novel Lineage. Viruses 2017, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- Farlow, J.; Ichou, M.A.; Huggins, J.; Ibrahim, S. Comparative Whole Genome Sequence Analysis of Wild-Type and Cidofovir-Resistant Monkeypoxvirus. Virol. J. 2010, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Babkin, I.V.; Babkina, I.N.; Tikunova, N.V. An Update of Orthopoxvirus Molecular Evolution. Viruses 2022, 14, 388. [Google Scholar] [CrossRef]
- Gigante, C.M.; Korber, B.; Seabolt, M.H.; Wilkins, K.; Davidson, W.; Rao, A.K.; Zhao, H.; Smith, T.G.; Hughes, C.M.; Minhaj, F.S.; et al. Multiple Lineages of Monkeypox Virus Detected in the United States, 2021–2022. Science 2022, 378, 560–565. [Google Scholar] [CrossRef]
- Mauldin, M.R.; McCollum, A.M.; Nakazawa, Y.J.; Mandra, A.; Whitehouse, E.R.; Davidson, W.; Zhao, H.; Gao, J.; Li, Y.; Doty, J.; et al. Exportation of Monkeypox Virus from the African Continent. J. Infect. Dis. 2022, 225, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Costello, V.; Sowash, M.; Gaur, A.; Cardis, M.; Pasieka, H.; Wortmann, G.; Ramdeen, S. Imported Monkeypox from International Traveler, Maryland, USA, 2021. Emerg. Infect. Dis. 2022, 28, 1002–1005. [Google Scholar] [CrossRef]
- McLysaght, A.; Baldi, P.F.; Gaut, B.S. Extensive Gene Gain Associated with Adaptive Evolution of Poxviruses. Proc. Natl. Acad. Sci. USA 2003, 100, 15655–15660. [Google Scholar] [CrossRef]
- Popova, A.Y.; Maksyutov, R.A.; Taranov, O.S.; Tregubchak, T.V.; Zaikovskaya, A.V.; Sergeev, A.A.; Vlashchenko, I.V.; Bodnev, S.A.; Ternovoi, V.A.; Alexandrova, N.S.; et al. Cowpox in a Human, Russia, 2015. Epidemiol. Infect. 2017, 145, 755–759. [Google Scholar] [CrossRef]
- Meng, X.; Krumm, B.; Li, Y.; Deng, J.; Xiang, Y. Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 14858–14863. [Google Scholar] [CrossRef]
- Strnadova, P.; Ren, H.; Valentine, R.; Mazzon, M.; Sweeney, T.R.; Brierley, I.; Smith, G.L. Inhibition of Translation Initiation by Protein 169: A Vaccinia Virus Strategy to Suppress Innate and Adaptive Immunity and Alter Virus Virulence. PLoS Pathog. 2015, 11, e1005151. [Google Scholar] [CrossRef] [PubMed]
- Herbert, M.H.; Squire, C.J.; Mercer, A.A. Poxviral Ankyrin Proteins. Viruses 2015, 7, 709–738. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.L.; Benfield, C.T.O.; Maluquer de Motes, C.; Mazzon, M.; Ember, S.W.J.; Ferguson, B.J.; Sumner, R.P. Vaccinia Virus Immune Evasion: Mechanisms, Virulence and Immunogenicity. J. Gen. Virol. 2013, 94, 2367–2392. [Google Scholar] [CrossRef]
- Goebel, S.J.; Johnson, G.P.; Perkus, M.E.; Davis, S.W.; Winslow, J.P.; Paoletti, E. The Complete DNA Sequence of Vaccinia Virus. Virology 1990, 179, 247–266. [Google Scholar] [CrossRef]
- Schröder, M.; Baran, M.; Bowie, A.G. Viral Targeting of DEAD Box Protein 3 Reveals Its Role in TBK1/IKKε-Mediated IRF Activation. EMBO J. 2008, 27, 2147–2157. [Google Scholar] [CrossRef]
- Zhai, D.; Yu, E.; Jin, C.; Welsh, K.; Shiau, C.W.; Chen, L.; Salvesen, G.S.; Liddington, R.; Reed, J.C. Vaccinia Virus Protein F1L Is a Caspase-9 Inhibitor. J. Biol. Chem. 2010, 285, 5569–5580. [Google Scholar] [CrossRef]
- Froggatt, G.C.; Smith, G.L.; Beard, P.M. Vaccinia Virus Gene F3L Encodes an Intracellular Protein That Affects the Innate Immune Response. J. Gen. Virol. 2007, 88, 1917–1921. [Google Scholar] [CrossRef]
- Leite, J.A.; da Fonseca, F.G.; de Souza Trindade, G.; Abrahão, J.S.; Arantes, R.M.; de Almeida-Leite, C.M.; dos Santos, J.R.; Guedes, M.I.; Ribeiro, B.M.; Bonjardim, C.A.; et al. A-Type Inclusion Bodies: A Factor Influencing Cowpox Virus Lesion Pathogenesis. Arch. Virol. 2011, 156, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Wolffe, E.J.; Weisberg, A.S.; Moss, B. The Vaccinia Virus A33R Protein Provides a Chaperone Function for Viral Membrane Localization and Tyrosine Phosphorylation of the A36R Protein. J. Virol. 2001, 75, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Rehm, K.E.; Connor, R.F.; Jones, G.J.; Yimbu, K.; Roper, R.L. Vaccinia Virus A35R Inhibits MHC Class II Antigen Presentation. Virology 2010, 397, 176–186. [Google Scholar] [CrossRef]
- Van Eijl, H.; Hollinshead, M.; Smith, G.L. The Vaccinia Virus A36R Protein Is a Type Ib Membrane Protein Present on Intracellular but Not Extracellular Enveloped Virus Particles. Virology 2000, 271, 26–36. [Google Scholar] [CrossRef]
- Sood, C.L.; Moss, B. Vaccinia Virus A43R Gene Encodes an Orthopoxvirus-Specific Late Non-Virion Type-1 Membrane Protein That Is Dispensable for Replication but Enhances Intradermal Lesion Formation. Virology 2010, 396, 160–168. [Google Scholar] [CrossRef] [PubMed]
- DeHaven, B.C.; Gupta, K.; Isaacs, S.N. The Vaccinia Virus A56 Protein: A Multifunctional Transmembrane Glycoprotein That Anchors Two Secreted Viral Proteins. J. Gen. Virol. 2011, 92, 1971–1980. [Google Scholar] [CrossRef]
- Rico, A.B.; Linville, A.C.; Olson, A.T.; Wang, Z.; Wiebe, M.S. The Vaccinia Virus B12 Pseudokinase Represses Viral Replication via Interaction with the Cellular Kinase VRK1 and Activation of the Antiviral Effector BAF. J. Virol. 2021, 95, e02114–e02120. [Google Scholar] [CrossRef]
- Alcamí, A.; Symons, J.A.; Smith, G.L. The Vaccinia Virus Solable Alpha/Beta Interferon (IFN) Receptor Binds to the Cell Surface and Protects Cells from the Antiviral Effects of IFN. J. Virol. 2000, 74, 11230–11239. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Moss, B. Repair of a Previously Uncharacterized Second Host-Range Gene Contributes to Full Replication of Modified Vaccinia Virus Ankara (MVA) in Human Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 3759–3767. [Google Scholar] [CrossRef] [PubMed]
- Isidro, J.; Borges, V.; Pinto, M.; Sobral, D.; Santos, J.D.; Nunes, A.; Mixão, V.; Ferreira, R.; Santos, D.; Duarte, S.; et al. Phylogenomic Characterization and Signs of Microevolution in the 2022 Multi-Country Outbreak of Monkeypox Virus. Nat. Med. 2022, 28, 1569–1572. [Google Scholar] [CrossRef]
- Nolen, L.D.; Osadebe, L.; Katomba, J.; Likofata, J.; Mukadi, D.; Monroe, B.; Doty, J.; Hughes, C.M.; Kabamba, J.; Malekani, J.; et al. Extended Human-to-Human Transmission during a Monkeypox Outbreak in the Democratic Republic of the Congo. Emerg. Infect. Dis. 2016, 22, 1014–1021. [Google Scholar] [CrossRef]
- Sereewit, J.; Lieberman, N.A.P.; Xie, H.; Bakhash, S.A.K.M.; Nunley, B.E.; Chung, B.; Mills, M.G.; Roychoudhury, P.; Greninger, A.L. ORF-Interrupting Mutations in Monkeypox Virus Genomes from Washington and Ohio, 2022. Viruses 2022, 14, 2393. [Google Scholar] [CrossRef]
- Molteni, C.; Forni, D.; Cagliani, R.; Mozzi, A.; Clerici, M.; Sironi, M. Evolution of the Orthopoxvirus Core Genome. Virus Res. 2022, 323, 198975. [Google Scholar] [CrossRef]
- Kryazhimskiy, S.; Plotkin, J.B. The Population Genetics of dN/dS. PLoS Genet. 2008, 4, e1000304. [Google Scholar] [CrossRef]
- Manes, N.P.; Estep, R.D.; Mottaz, H.M.; Moore, R.J.; Clauss, T.R.; Monroe, M.E.; Du, X.; Adkins, J.N.; Wong, S.W.; Smith, R.D. Comparative proteomics of human monkeypox and vaccinia intracellular mature and extracellular enveloped virions. J. Proteome Res. 2008, 7, 960–968. [Google Scholar] [CrossRef]
- Sagdat, K.; Batyrkhan, A.; Kanayeva, D. Exploring monkeypox virus proteins and rapid detection techniques. Front. Cell. Infect. Microbiol. 2024, 14, 1414224. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.K.; Turner, P.C.; Moyer, R.W. Molecular characterization of the vaccinia virus hemagglutinin gene. J. Virol. 1991, 65, 3598–3606. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Oie, M.; Ichihashi, Y.; Shida, H. Hemadsorption and fusion inhibition activities of hemagglutinin analyzed by vaccinia virus mutants. Virology 1990, 175, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D.S.; Emerson, G.L.; Li, Y.; Sammons, S.; Olson, V.; Frace, M.; Nakazawa, Y.; Czerny, C.P.; Tryland, M.; Kolodziejek, J.; et al. Chasing Jenner’s Vaccine: Revisiting Cowpox Virus Classification. PLoS ONE 2011, 6, e23086. [Google Scholar] [CrossRef] [PubMed]

| # | Gene Name a | Gene Name b | Function | The Gene Is Missing in MPXV |
|---|---|---|---|---|
| 1. | D3L | OPG003 | ankyrin repeat-containing protein | clade I |
| 2. | D4L | OPG004 | ankyrin repeat-containing protein | all strains |
| 3. | D5L | OPG005 | Bcl-2-like protein | all strains |
| 4. | D6L | OPG006 | alpha-amanitin target protein | all strains |
| 5. | D7L | OPG008 | kelch-like protein | all strains |
| 6. | D8L | OPG009 | ankyrin repeat-containing protein | all strains |
| 7. | D9L | - | C-type lectin-like protein | all strains |
| 8. | D10L | - | C-type lectin domain-containing protein | all strains |
| 9. | D11L | OPG011 | kelch-like protein | all strains |
| 10. | D12L | OPG012 | TNF-alpha-receptor-like protein | all strains |
| 11. | D13L | OPG013 | TNF-alpha-receptor-like protein | all strains |
| 12. | D14L | OPG014 | ankyrin repeat-containing protein | all strains |
| 13. | C2L | OPG016 | MHC class I-like protein | clade I and IIb |
| 14. | C3L | OPG017 | ankyrin repeat-containing protein | all strains |
| 15. | C4L | OPG018 | host-range protein | all strains |
| 16. | C6L | OPG020 | IL-1 receptor antagonist | all strains |
| 17. | C12L | OPG026 | TNF-rcpt-II_C domain containing protein | all strains |
| 18. | C17L | OPG032 | secreted complement-binding protein | clade II |
| 19. | C18L | OPG033 | kelch-like protein | all strains |
| 20. | C19L | OPG034 | Bcl-2-like protein | all strains |
| 21. | P1L | OPG037 | ankyrin-like protein | all strains |
| 22. | M1L | OPG039 | ankyrin-like protein | strain UTC |
| 23. | A41R | OPG168 | semaphorin-like protein | all strains |
| 24. | A42R | OPG169 | C-type lectin-like type-II membrane protein | all strains |
| 25. | A50L | OPG177 | immunoprevalent protein | all strains |
| 26. | A52R | OPG179 | Bcl-2-like protein | all strains |
| 27. | A55R | OPG182 | Toll-IL receptor-like protein | all strains |
| 28. | A56R | OPG183 | secreted TNF-receptor-like protein | all strains |
| 29. | A57R | OPG184 | kelch-like protein | all strains |
| 30. | A59R | OPG186 | guanylate kinase | all strains |
| 31. | B8R | OPG195 | ER-localized apoptosis regulator | clade IIa |
| 32. | B9R | OPG196 | Kelch-like protein | all strains |
| 33. | B14R | OPG201 | IL-beta-binding protein | clade IIa |
| 34. | B15L | OPG202 | IL-1-beta-inhibitor | all strains |
| 35. | B16R | OPG203 | ankyrin-like protein | all strains |
| 36. | B19R | OPG206 | Kelch-like protein | all strains |
| 37. | K1R | OPG211 | ankyrin repeat containing protein | all strains |
| 38. | K2R | OPG212 | TNF-alpha-receptor-like protein | all strains |
| 39. | K3R | OPG213 | TNF-alpha-receptor-like protein | all strains |
| 40. | T1R | OPG214 | Golgi antiapoptotic protein | all strains |
| 41. | I2R | OPG004 | ankyrin repeat-containing protein | all strains |
| # | Gene Name a | Gene Name b | Function | Strains for Which Adaptive Selection Is Shown | Gene Name c/Ref. |
|---|---|---|---|---|---|
| 1. | C9L | OPG023 | Host range, ankyrin repeat-containing protein | some isolates of the clade IIa MPXV | -/[61] |
| 2. | C11L | OPG025 | Host range, ankyrin repeat-containing protein | some isolates of the clade IIa MPXV | C9L/[61] |
| 3. | C13L | OPG027 | Interferon antagonist, host-range protein, likely involved in host immune evasion | CPXV when compared with clade II MPXV | C7L/[62] |
| 4. | C16L | OPG031 | IL-1 receptor antagonist | W-Nigeria when compared with clade I MPXV | C4L/[63] |
| 5. | M6R | OPG044 | Bcl-2-like protein which, through its interaction with the DEAD box RNA helicase DDX3X/DDX3, prevents TBK1/IKKepsilon-mediated IRF3 activation, contributes to virulence by binding to the host TRAF6 and IRAK2 and preventing host NF-kappa-B activation | clade I when compared with clade II MPXV | K7R/[64] |
| 6. | G1L | OPG045 | Caspase-9 inhibitor, protein with a BCL2-like fold which is essential for survival of infected cells. | some isolates of the clade IIa MPXV | F1L/[65] |
| 7. | G3L | OPG047 | Kelch-like protein, intracellular protein that affects the innate immune response | some isolates of the clade I MPXV | F3L/[66] |
| 8. | A26L | OPG152 | cowpox A-type inclusion protein | some isolates of the clade IIa MPXV | -/[67] |
| 9. | A27L | OPG153 | cowpox A-type inclusion protein | some isolates of the clade IIa MPXV | A26L/[67] |
| 10. | A34R | OPG161 | EEV membrane phosphoglycoprotein coordinates the incorporation of A36 into intracellular enveloped virion (IEV) membranes and, subsequently, the production of actin tails, therefore plays an essential role in efficient cell-to-cell spread of viral particles | some isolates of the clade IIa when compared with clade IIb MPXV | A33R/[68] |
| 11. | A36R | OPG163 | MHC class II antigen presentation inhibitor | some isolates of the clade IIa when compared with clade I MPXV | A35R/[69] |
| 12. | A37R | OPG164 | IEV transmembrane phosphoprotein | strain Zaire-96-I-16 when compared with clade I MPXV | A36R/[70] |
| 13. | A45R | OPG172 | Type-I membrane glycoprotein | some isolates of the clade IIa MPXV | A43R/[71] |
| 14. | A46R | OPG173 | Plays a role in the inhibition of host protein synthesis. Specifically, it inhibits the initiation of cap-dependent and cap-independent translation. In turn, it affects the outcome of infection by decreasing recruitment of inflammatory leukocytes and reducing the memory CD8+ T-cell response. Localizes in cytoplasmic structures that differ from virus factories. | CPXV when compared with clades I и IIa MPXV | -/[60] |
| 15. | A58R | OPG185 | Hemagglutinin protein is capable of binding two viral proteins, a serine protease inhibitor (K2) and the vaccinia virus complement control protein (VCP), and anchoring them to the surface of infected cells. | clade IIa when compared with clade IIb MPXV | A56R/[72] |
| 16. | B5R | OPG191 | Ankyrin repeat-containing protein | some isolates of the clade II when compared with clade I MPXV | B6R/[61] |
| 17. | B11R | OPG198 | Ser-Thr kinase protein, pseudokinase that plays a role in viral DNA replication repression by activating the antiviral protein BANF1 and inhibiting the activity of host VRK1, a cellular modulator of BANF1. | some isolates of the clade II when compared with clade I MPXV | B12R/[73] |
| 18. | B17R | OPG204 | Soluble interferon alpha/beta receptor protein | clade IIa when compared with clade IIb MPXV | B19R/[74] |
| 19. | B20R | OPG208 | Serine protease inhibitor (SPI-1), host-range protein, this viral protein may be involved in the regulation of the complement cascade | some isolates of the clade IIa when compared with clade IIb MPXV | C12L/[75] |
| 20. | B21R | OPG209 | Virulence protein, soluble TNF receptor II | clade IIa when compared with clade I MPXV | C13L/[63] |
| 21. | B22R | OPG210 | Putative membrane-associated glycoprotein | some isolates of the clades IIb и I MPXV | C14L/[63] |
| 22. | I3R | OPG003 | Ankyrin repeat-containing protein | some isolates of the clade IIa when compared with clade I MPXV | C19/[61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babkin, I.V.; Babkina, I.N.; Tikunova, N.V. Molecular Aspects of the Emergence of Monkeypox Virus Clades. Viruses 2025, 17, 1549. https://doi.org/10.3390/v17121549
Babkin IV, Babkina IN, Tikunova NV. Molecular Aspects of the Emergence of Monkeypox Virus Clades. Viruses. 2025; 17(12):1549. https://doi.org/10.3390/v17121549
Chicago/Turabian StyleBabkin, Igor V., Irina N. Babkina, and Nina V. Tikunova. 2025. "Molecular Aspects of the Emergence of Monkeypox Virus Clades" Viruses 17, no. 12: 1549. https://doi.org/10.3390/v17121549
APA StyleBabkin, I. V., Babkina, I. N., & Tikunova, N. V. (2025). Molecular Aspects of the Emergence of Monkeypox Virus Clades. Viruses, 17(12), 1549. https://doi.org/10.3390/v17121549






