Molecular and Phylogenetic Analyses of Lumpy Skin Disease Virus (LSDV) Outbreak (2021/22) in Pakistan Indicate Involvement of a Clade 1.2 LSDV Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval and Study Setting
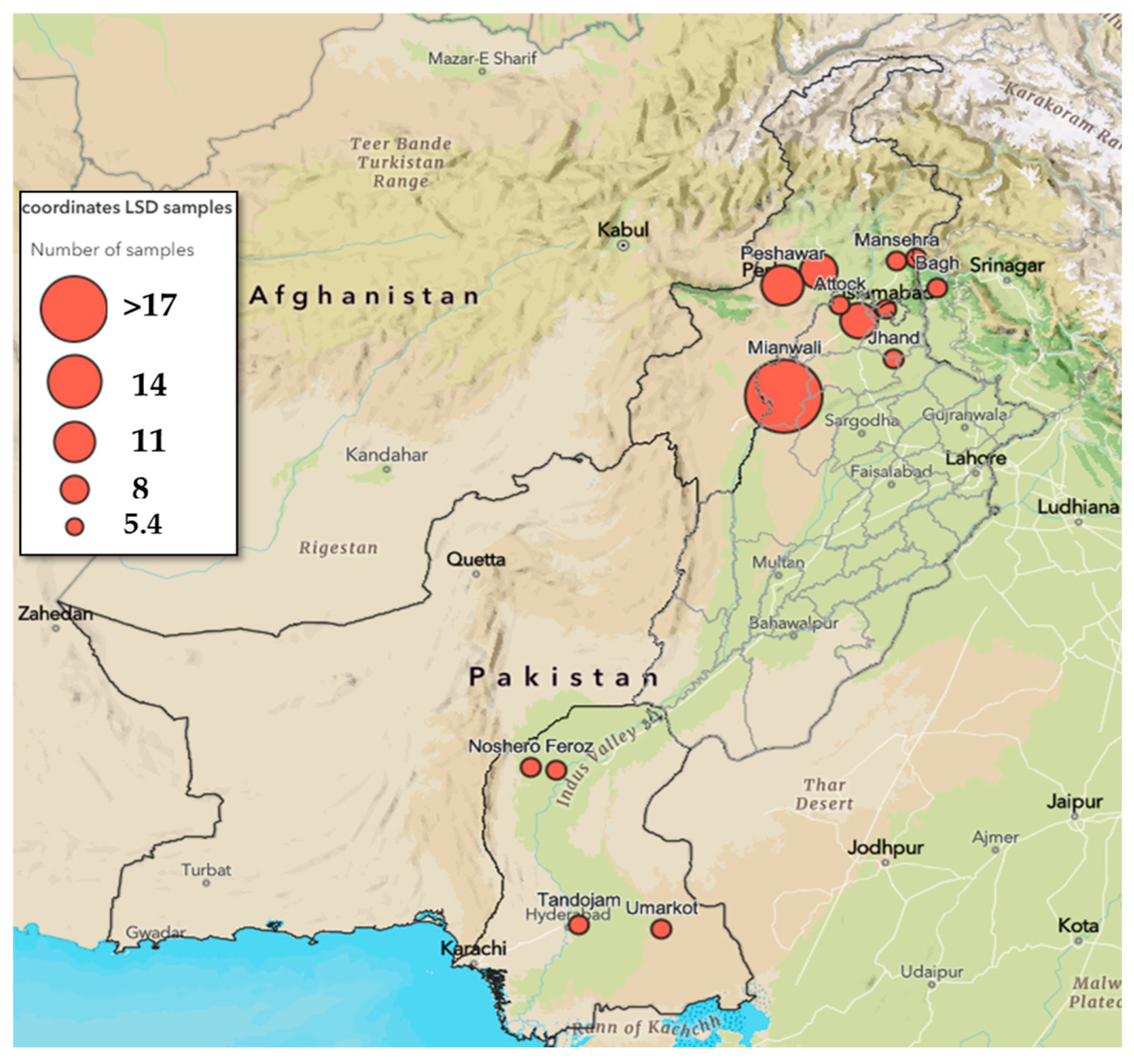
2.2. Sample Collection and Nucleic Acid Extraction
2.3. Application of Real-Time PCR Assays for Genus, Species, and Clade Identifications
2.4. Molecular Detection and Sequence Analysis of Ten Genomic Regions
2.4.1. Amplification of Selected Regions for Sequence Analysis
2.4.2. Differentiation Between KSGP0240-like Vaccine Strain and EUROPEAN/Middle Eastern LSDV Strains
2.4.3. Gel Electrophoresis, Extraction, and Sequencing
2.4.4. Phylogenetic Analysis
3. Results
3.1. Sample Quality Control and Genus Confirmation
3.2. Sequence Analysis of 10 Genomic Regions
3.3. Differentiation Between Genotype 1.2 Strains by Gel-Based PCR
3.4. Amino Acid Sequence Comparison of Amplified Regions
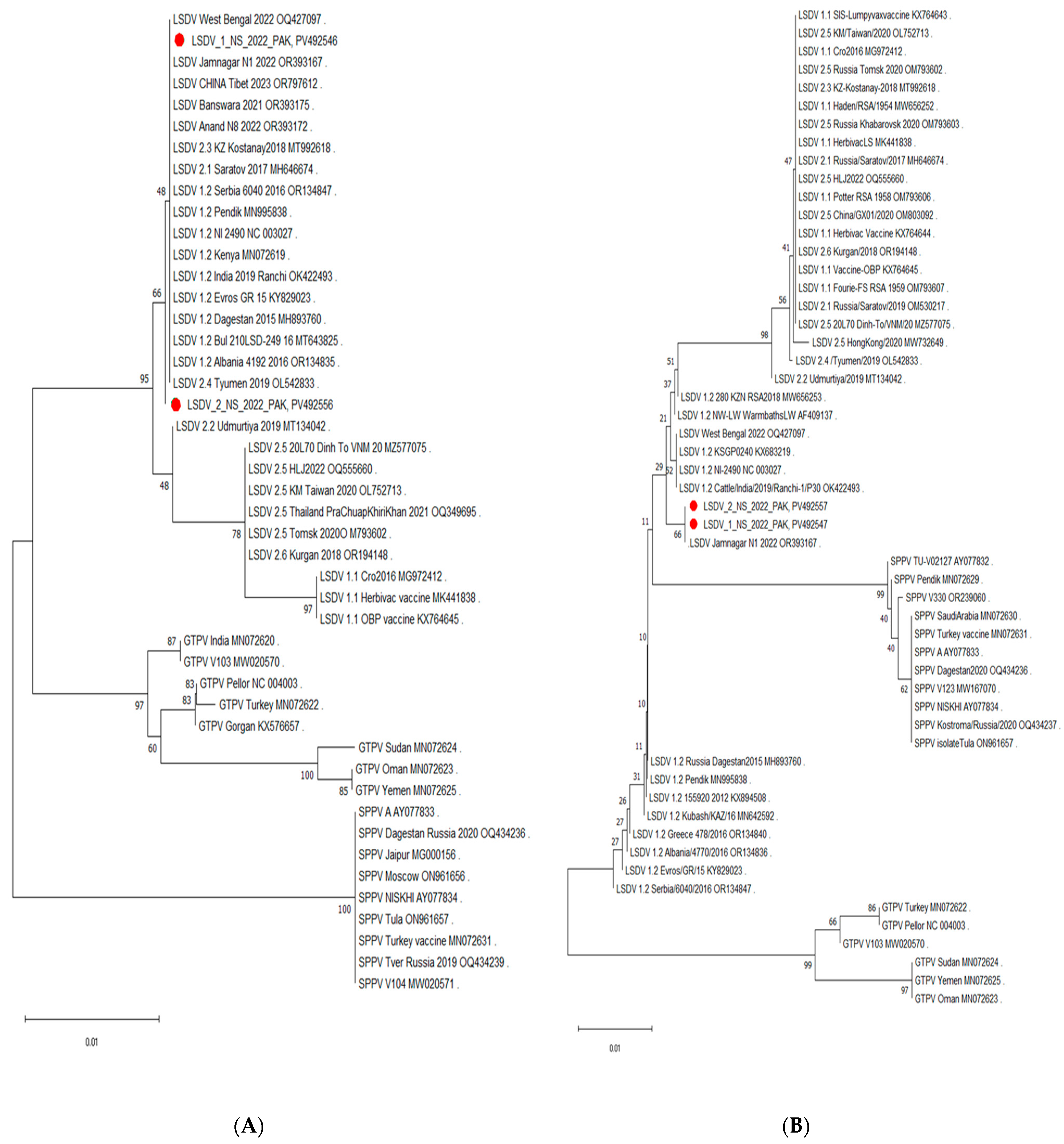
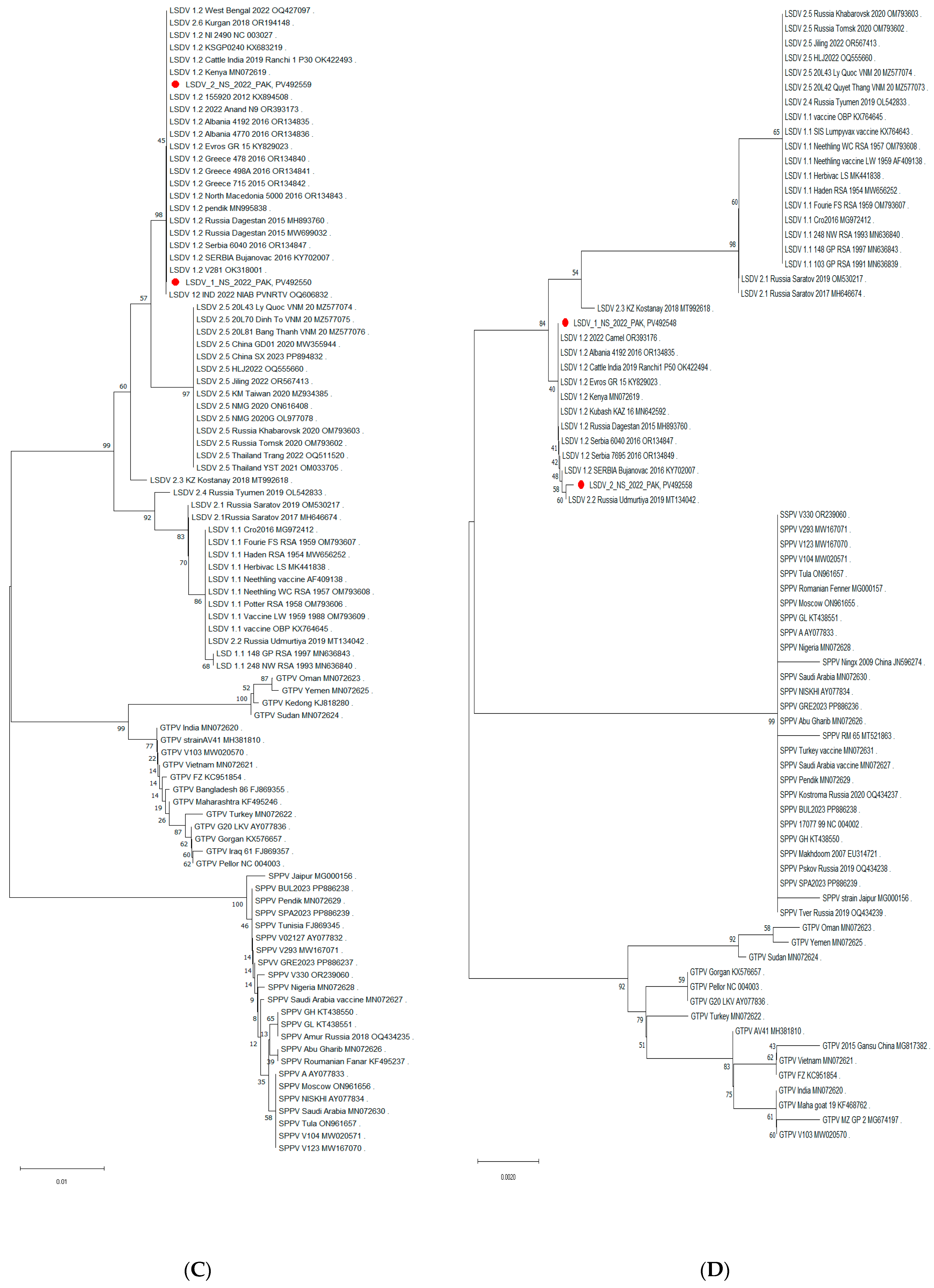
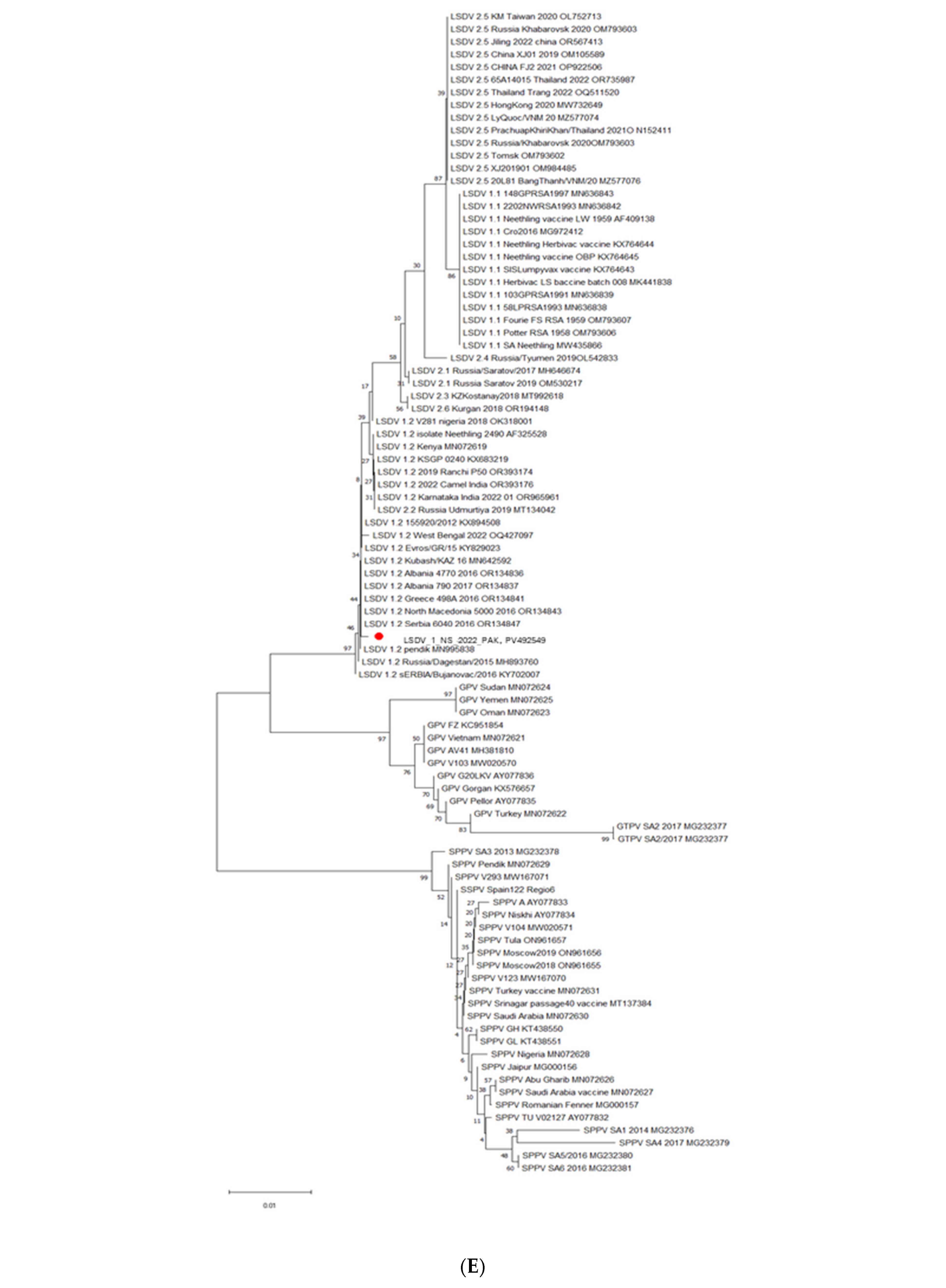
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LSDV | Lumpy Skin Disease Virus |
| LSD | Lumpy Skin Disease |
| B22R | LSDV Immunomodulatory Protein |
| GPCR | G Protein-Coupled Chemokine Receptor Gene |
| EEV | Extracellular Enveloped Virion (EEV) Proteins |
| VLTF-1 | Viral Late Transcription Factor 1 |
| RPO132 | Orf132 Gene Encodes the LSDV RNA Polymerase 132 kDa Subunit |
| RPO30 | RNA-Dependent RNA Polymerase Subunit 30 Gene |
| P32 | Protein Envelope Protein |
| KPK | Khyber Pakhtunkhwa |
| AJ and K | Azad Jammu and Kashmir |
References
- Haider, A.; Farhan, A.; Nawaz, A.; Ali, A.; Abbas, Z.; Mehmood, A. The financial toll of lumpy skin disease in Pakistan, and whether or not vaccination is worth it for preventing future outbreaks. Ann. PIMS-Shaheed Zulfiqar Ali Bhutto Med Univ. 2023, 19, 187–193. [Google Scholar] [CrossRef]
- Rehman, A.; Jingdong, L.; Chandio, A.A.; Hussain, I. Livestock production and population census in Pakistan: Determining their relationship with agricultural GDP using econometric analysis. Inf. Process. Agric. 2017, 4, 168–177. [Google Scholar] [CrossRef]
- Khan, F.A.; Mukhtar, N.; Bin Aslam, H.; Pervaiz, H.; Aziz, M.W.; Shahid, M.F.; Nawaz, M.; Durrani, A.Z.; Yaqub, T. Sequence Based Characterization of Lumpy Skin Disease Virus from Punjab, Pakistan. Pak. J. Zool. 2025, 56. [Google Scholar] [CrossRef]
- Khatri, G.; Rai, A.; Aashish; Shahzaib; Hyder, S.; Priya; Hasan, M.M. Epidemic of lumpy skin disease in Pakistan. Vet. Med. Sci. 2023, 9, 982–984. [Google Scholar] [CrossRef] [PubMed]
- Khadim, G.; Shah, A.A.; Riaz, A.; Yousaf, A.; Ali, M.; Awais, M.M.; Zafar, M.A.; Khadim, B.; Riaz, N. Epidemiology of Lumpy Skin Disease virus in large ruminants of Potohar region, Pakistan. Trop. Anim. Health Prod. 2025, 57, 310. [Google Scholar] [CrossRef]
- Badhy, S.C.; Chowdhury, M.G.A.; Settypalli, T.B.K.; Cattoli, G.; Lamien, C.E.; Fakir, M.A.U.; Akter, S.; Osmani, M.G.; Talukdar, F.; Begum, N.; et al. Molecular characterization of lumpy skin disease virus (LSDV) emerged in Bangladesh reveals unique genetic features compared to contemporary field strains. BMC Vet. Res. 2021, 17, 61. [Google Scholar] [CrossRef]
- Sprygin, A.; Babin, Y.; Pestova, Y.; Kononova, S.; Wallace, D.B.; Van Schalkwyk, A.; Byadovskaya, O.; Diev, V.; Lozovoy, D.; Kononov, A. Analysis and insights into recombination signals in lumpy skin disease virus recovered in the field. PLOS ONE 2018, 13, e0207480. [Google Scholar] [CrossRef]
- Sohier, C.; Haegeman, A.; Mostin, L.; De Leeuw, I.; Van Campe, W.; De Vleeschauwer, A.; Tuppurainen, E.S.M.; van den Berg, T.; De Regge, N.; De Clercq, K. Experimental evidence of mechanical lumpy skin disease virus transmission by Stomoxys calcitrans biting flies and Haematopota spp. horseflies. Sci. Rep. 2019, 9, 20076. [Google Scholar] [CrossRef]
- Namazi, F.; Tafti, A.K. Lumpy skin disease, an emerging transboundary viral disease: A review. Vet. Med. Sci. 2021, 7, 888–896. [Google Scholar] [CrossRef]
- Gupta, T.; Patial, V.; Bali, D.; Angaria, S.; Sharma, M.; Chahota, R. A review: Lumpy skin disease and its emergence in India. Vet. Res. Commun. 2020, 44, 111–118. [Google Scholar] [CrossRef]
- Das, M.; Chowdhury, M.S.R.; Akter, S.; Mondal, A.K.; Uddin, M.J.; Rahman, M.M. An updated review on lumpy skin disease: Perspective of southeast asian countries. J. Adv. Biotechnol. Exp. Ther. 2021, 4, 322. [Google Scholar] [CrossRef]
- Şevik, M.; Doğan, M. Epidemiological and Molecular Studies on Lumpy Skin Disease Outbreaks in Turkey During 2014–2015. Transbound. Emerg. Dis. 2017, 64, 1268–1279. [Google Scholar] [CrossRef] [PubMed]
- Akther, M.; Akter, S.H.; Sarker, S.; Aleri, J.W.; Annandale, H.; Abraham, S.; Uddin, J.M. Global Burden of Lumpy Skin Disease, Outbreaks, and Future Challenges. Viruses 2023, 15, 1861. [Google Scholar] [CrossRef] [PubMed]
- Saegerman, C.; Bertagnoli, S.; Meyer, G.; Ganière, J.; Caufour, P.; De Clercq, K.; Jacquiet, P.; Hautefeuille, C.; Etore, F.; Casal, J. Risk of introduction of Lumpy Skin Disease into France through imports of cattle. Transbound. Emerg. Dis. 2018, 66, 957–967. [Google Scholar] [CrossRef]
- Kononov, A.; Prutnikov, P.; Shumilova, I.; Kononova, S.; Nesterov, A.; Byadovskaya, O.; Pestova, Y.; Diev, V.; Sprygin, A. Determination of lumpy skin disease virus in bovine meat and offal products following experimental infection. Transbound. Emerg. Dis. 2019, 66, 1332–1340. [Google Scholar] [CrossRef]
- Maw, M.T.; Khin, M.M.; Hadrill, D.; Meki, I.K.; Settypalli, T.B.K.; Kyin, M.M.; Myint, W.W.; Thein, W.Z.; Aye, O.; Palamara, E.; et al. First Report of Lumpy Skin Disease in Myanmar and Molecular Analysis of the Field Virus Isolates. Microorganisms 2022, 10, 897. [Google Scholar] [CrossRef]
- Mazloum, A.; Van Schalkwyk, A.; Babiuk, S.; Venter, E.; Wallace, D.B.; Sprygin, A. Lumpy skin disease: History, current understanding and research gaps in the context of recent geographic expansion. Front. Microbiol. 2023, 14, 1266759. [Google Scholar] [CrossRef]
- van Schalkwyk, A.; Kara, P.; Heath, L. Phylogenomic characterization of historic lumpy skin disease virus isolates from South Africa. Arch. Virol. 2022, 167, 2063–2070. [Google Scholar] [CrossRef]
- Xie, S.; Cui, L.; Liao, Z.; Zhu, J.; Ren, S.; Niu, K.; Li, H.; Jiang, F.; Wu, J.; Wang, J.; et al. Genomic analysis of lumpy skin disease virus asian variants and evaluation of its cellular tropism. npj Vaccines 2024, 9, 65. [Google Scholar] [CrossRef]
- Mathijs, E.; Vandenbussche, F.; Nguyen, L.; Aerts, L.; Nguyen, T.; De Leeuw, I.; Quang, M.; Nguyen, H.D.; Philips, W.; Dam, T.V.; et al. Coding-Complete Sequences of Recombinant Lumpy Skin Disease Viruses Collected in 2020 from Four Outbreaks in Northern Vietnam. Microbiol. Resour. Announc. 2021, 10, e0089721. [Google Scholar] [CrossRef]
- Breman, F.C.; Haegeman, A.; Krešić, N.; Philips, W.; De Regge, N. Lumpy Skin Disease Virus Genome Sequence Analysis: Putative Spatio-Temporal Epidemiology, Single Gene versus Whole Genome Phylogeny and Genomic Evolution. Viruses 2023, 15, 1471. [Google Scholar] [CrossRef]
- Zan, X.; Huang, H.; Guo, Y.; Di, D.; Fu, C.; Wang, S.; Wu, Y.; Wang, J.; Wang, Y.; Ma, Y.; et al. Molecular characterization of a novel subgenotype of lumpy skin disease virus strain isolated in Inner Mongolia of China. BMC Vet. Res. 2022, 18, 295. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.B.N.; Nguyen, V.T.; Nguyen, T.T.H.; Mai, N.T.A.; Le, P.N.; Lai, T.N.H.; Phan, T.H.; Tran, D.H.; Pham, N.T.; Dam, V.P.; et al. Molecular and histopathological characterization of lumpy skin disease in cattle in northern Vietnam during the 2020–2021 outbreaks. Arch. Virol. 2022, 167, 2143–2149. [Google Scholar] [CrossRef] [PubMed]
- Paungpin, W.; Sariya, L.; Chaiwattanarungruengpaisan, S.; Thongdee, M.; Kornmatitsuk, B.; Jitwongwai, A.; Taksinoros, S.; Sutummaporn, K.; Boonmasawai, S.; Nakthong, C. Coding-Complete Genome Sequence of a Lumpy Skin Disease Virus Isolated during the 2021 Thailand Outbreak. Genome Announc. 2022, 11, e0037522. [Google Scholar] [CrossRef]
- Krotova, A.; Byadovskaya, O.; Shumilova, I.; Zinyakov, N.; van Schalkwyk, A.; Sprygin, A. Molecular characterization of a novel recombinant lumpy skin disease virus isolated during an outbreak in Tyumen, Russia, in 2019. Transbound. Emerg. Dis. 2022, 69, e2312–e2317. [Google Scholar] [CrossRef]
- Aleksandr, K.; Olga, B.; David, W.B.; Pavel, P.; Yana, P.; Svetlana, K.; Alexander, N.; Vladimir, R.; Dmitriy, L.; Alexander, S. Non-vector-borne transmission of lumpy skin disease virus. Sci. Rep. 2020, 10, 7436. [Google Scholar] [CrossRef]
- Saltykov, Y.V.; Kolosova, A.A.; Filonova, N.N.; Chichkin, A.N.; Feodorova, V.A. Genetic Evidence of Multiple Introductions of Lumpy Skin Disease Virus into Saratov Region, Russia. Pathogens 2021, 10, 716. [Google Scholar] [CrossRef]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Kutish, G.F.; Rock, D.L. Genome of lumpy skin disease virus. J. Virol. 2001, 75, 7122–7130. [Google Scholar] [CrossRef]
- Douglass, N.; Van Der Walt, A.; Omar, R.; Munyanduki, H.; Williamson, A.-L. The complete genome sequence of the lumpy skin disease virus vaccine Herbivac LS reveals a mutation in the superoxide dismutase gene homolog. Arch. Virol. 2019, 164, 3107–3109. [Google Scholar] [CrossRef]
- Koirala, P.; Meki, I.K.; Maharjan, M.; Settypalli, B.K.; Manandhar, S.; Yadav, S.K.; Cattoli, G.; Lamien, C.E. Molecular Characterization of the 2020 Outbreak of Lumpy Skin Disease in Nepal. Microorganisms 2022, 10, 539. [Google Scholar] [CrossRef]
- Sprygin, A.; Pestova, Y.; Bjadovskaya, O.; Prutnikov, P.; Zinyakov, N.; Kononova, S.; Ruchnova, O.; Lozovoy, D.; Chvala, I.; Kononov, A. Evidence of recombination of vaccine strains of lumpy skin disease virus with field strains, causing disease. PLoS ONE 2020, 15, e0232584. [Google Scholar] [CrossRef]
- Haegeman, A.; De Leeuw, I.; Saduakassova, M.; Van Campe, W.; Aerts, L.; Philips, W.; Sultanov, A.; Mostin, L.; De Clercq, K. The importance of quality control of lsdv live attenuated vaccines for its safe application in the field. Vaccines 2021, 9, 1019. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, L.; Ward, M.P. The spread of lumpy skin disease virus across southeast asia: Insights from surveillance. Transbound. Emerg. Dis. 2023, 2023, 3972359. [Google Scholar] [CrossRef] [PubMed]
- Ochwo, S.; VanderWaal, K.; Ndekezi, C.; Nkamwesiga, J.; Munsey, A.; Witto, S.G.; Nantima, N.; Mayanja, F.; Okurut, A.R.A.; Atuhaire, D.K.; et al. Molecular detection and phylogenetic analysis of lumpy skin disease virus from outbreaks in Uganda 2017–2018. BMC Vet. Res. 2020, 16, 66. [Google Scholar] [CrossRef] [PubMed]
- Singhla, T.; Boonsri, K.; Kreausukon, K.; Modethed, W.; Pringproa, K.; Sthitmatee, N.; Punyapornwithaya, V.; Vinitchaikul, P. Molecular Characterization and Phylogenetic Analysis of Lumpy Skin Disease Virus Collected from Outbreaks in Northern Thailand in 2021. Vet. Sci. 2022, 9, 194. [Google Scholar] [CrossRef]
- Menasherow, S.; Erster, O.; Rubinstein-Giuni, M.; Kovtunenko, A.; Eyngor, E.; Gelman, B.; Khinich, E.; Stram, Y. A high-resolution melting (HRM) assay for the differentiation between Israeli field and Neethling vaccine lumpy skin disease viruses. J. Virol. Methods 2016, 232, 12–15. [Google Scholar] [CrossRef]
- Le Goff, C.; Lamien, C.E.; Fakhfakh, E.; Chadeyras, A.; Aba-Adulugba, E.; Libeau, G.; Tuppurainen, E.; Wallace, D.B.; Adam, T.; Silber, R.; et al. Capripoxvirus G-protein-coupled chemokine receptor: A host-range gene suitable for virus animal origin discrimination. J. Gen. Virol. 2009, 90, 1967–1977. [Google Scholar] [CrossRef]
- Lamien, C.E.; Lelenta, M.; Goger, W.; Silber, R.; Tuppurainen, E.; Matijevic, M.; Luckins, A.G.; Diallo, A. Real time PCR method for simultaneous detection, quantitation and differentiation of capripoxviruses. J. Virol. Methods 2011, 171, 134–140. [Google Scholar] [CrossRef]
- Haegeman, A.; Zro, K.; Vandenbussche, F.; Demeestere, L.; Van Campe, W.; Ennaji, M.; De Clercq, K. Development and validation of three Capripoxvirus real-time PCRs for parallel testing. J. Virol. Methods 2013, 193, 446–451. [Google Scholar] [CrossRef]
- Wolff, J.; Beer, M.; Hoffmann, B. Probe-based real-time qPCR assays for a reliable differentiation of Capripox virus species. Microorganisms 2021, 9, 765. [Google Scholar] [CrossRef]
- Haegeman, A.; De Leeuw, I.; Philips, W.; De Regge, N. Development and validation of a new DIVA real-time PCR allowing to differentiate wild-type lumpy skin disease virus strains, including the Asian recombinant strains, from Neethling-based vaccine strains. Viruses 2023, 15, 870. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, P.S.; Dalir-Naghadeh, B.; Mardani, K.; Jalilzadeh-Amin, G. Phylogenetic analysis of the lumpy skin disease viruses in northwest of Iran. Trop. Anim. Health Prod. 2018, 50, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Chibssa, T.R.; Sombo, M.; Lichoti, J.K.; Adam, T.I.B.; Liu, Y.; Elraouf, Y.A.; Grabherr, R.; Settypalli, T.B.K.; Berguido, F.J.; Loitsch, A.; et al. Molecular analysis of east african lumpy skin disease viruses reveals a mixed isolate with features of both vaccine and field isolates. Microorganisms 2021, 9, 1142. [Google Scholar] [CrossRef] [PubMed]
- Lamien, C.E.; Le Goff, C.; Silber, R.; Wallace, D.B.; Gulyaz, V.; Tuppurainen, E.; Madani, H.; Caufour, P.; Adam, T.; El Harrak, M.; et al. Use of the Capripoxvirus homologue of Vaccinia virus 30kDa RNA polymerase subunit (RPO30) gene as a novel diagnostic and genotyping target: Development of a classical PCR method to differentiate Goat poxvirus from Sheep poxvirus. Vet. Microbiol. 2011, 149, 30–39. [Google Scholar] [CrossRef]
- Haegeman, A.; Zro, K.; Sammin, D.; Vandenbussche, F.; Ennaji, M.M.; De Clercq, K. Investigation of a Possible Link between Vaccination and the 2010 Sheep Pox Epizootic in Morocco. Transbound. Emerg. Dis. 2016, 63, e278–e287. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols; Springer: Berlin, Germany, 2000; pp. 365–386. [Google Scholar]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Prabhu, M.; Malmarugan, S.; Rajagunalan, S.; Govindan, B.; Thangavelu, L.P.; Palanisamy, G.; Yogisharadhya, R.; Karthik, K. Isolation and molecular characterization of lumpy skin disease virus from Tamil Nadu, India during the outbreaks from 2020 to 2022. Virus Genes 2024, 60, 159–172. [Google Scholar] [CrossRef]
- Turan, N.; Yilmaz, A.; Tekelioglu, B.K.; Yilmaz, H. Lumpy skin disease: Global and Turkish perspectives, Approaches in Poultry. Dairy Vet. Sci. 2017, 1, APDV.000504. [Google Scholar]
- Bhat, S.; Chaudry, W.; Mittal, P.; Ahmad, B.; Haroon, Z.; Shaheen, S.; Mir, M.; Reddy, M.; Wani, M. Complete genome sequence of the lumpy skin disease virus reported from Jammu and Kashmir, India, during 2022 outbreak. Genome Announc. 2023, 12, e0031723. [Google Scholar] [CrossRef]
- Song, Y.; Zuo, O.; Zhang, G.; Hu, J.; Tian, Z.; Guan, G.; Luo, J.; Yin, H.; Shang, Y.; Du, J. Emergence of Lumpy Skin Disease Virus Infection in Yaks, Cattle-Yaks, and Cattle on the Qinghai–Xizang Plateau of China. Transbound. Emerg. Dis. 2024, 2024, 2383886. [Google Scholar] [CrossRef]
- Reddy, G.B.M.; Mounica, P.S.; Sudeep, N.; Vikram, R.; Garam, G.B.; Lalzampuia, H.; Ragulraj, S.; Pal, S.; Khate, K.; Bijalwan, S.; et al. First evidence of lumpy skin disease in mithun (Bos frontalis) in India. Arch. Virol. 2024, 169, 65. [Google Scholar] [CrossRef]
- Li, Y.; An, Q.; Sun, Z.; Gao, X.; Wang, H. Risk factors and spatiotemporal distribution of lumpy skin disease occurrence in the asian continent during 2012–2022: An ecological niche model. Transbound. Emerg. Dis. 2023, 2023, 6207149. [Google Scholar] [CrossRef]
- Jabbar, M.H.; Atif, F.A.; Kashif, M.; Ahmed, I.; Iarussi, F.; A Swelum, A. Molecular epidemiology and phylogenetic insights of lumpy skin disease in cattle from diverse agro-ecological regions of Punjab, Pakistan. PLoS ONE 2025, 20, e0315532. [Google Scholar] [CrossRef]


| Serial No. | Targeted Genes | Primer Sequence (Forward) | Amplicon Size (bp) and Annealing Temperature |
|---|---|---|---|
| 1 | P32 | F-5′TCGTTGGTCGCGAAATTTCAG3′ R-5′GAGCCATCCATTTTCCAACTCT3′ | 759, 56 °C [42] |
| 2 | EEV | F-5′ATGGGAATAGTATCTGTTGTATACG3′ R-5′CGAACCCCTATTTACTTGAGAA3′ | 930, 55 °C [43] |
| 3 | B22R | F-5′TCATTTTCTTCTAGTTCCGACGA3′ R-5′TTCGTTGATGATAAATAACTGGAAA3 | 863, 58 °C [30] |
| 4 | RPO30 | F-5′ATTCGTTATCGCAGAACAAGG3′ R-5′CACCAACCATAGAATAGTATTGAGAC3′ | 1234, 55 °C [44] |
| 5 | GPCR GPCR internal sequencing primers | F5′TTAAGTAAAGCATAACTCCAACAAAAATG3′ R5′TTTTTTTATTTTTTATCCAATGCTAATACT3′ F5′GATGAGTATTGATAGATACCTAGCTGTAGTT3′ R5′TTAAGTAAAGCATAACTCCAACAAAAATG3′ | 1158, 50 °C [28,37] |
| 6 | NTPase (ORF83) | F-5′GAGAAACCGCAACAGGAAAA3′ R-5′GGATGAGCAACGAACCAACT3′ | 614, 60 °C [32] |
| 7 | RPO132 (ORF116/117) | F-5′TGGAGAAATGGAAAGGGATTG3′ R-5′CAGGCGACGATGATGAAAC3′ | 750, 60 °C [32] |
| 8 | VLTF-1 (ORF58/59) | F-5′TTTTATGGCGTTCCACGATT3′ R-5′CCCAACACTCTCTCGCTTCA3′ | 755, 60 °C [32] |
| 9 | Finger Protein (ORF10) | F-5′ACCCAACAACACAAGGAAGG3′ R-5′CATCGCAAACAAAGAATAAGAAAG3′ | 708, 60 °C [32] |
| 10 | Ser-Thr kinase (ORF25/26) | F-5′TTCGTTTTCAGCGATTTTATTT3′ R-5′AGGAGATTTTATTATGAGTGGCTT3′ | 739, 57 °C [45] |
| Area of Sample Collection | Sample No. (n) | D5r (Cp Values) % | IC % | ||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| Azad Jammu and Kashmir (AJ&K) | 8 | 75 | 25 | 87.5 | 12.5 |
| Islamabad | 1 | 100 | 0 | 100 | 0 |
| Khaibar Pukhtoon Khuva (KPK) | 14 | 35.7 | 64.3 | 42.9 | 57.1 |
| Punjab | 17 | 64.7 | 35.3 | 70.6 | 29.4 |
| 40 | 57.5 | 42.5 | 57.5 | 42.5 | |
| Sample Id | Location | PanCapripox | DIVA Recombinant Assay | Wolff Assay | ||||
|---|---|---|---|---|---|---|---|---|
| D5r | Internal Control | External Control | Wild-Type and Recombinant | Neethling Vaccine | Wild-Type | Neethling Vaccine | ||
| LSDV_1_NS_2022_PAK | Punjab | 31.56 | 36.85 | 29.03 | 29.78 | Negative | 31.04 | Negative |
| LSDV_2_NS_2022_PAK | KPK * | 29.79 | 34.68 | 28.45 | 33.18 | Negative | Not Performed | Not Performed |
| Reference Strains and Subclades | Accession Number | Regions (Sample 1.4 (LSDV_1_NS_2022_PAK) and 3.29 (LSDV_2_NS_2022_PAK) PCR Fragment Size in Nucleotides) Number of Different Nucleotides in Isolates from Pakistan | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B22R (773 *, 765 ǂ) | EEV (877, 878) | P32 (716, 678) | ◊RPO30 (1146) | GPCR (1083, 1098) | NTPase (575, 575) | RPO132 (711, 729) | VLTF-1 (735, 735) | LAP/PHD (692, 692) | Ser-Thr Kinase (657, 677) | ||
| Herbivac LS (1.1) | MK441838 | 11, 10 | 39, 39 | 6, 6 | 14 | 32, 33 | 2, 2 | 9, 9 | 4, 4 | 13, 13 | 7, 7 |
| Cro2016 (1.1) | MG972412 | 11, 10 | 39, 39 | 6, 6 | 14 | 32, 33 | 2, 2 | 9, 9 | 4, 4 | 13, 13 | 7, 7 |
| Neethling-LSD vaccine-OBP (1.1) | KX764645 | 11, 10 | 39, 39 | 6, 6 | 15 | 32, 33 | 2, 2 | 9, 9 | 4, 4 | 13, 13 | 7, 7 |
| LSDV/Serbia/6040/2016 (1.2) | OR134847 | 0, 0 | 3, 3 | 0, 0 | 1 | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1, 1 | 2, 2 |
| Kenya (1.2) | MN072619 | 0, 0 | 3, 3 | 0, 0 | 4 | 12, 12 | 1, 1 | 2, 2 | 0, 0 | 1, 1 | 1, 1 |
| Evros GR/15 (1.2) | KY829023 | 0, 0 | 3, 3 | 0, 0 | 1 | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1, 1 | 2, 2 |
| Saratov 2017 (2.1) | MH646674 | 0, 0 | 39, 39 | 5, 5 | 10 | 30, 31 | 2, 2 | 9, 9 | 4, 4 | 13, 13 | 7, 7 |
| Udmurtiya/2019 (2.2) | MT134042 | 2, 2 | 37, 37 | 0, 0 | 4 | 32, 33 | 1, 1 | 3, 3 | 0, 0 | 13, 13 | 4, 4 |
| KZ-Kostanay-2018 (2.3) | MT992618 | 0, 0 | 39, 39 | 1, 1 | 9 | 17, 17 | 2, 2 | 9, 9 | 4, 4 | 1, 1 | 7, 7 |
| Tyumen/2019 (2.4) | OL542833 | 1, 1 | 38, 38 | 6, 6 | 8 | 27, 27 | 1, 1 | 9, 9 | 4, 4 | 10, 10 | 7, 7 |
| CHINA/HLJ/2022 (2.5) | OQ555660 | 6, 6 | 39, 39 | 6, 6 | 13 | 20, 20 | 1, 1 | 2, 2 | 1, 1 | 13, 13 | 7, 7 |
| Tomsk_2020 (2.5) | OM793602 | 6, 6 | 39, 39 | 6, 6 | 13 | 20, 20 | 1, 1 | 2, 2 | 1, 1 | 13, 13 | 7, 7 |
| Kurgan 2018 (2.6) | OR194148 | 6, 6 | 39, 39 | 1, 1 | 9 | 12, 12 | 2, 2 | 9, 9 | 4, 4 | 1, 1 | 7, 7 |
| KSGP0240 (1.2) | KX683219 | 1, 1 | 3, 3 | 0, 0 | 4 | 12, 12 | 1, 1 | 2, 2 | 0, 0 | 1, 1 | 1, 1 |
| SPPV | MN072627 | 24, 23 | 48, 48 | 12, 12 | 61 | 51, 52 | 8, 8 | 36, 36 | 20, 20 | 22, 22 | 59, 59 |
| GTPV | MN072622 | 16, 16 | 53, 53 | 7, 7 | 27 | 49, 51 | 8, 8 | 16, 16 | 24, 24 | 26, 26 | 34, 34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferdoos, S.; Haegeman, A.; Sattar, S.; Ahmed, I.; Javed, S.; Tariq, A.; Regge, N.D.; Bostan, N. Molecular and Phylogenetic Analyses of Lumpy Skin Disease Virus (LSDV) Outbreak (2021/22) in Pakistan Indicate Involvement of a Clade 1.2 LSDV Strain. Viruses 2025, 17, 1546. https://doi.org/10.3390/v17121546
Ferdoos S, Haegeman A, Sattar S, Ahmed I, Javed S, Tariq A, Regge ND, Bostan N. Molecular and Phylogenetic Analyses of Lumpy Skin Disease Virus (LSDV) Outbreak (2021/22) in Pakistan Indicate Involvement of a Clade 1.2 LSDV Strain. Viruses. 2025; 17(12):1546. https://doi.org/10.3390/v17121546
Chicago/Turabian StyleFerdoos, Saiba, Andy Haegeman, Sadia Sattar, Ibrar Ahmed, Sundus Javed, Aamira Tariq, Nick De Regge, and Nazish Bostan. 2025. "Molecular and Phylogenetic Analyses of Lumpy Skin Disease Virus (LSDV) Outbreak (2021/22) in Pakistan Indicate Involvement of a Clade 1.2 LSDV Strain" Viruses 17, no. 12: 1546. https://doi.org/10.3390/v17121546
APA StyleFerdoos, S., Haegeman, A., Sattar, S., Ahmed, I., Javed, S., Tariq, A., Regge, N. D., & Bostan, N. (2025). Molecular and Phylogenetic Analyses of Lumpy Skin Disease Virus (LSDV) Outbreak (2021/22) in Pakistan Indicate Involvement of a Clade 1.2 LSDV Strain. Viruses, 17(12), 1546. https://doi.org/10.3390/v17121546







