Bioinformatics Tools and Approaches for Virus Discovery in Genomic Data: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
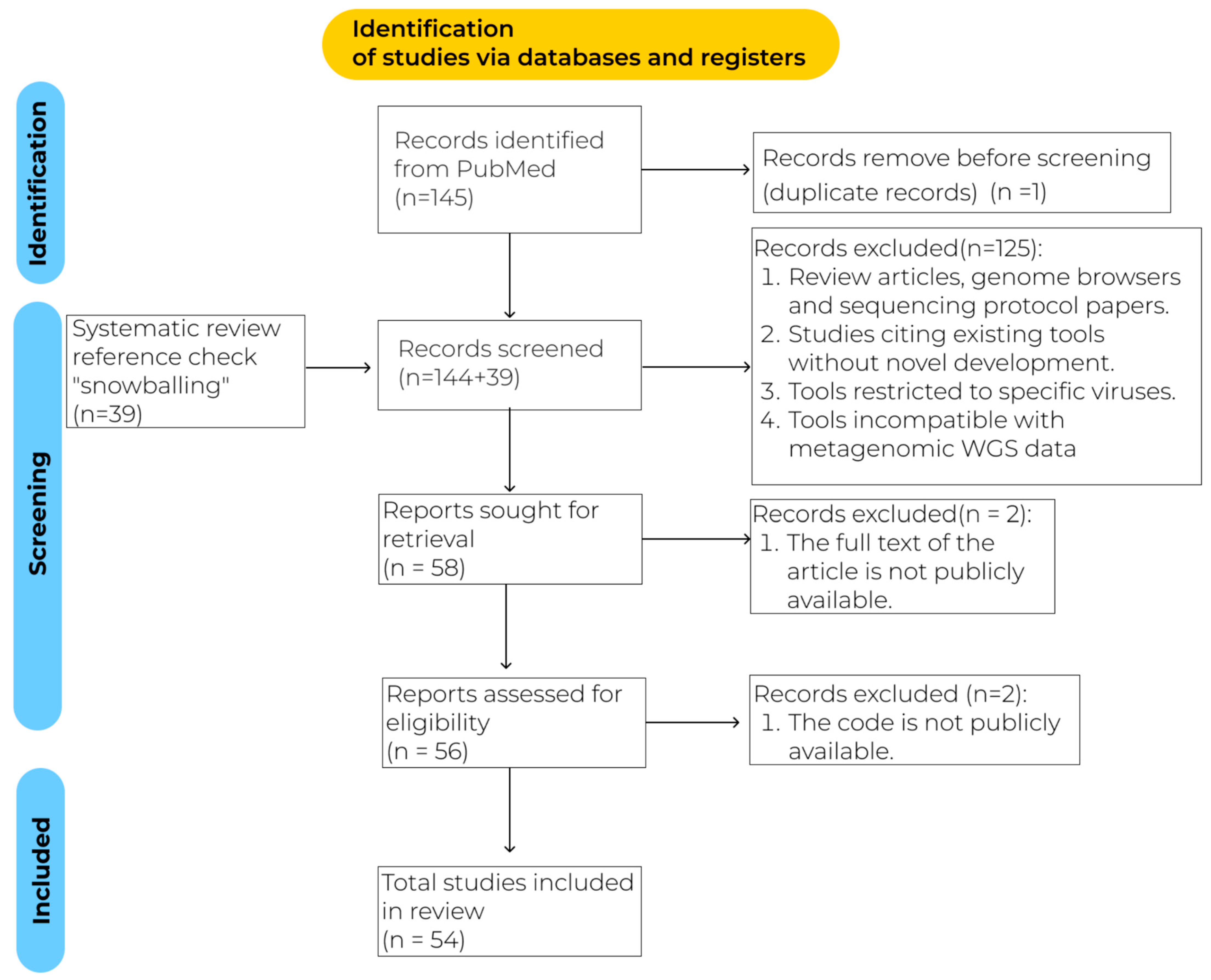
3. Results
3.1. Alignment-Based Approaches for Virus Sequence Identification
3.1.1. Pairwise Alignment Methods
3.1.2. Multiple Sequence Alignment Methods
3.1.3. Rapid Similarity Estimation Methods
3.2. Profile Hidden Markov Models Methods
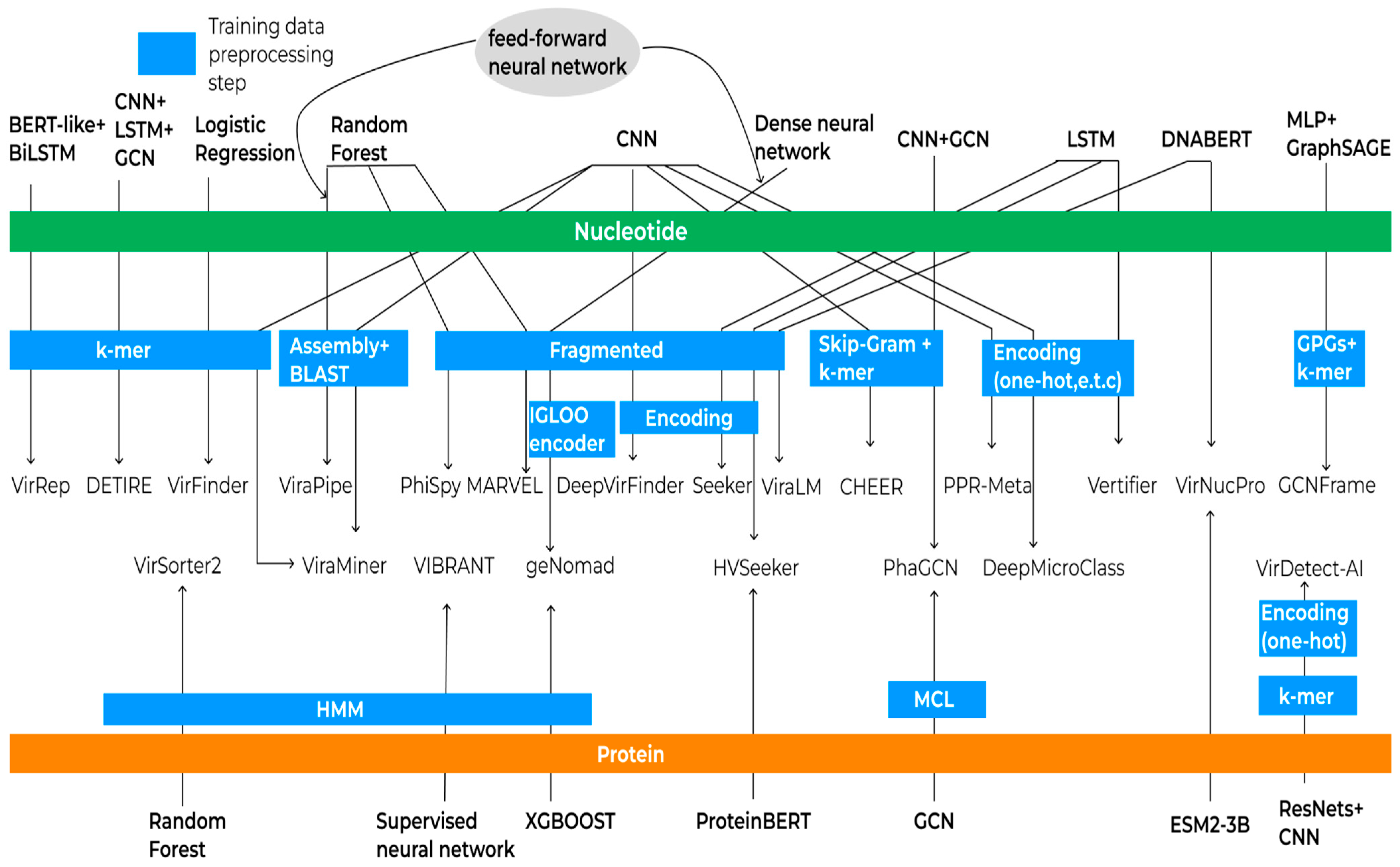
3.3. Machine-Learning-Based Approach
3.4. K-Mer-Based Approach
4. Discussion
5. Summary
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| VLPs | Virus-Like Particles |
| ICTV | International Committee on Taxonomy of Viruses |
| HMMs | Hidden Markov Models |
| LLM | Large Language Model |
| LCA | Lowest Common Ancestor |
| MSPs | Maximal Segment Pairs |
| ORFs | Open Reading Frames |
| GBDP | Genome BLAST Distance Phylogeny |
| MCL | Markov Cluster Algorithm |
| MLCA | Maximum Likelihood Clade Assignment |
| PC | Protein Clusters |
| MSA | Multiple Sequence Alignment |
| PSSM | Position-Specific Scoring Matrix |
| FFT | Fast Fourier Transform |
| EPA | Evolutionary Placement Algorithm |
| ANI | Average Nucleotide Identity |
| NCLDVs | Nucleo-Cytoplasmic Large DNA Viruses |
| GVOGs | Giant Virus Orthologous Groups |
| RdRp | RNA-dependent RNA polymerase |
| dsDNA phages | Double-Stranded DNA bacteriophages |
| FFNN | Feed-Forward Neural Network |
| RSCU | Relative Synonymous Codon Usage |
| CNNs | Convolutional Neural Network |
| BiPathCNN | Bi-Path Convolutional Neural Network |
| LSTM | Long Short-Term Memory |
| RNN | Recurrent Neural Network |
| GCN | Graph Convolutional Network |
| BiLSTM | Bidirectional LSTM |
| MLPs | Multilayer Perceptrons |
| GPGs | Gapped Pattern Graphs |
| SNPs | Single-Nucleotide Polymorphisms |
| MLM | Masked Language Modeling |
| ERT | Extremely Randomized Trees |
| GVP | Global Virome Project |
References
- Mushegian, A.R. Are There 10 Virus Particles on Earth, or More, or Fewer? J. Bacteriol. 2020, 202, e00052-20. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, C.; Salles, S.; Mercado, J.M.; Cortés, D.; Yebra, L.; Gómez-Jakobsen, F.; Sánchez, A.; Alonso, A.; Valcárcel-Pérez, N. Abundance of Virus-like Particles (VLPs) and Microbial Plankton Community Composition in a Mediterranean Sea Coastal Area. Aquat. Microb. Ecol. 2018, 81, 137–148. [Google Scholar] [CrossRef]
- Cornell, C.R.; Zhang, Y.; Van Nostrand, J.D.; Wagle, P.; Xiao, X.; Zhou, J. Temporal Changes of Virus-Like Particle Abundance and Metagenomic Comparison of Viral Communities in Cropland and Prairie Soils. mSphere 2021, 6, e0116020. [Google Scholar] [CrossRef] [PubMed]
- Takada, K.; Holmes, E.C. Genome Sizes of Animal RNA Viruses Reflect Phylogenetic Constraints. Virus Evol. 2025, 11, veaf005. [Google Scholar] [CrossRef]
- Ain, Q.u.; Wu, K.; Wu, X.; Bai, Q.; Li, Q.; Zhou, C.-Z.; Wu, Q. Cyanophage-Encoded Auxiliary Metabolic Genes in Modulating Cyanobacterial Metabolism and Algal Bloom Dynamics. Front. Virol. 2024, 4, 1461375. [Google Scholar] [CrossRef]
- Camargo, A.P.; Roux, S.; Schulz, F.; Babinski, M.; Xu, Y.; Hu, B.; Chain, P.S.G.; Nayfach, S.; Kyrpides, N.C. Identification of Mobile Genetic Elements with geNomad. Nat. Biotechnol. 2024, 42, 1303–1312. [Google Scholar] [CrossRef]
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral Mutation Rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef]
- Irwin, N.A.T.; Pittis, A.A.; Richards, T.A.; Keeling, P.J. Systematic Evaluation of Horizontal Gene Transfer between Eukaryotes and Viruses. Nat. Microbiol. 2022, 7, 327–336. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and Evolution of Viruses of Eukaryotes: The Ultimate Modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef]
- ICTV. Available online: https://ictv.global/ (accessed on 19 November 2025).
- Santiago-Rodriguez, T.M.; Hollister, E.B. Unraveling the Viral Dark Matter through Viral Metagenomics. Front. Immunol. 2022, 13, 1005107. [Google Scholar] [CrossRef]
- Krishnamurthy, S.R.; Wang, D. Origins and Challenges of Viral Dark Matter. Virus Res. 2017, 239, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E. Phage_Finder: Automated Identification and Classification of Prophage Regions in Complete Bacterial Genome Sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef]
- Ren, J.; Ahlgren, N.A.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. VirFinder: A Novel K-Mer Based Tool for Identifying Viral Sequences from Assembled Metagenomic Data. Microbiome 2017, 5, 69. [Google Scholar] [CrossRef]
- Ren, J.; Song, K.; Deng, C.; Ahlgren, N.A.; Fuhrman, J.A.; Li, Y.; Xie, X.; Poplin, R.; Sun, F. Identifying Viruses from Metagenomic Data Using Deep Learning. Quant. Biol. 2020, 8, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated Recovery, Annotation and Curation of Microbial Viruses, and Evaluation of Viral Community Function from Genomic Sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef]
- Peng, C.; Shang, J.; Guan, J.; Wang, D.; Sun, Y. ViraLM: Empowering Virus Discovery through the Genome Foundation Model. Bioinformatics 2024, 40, btae704. [Google Scholar] [CrossRef]
- Home. Available online: https://pubmed.ncbi.nlm.nih.gov (accessed on 21 November 2025).
- OSF. Available online: https://osf.io (accessed on 21 November 2025).
- Christensen, H.; Olsen, J.E. Pairwise Alignment, Multiple Alignment, and BLAST. In Introduction to Bioinformatics in Microbiology; Springer International Publishing: Cham, Switzerland, 2018; pp. 51–79. ISBN 9783319992792. [Google Scholar]
- Pearson, W.R. An Introduction to Sequence Similarity (“homology”) Searching. Curr. Protoc. Bioinform. 2013, 42, 3.1.1–3.1.8. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, F.M.; Crane, A.; Kuhn, J.H.; Simmonds, P.; Lefkowitz, E.J.; ICTV Taxonomy Summary Consortium. Summary of Taxonomy Changes Ratified by the International Committee on Taxonomy of Viruses (ICTV)—General Taxonomy Proposals, 2025. J. Gen. Virol. 2025, 106, 002116. [Google Scholar] [CrossRef]
- Home. Available online: https://ictv.global/news/taxablast (accessed on 20 November 2025).
- NCBI Virus. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/ (accessed on 19 November 2025).
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database Indexing for Production MegaBLAST Searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef]
- Jurtz, V.I.; Villarroel, J.; Lund, O.; Voldby Larsen, M.; Nielsen, M. MetaPhinder-Identifying Bacteriophage Sequences in Metagenomic Data Sets. PLoS ONE 2016, 11, e0163111. [Google Scholar] [CrossRef]
- Bao, Y.; Chetvernin, V.; Tatusova, T. PAirwise Sequence Comparison (PASC) and Its Application in the Classification of Filoviruses. Viruses 2012, 4, 1318–1327. [Google Scholar] [CrossRef]
- Muhire, B.M.; Roumagnac, P.; Varsani, A.; Martin, D.P. Sequence Demarcation Tool (SDT), a Free User-Friendly Computer Program Using Pairwise Genetic Identity Calculations to Classify Nucleotide or Amino Acid Sequences. Methods Mol. Biol. 2025, 2912, 71–79. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-Based Phylogeny and Classification of Prokaryotic Viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Bin Jang, H.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Brister, J.R.; Kropinski, A.M.; Krupovic, M.; Lavigne, R.; et al. Taxonomic Assignment of Uncultivated Prokaryotic Virus Genomes Is Enabled by Gene-Sharing Networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, B.; Jang, H.B.; Doulcier, G.; You, Z.-Q.; Roux, S.; Sullivan, M.B. vConTACT: An iVirus Tool to Classify Double-Stranded DNA Viruses That Infect and. PeerJ 2017, 5, e3243. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.-I.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega for Making Accurate Alignments of Many Protein Sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Libin, P.J.K.; Deforche, K.; Abecasis, A.B.; Theys, K. VIRULIGN: Fast Codon-Correct Alignment and Annotation of Viral Genomes. Bioinformatics 2019, 35, 1763–1765. [Google Scholar] [CrossRef]
- Moshiri, N. ViralMSA: Massively Scalable Reference-Guided Multiple Sequence Alignment of Viral Genomes. Bioinformatics 2021, 37, 714–716. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Ranwez, V.; Harispe, S.; Delsuc, F.; Douzery, E.J.P. MACSE: Multiple Alignment of Coding SEquences Accounting for Frameshifts and Stop Codons. PLoS ONE 2011, 6, e22594. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple Alignment of Nucleotide Sequences Guided by Amino Acid Translations. Nucleic Acids Res. 2010, 38, W7–W13. [Google Scholar] [CrossRef] [PubMed]
- Singer, J.B.; Thomson, E.C.; McLauchlan, J.; Hughes, J.; Gifford, R.J. GLUE: A Flexible Software System for Virus Sequence Data. BMC Bioinform. 2018, 19, 532. [Google Scholar] [CrossRef]
- Jain, C.; Dilthey, A.; Koren, S.; Aluru, S.; Phillippy, A.M. A Fast Approximate Algorithm for Mapping Long Reads to Large Reference Databases. J. Comput. Biol. 2018, 25, 766–779. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R., L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Zielezinski, A.; Gudyś, A.; Barylski, J.; Siminski, K.; Rozwalak, P.; Dutilh, B.E.; Deorowicz, S. Ultrafast and Accurate Sequence Alignment and Clustering of Viral Genomes. Nat. Methods 2025, 22, 1191–1194. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Nguyen, V.-A.; Boyd-Graber, J.; Altschul, S.F. Dirichlet Mixtures, the Dirichlet Process, and the Structure of Protein Space. J. Comput. Biol. 2013, 20, 1–18. [Google Scholar] [CrossRef]
- Eddy, S.R. A Probabilistic Model of Local Sequence Alignment That Simplifies Statistical Significance Estimation. PLoS Comput. Biol. 2008, 4, e1000069. [Google Scholar] [CrossRef]
- Aylward, F.O.; Moniruzzaman, M. ViralRecall-A Flexible Command-Line Tool for the Detection of Giant Virus Signatures in ’Omic Data. Viruses 2021, 13, 150. [Google Scholar] [CrossRef] [PubMed]
- Tisza, M.J.; Belford, A.K.; Domínguez-Huerta, G.; Bolduc, B.; Buck, C.B. Cenote-Taker 2 Democratizes Virus Discovery and Sequence Annotation. Virus Evol. 2021, 7, veaa100. [Google Scholar] [CrossRef]
- Starikova, E.V.; Tikhonova, P.O.; Prianichnikov, N.A.; Rands, C.M.; Zdobnov, E.M.; Ilina, E.N.; Govorun, V.M. Phigaro: High-Throughput Prophage Sequence Annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol. Mol. Biol. Rev. 2020, 84, e00061-19. [Google Scholar] [CrossRef]
- Edgar, R.C.; Taylor, B.; Lin, V.; Altman, T.; Barbera, P.; Meleshko, D.; Lohr, D.; Novakovsky, G.; Buchfink, B.; Al-Shayeb, B.; et al. Petabase-Scale Sequence Alignment Catalyses Viral Discovery. Nature 2022, 602, 142–147. [Google Scholar] [CrossRef]
- Charon, J.; Buchmann, J.P.; Sadiq, S.; Holmes, E.C. RdRp-Scan: A Bioinformatic Resource to Identify and Annotate Divergent RNA Viruses in Metagenomic Sequence Data. Virus Evol. 2022, 8, veac082. [Google Scholar] [CrossRef]
- Sakaguchi, S.; Nakano, T.; Nakagawa, S. NeoRdRp2 with Improved Seed Data, Annotations, and Scoring. Front. Virol. 2024, 4, 1378695. [Google Scholar] [CrossRef]
- Amgarten, D.; Braga, L.P.P.; da Silva, A.M.; Setubal, J.C. MARVEL, a Tool for Prediction of Bacteriophage Sequences in Metagenomic Bins. Front. Genet. 2018, 9, 304. [Google Scholar] [CrossRef] [PubMed]
- Bzhalava, Z.; Tampuu, A.; Bała, P.; Vicente, R.; Dillner, J. Machine Learning for Detection of Viral Sequences in Human Metagenomic Datasets. BMC Bioinform. 2018, 19, 336. [Google Scholar] [CrossRef]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitúa, M.C.; Vik, D.; Sullivan, M.B.; et al. VirSorter2: A Multi-Classifier, Expert-Guided Approach to Detect Diverse DNA and RNA Viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; McNair, K.; Cuevas, D.A.; Bailey, B.A.; Segall, A.M.; Edwards, R.A. Prophage Genomics Reveals Patterns in Phage Genome Organization and Replication. bioRxiv 2017. [Google Scholar] [CrossRef]
- Shang, J.; Sun, Y. CHEER: HierarCHical Taxonomic Classification for Viral mEtagEnomic Data via Deep leaRning. Methods 2021, 189, 95–103. [Google Scholar] [CrossRef]
- Hou, S.; Tang, T.; Cheng, S.; Liu, Y.; Xia, T.; Chen, T.; Fuhrman, J.A.; Sun, F. DeepMicroClass Sorts Metagenomic Contigs into Prokaryotes, Eukaryotes and Viruses. NAR Genom. Bioinform. 2024, 6, lqae044. [Google Scholar] [CrossRef]
- Zárate, A.; Díaz-González, L.; Taboada, B. VirDetect-AI: A Residual and Convolutional Neural Network-Based Metagenomic Tool for Eukaryotic Viral Protein Identification. Brief. Bioinform. 2024, 26, bbaf001. [Google Scholar] [CrossRef]
- Auslander, N.; Gussow, A.B.; Benler, S.; Wolf, Y.I.; Koonin, E.V. Seeker: Alignment-Free Identification of Bacteriophage Genomes by Deep Learning. Nucleic Acids Res. 2020, 48, e121. [Google Scholar] [CrossRef]
- Al-Najim, A.; Hauns, S.; Tran, V.D.; Backofen, R.; Alkhnbashi, O.S. HVSeeker: A Deep-Learning-Based Method for Identification of Host and Viral DNA Sequences. Gigascience 2025, 14, giaf037. [Google Scholar] [CrossRef]
- Miao, Y.; Liu, F.; Hou, T.; Liu, Y. Virtifier: A Deep Learning-Based Identifier for Viral Sequences from Metagenomes. Bioinformatics 2022, 38, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Bian, J.; Dong, G.; Dai, T. DETIRE: A Hybrid Deep Learning Model for Identifying Viral Sequences from Metagenomes. Front. Microbiol. 2023, 14, 1169791. [Google Scholar] [CrossRef]
- Shang, J.; Jiang, J.; Sun, Y. Bacteriophage Classification for Assembled Contigs Using Graph Convolutional Network. Bioinformatics 2021, 37, i25–i33. [Google Scholar] [CrossRef]
- Sourkov, V. IGLOO: Slicing the Features Space to Represent Sequences. arXiv 2018, arXiv:1807.03402. [Google Scholar]
- Li, J.; Mi, J.; Lin, W.; Tian, F.; Wan, J.; Gao, J.; Tong, Y. VirNucPro: An Identifier for the Identification of Viral Short Sequences Using Six-Frame Translation and Large Language Models. Brief. Bioinform. 2025, 26, bbaf224. [Google Scholar] [CrossRef]
- Wang, R.H.; Ng, Y.K.; Zhang, X.; Wang, J.; Li, S.C. Coding Genomes with Gapped Pattern Graph Convolutional Network. Bioinformatics 2024, 40, btae188. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, W.-H.; Zhao, X.-M. VirRep: A Hybrid Language Representation Learning Framework for Identifying Viruses from Human Gut Metagenomes. Genome Biol. 2024, 25, 177. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, M.L.; Polson, S.W.; Lee, K.H. k-mer-Based Metagenomics Tools Provide a Fast and Sensitive Approach for the Detection of Viral Contaminants in Biopharmaceutical and Vaccine Manufacturing Applications Using Next-Generation Sequencing. mSphere 2021, 6, e01336-20. [Google Scholar] [CrossRef]
- Maabar, M.; Davison, A.J.; Vučak, M.; Thorburn, F.; Murcia, P.R.; Gunson, R.; Palmarini, M.; Hughes, J. DisCVR: Rapid Viral Diagnosis from High-Throughput Sequencing Data. Virus Evol. 2019, 5, vez033. [Google Scholar] [CrossRef]
- Audano, P.; Vannberg, F. KAnalyze: A Fast Versatile Pipelined K-Mer Toolkit. Bioinformatics 2014, 30, 2070–2072. [Google Scholar] [CrossRef] [PubMed]
- Ounit, R.; Wanamaker, S.; Close, T.J.; Lonardi, S. CLARK: Fast and Accurate Classification of Metagenomic and Genomic Sequences Using Discriminative K-Mers. BMC Genom. 2015, 16, 236. [Google Scholar] [CrossRef]
- Popov, N.; Sonets, I.; Evdokimova, A.; Molchanova, M.; Panova, V.; Korneenko, E.; Manolov, A.; Ilina, E. AliMarko: A Pipeline for Virus Identification Using an Expert-Guided Approach. Viruses 2025, 17, 355. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Guo, X.; Zhang, X.; Zheng, X.; Zhang, M.; Bao, Y. VISTA: A Tool for Fast Taxonomic Assignment of Viral Genome Sequences. Genom. Proteom. Bioinform. 2025, 23, qzae082. [Google Scholar] [CrossRef]
- Carroll, D.; Daszak, P.; Wolfe, N.D.; Gao, G.F.; Morel, C.M.; Morzaria, S.; Pablos-Méndez, A.; Tomori, O.; Mazet, J.A.K. The Global Virome Project. Science 2018, 359, 872–874. [Google Scholar] [CrossRef]
- Wu, Y.; Peng, Y. Ten Computational Challenges in Human Virome Studies. Virol. Sin. 2024, 39, 845–850. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Mirdita, M.; Steinegger, M.; Söding, J. MMseqs2 Desktop and Local Web Server App for Fast, Interactive Sequence Searches. Bioinformatics 2019, 35, 2856–2858. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Hu, B.; Zhang, L.; Gan, M.; Ding, Q.; Pan, K.; Wei, J.; Xu, W.; Chen, D.; Zheng, S.; et al. Virome Landscape of Wild Rodents and Shrews in Central China. Microbiome 2025, 13, 63. [Google Scholar] [CrossRef]
- Guo, J.; Huang, X.; Zhang, C.; Huang, P.; Li, Y.; Wen, F.; Wang, X.; Yang, N.; Xu, M.; Bi, Y.; et al. The Blood Virome of 10,585 Individuals from the ChinaMAP. Cell Discov. 2022, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Páez-Espino, D.; Call, L.; Low, S.J.; Sberro, H.; Ivanova, N.N.; Proal, A.D.; Fischbach, M.A.; Bhatt, A.S.; Hugenholtz, P.; et al. Metagenomic Compendium of 189,680 DNA Viruses from the Human Gut Microbiome. Nat. Microbiol. 2021, 6, 960–970. [Google Scholar] [CrossRef]
- Zeng, S.; Almeida, A.; Li, S.; Ying, J.; Wang, H.; Qu, Y.; Paul Ross, R.; Stanton, C.; Zhou, Z.; Niu, X.; et al. A Metagenomic Catalog of the Early-Life Human Gut Virome. Nat. Commun. 2024, 15, 1864. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, S.; Nagata, N.; Kiguchi, Y.; Kojima, Y.; Miyoshi-Akiyama, T.; Kimura, M.; Ohsugi, M.; Ueki, K.; Oka, S.; Mizokami, M.; et al. Extensive Gut Virome Variation and Its Associations with Host and Environmental Factors in a Population-Level Cohort. Nat. Commun. 2022, 13, 5252. [Google Scholar] [CrossRef]
- Yan, Q.; Huang, L.; Li, S.; Zhang, Y.; Guo, R.; Zhang, P.; Lei, Z.; Lv, Q.; Chen, F.; Li, Z.; et al. The Chinese Gut Virus Catalogue Reveals Gut Virome Diversity and Disease-Related Viral Signatures. Genome Med. 2025, 17, 30. [Google Scholar] [CrossRef]
- Galperina, A.; Lugli, G.A.; Milani, C.; De Vos, W.M.; Ventura, M.; Salonen, A.; Hurwitz, B.; Ponsero, A.J. The Aggregated Gut Viral Catalogue (AVrC): A Unified Resource for Exploring the Viral Diversity of the Human Gut. PLoS Comput. Biol. 2025, 21, e1012268. [Google Scholar] [CrossRef]
- Alves, J.M.P.; de Oliveira, A.L.; Sandberg, T.O.M.; Moreno-Gallego, J.L.; de Toledo, M.A.F.; de Moura, E.M.M.; Oliveira, L.S.; Durham, A.M.; Mehnert, D.U.; Zanotto, P.M.d.A.; et al. GenSeed-HMM: A Tool for Progressive Assembly Using Profile HMMs as Seeds and Its Application in Alpavirinae Viral Discovery from Metagenomic Data. Front. Microbiol. 2016, 7, 269. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.; Zhang, X.; Vaas, J.; Klingler, F.; Mutz, P.; Dubin, A.; Pietschmann, T.; Roth, O.; Neuman, B.W.; Gorbalenya, A.E.; et al. Deep Mining of the Sequence Read Archive Reveals Major Genetic Innovations in Coronaviruses and Other Nidoviruses of Aquatic Vertebrates. PLoS Pathog. 2024, 20, e1012163. [Google Scholar] [CrossRef] [PubMed]
- Taxonomy Release History. Available online: https://ictv.global/taxonomy/history (accessed on 19 November 2025).
- Goldfarb, T.; Kodali, V.K.; Pujar, S.; Brover, V.; Robbertse, B.; Farrell, C.M.; Oh, D.-H.; Astashyn, A.; Ermolaeva, O.; Haddad, D.; et al. NCBI RefSeq: Reference Sequence Standards through 25 Years of Curation and Annotation. Nucleic Acids Res. 2025, 53, D243–D257. [Google Scholar] [CrossRef] [PubMed]


| No. | Tool | Methodology | Database Source | Viral Specialization | CI | Limitations |
|---|---|---|---|---|---|---|
| 1 | BLAST | Pairwise alignment | NCBI RefSeq Viral | DNA viruses, RNA, eukaryotic viruses, phages, archaeal viruses, and protein queries across all viral taxa | 120,903 | - Slower than MegaBLAST for near-identical sequences - Requires pre-formatted databases - May miss extremely weak similarities - Computationally heavy for very large datasets |
| 2 | MegaBLAST | Pairwise alignment (fast) | NCBI RefSeq Viral | DNA viruses, eukaryotic viruses, phages, and archaeal viruses | 1620 | - Performance degrades with very long queries (>100 kb) or highly repetitive regions - Memory-intensive indexing - Limited sensitivity for highly divergent (low-identity < 70%) or RNA virus sequences |
| 3 | PSI-BLAST | Multiple sequence alignment (iterative) | NCBI RefSeq Viral | DNA viruses, RNA viruses, eukaryotic viruses, phages (bacteriophages), and archaeal viruses | 89,349 | - Deceptive alignments caused by highly biased amino acid regions, which can lead to a high false positive rate |
| 4 | PASC | Pairwise alignment | NCBI | DNA viruses, RNA viruses, eukaryotic viruses, phages, and archaeal viruses | 196 | - Some virus families show overlapping peaks, making demarcations difficult - Global alignment is less reliable for large or distantly related genomes |
| 5 | MetaPhinder | Pairwise alignment | NCBI RefSeq Viral, EMBL EBI, phagesdb | dsDNA bacteriophages and archaeal viruses | 78 | - Requires similarity to known phage genomes - Drop in accuracy for short contigs (<5 kb) |
| 5 | SDT | Pairwise alignment | ICTV | Plant ssDNA viruses and novel circular DNA viruses from metagenomic studies optimized for small circular DNA viruses, but applicable to any virus family with ICTV identity thresholds | 1817 | - Alignment quality affects results for highly divergent genomes |
| 6 | VICTOR | Pairwise alignment | INSDC | Prokaryotic (bacterial and archaeal) DNA viruses, mainly tailed dsDNA phages (order Caudovirales) | 617 | - Requires nearly complete genomes - Lower resolution at family rank - Computationally heavy for very large datasets |
| 7 | VIRDIC | Pairwise alignment | GenBank | Prokaryote-infecting dsDNA viral genomes (bacteriophages, archaeal viruses) | 669 | - BLASTN sensitivity limit (65%) can miss distant relationships - Draft/scaffold genomes with gaps cause underestimation - Repetitive regions may bias results |
| 8 | vConTACT v.2.0 | Pairwise alignment | NCBI RefSeq Viral | dsDNA viruses infecting Bacteria and Archaea | 295 | - Requires rebuilding reference networks when adding new data - Struggles with short contigs, overlapping genomes, and highly mosaic viruses |
| 9 | ViralMSA | Multiple sequence alignment | GenBank | RNA, DNA, eukaryotic, and phage viruses | 86 | - Discards insertions relative to the reference genome - Not suitable for viruses lacking a reliable reference genome - Dependent on external mapper performance |
| 10 | VIRULIGN | Multiple sequence alignment | - | RNA and DNA viruses, including eukaryotic viruses, phages | 73 | - Requires a reliable reference sequence and annotation - Discards sequences with excessive frame-shifts |
| 11 | MAFFT | Multiple sequence alignment | - | RNA viruses, DNA viruses, phages (bacteriophages), archaeal viruses, and eukaryotic viral genes | 17,706 | - Slightly less accurate than structural aligners for very distantly related proteins - Requires parameter tuning for low-similarity sequences |
| 12 | ClustalW | Multiple sequence alignment | - | DNA, RNA, phage, archaeal, and eukaryotic viruses | 75,099 | - Early misalignments in progressive steps are not re-optimized (local-minimum problem) - Accuracy declines for extremely divergent sequences |
| 13 | ClustalX | Multiple sequence alignment | - | DNA, RNA, phage, archaeal, and eukaryotic viruses | 3262 | - Alignment quality can be sensitive to the initial choice of parameters |
| 14 | Clustal Omega | Multiple sequence alignment | - | DNA, RNA, phage, archaeal, and eukaryotic viruses | 1458 | - Initial misalignments may remain uncorrected in the final alignment - Uses the MAC algorithm for profile alignment (demands high memory for large datasets) |
| 15 | GLUE | Multiple sequence alignment | GenBank | Hepatitis C virus (HCV), Rabies virus, Bluetongue virus, Hepatitis B virus, Circoviridae, Parvoviridae, Flaviviridae, Retroviridae | 118 | - Assumes a single evolutionary history per alignment tree - Insertions relative to the reference lose homology information |
| 16 | MUSCLE | Multiple sequence alignment | - | DNA viruses, RNA viruses, bacteriophages, archaeal viruses, and eukaryotic viruses | 50,759 | - Performance drops for extremely divergent sequences (<15% identity) - Progressive alignment errors are irreversible (no global re-optimization) |
| 17 | MACSE | Multiple sequence alignment | - | RNA Viruses (ssRNA, dsRNA), Plant RNA Viruses (Tobamovirus, Potyvirus), Archaeal and Eukaryotic DNA Viruses (NCLDVs, Adenoviridae) | 693 | - Slower than standard tools like MUSCLE or TranslatorX due to algorithm complexity - Ignores certain frameshift event types, leading to approximate solutions - Requires parameter tuning for optimal performance |
| 18 | TranslatorX | Multiple sequence alignment | - | RNA viruses, DNA viruses, phages (bacteriophages), archaeal viruses, and eukaryotic viral genes, Plant and archaeal viruses | 1505 | - Cannot accommodate frameshifts - Cannot explicitly handle frameshifts or true pseudogene sequences - Relies on external aligners for performance and accuracy - Limited performance on extremely fragmented contigs (<100 bp) |
| 19 | FastANI | Rapid similarity estimation (ANI) | - | large dsDNA viruses (>200 kbp, NCLDVs, giant phages, poxviruses) | 4694 | - Performance drops significantly below ~80% ANI for divergent sequences - Sensitive to poor assembly - Computationally intensive for large databases |
| 20 | Vclust | Rapid similarity estimation (ANI) + k-mer + clustering | RefSeq, GenBank, IMG/VR v4.1, Kmer-db2 | DNA viruses, RNA viruses, bacteriophages, archaeal viruses, and eukaryotic viruses | 8 | - Performance may decrease with large datasets of highly similar genomes |
| No. | Tool | Methodology | Database Source | Viral Specialization | CI | Limitations |
|---|---|---|---|---|---|---|
| 1 | VirSorter | Profile hidden Markov models | RefSeq Virus genomes, data from public studies | Bacterial and archaeal dsDNA viruses (Caudovirales and non-Caudovirales); detects integrated prophages and free lytic viruses | 1149 | - Limited detection of eukaryotic viruses (database bias) - Inefficient for short contigs (<3 kb) or non-assembled reads - May include false positives (genomic islands, MGEs) - Does not analyze integrase/att sites, less accurate for complete prophage boundaries |
| 2 | Cenote-Taker 2 | Profile hidden Markov models + BLAST | Hallmark gene HMM (Hmmer) database, CDD, Pfam, PDB, GenBank, RefSeq | All virus classes with DNA or RNA genomes, Prokaryotic, Prophages, Archaeal, Eukaryotic | 141 | - Does not allow automatic identification of ribosomal frameshifts and intron-containing genes - Performance depends on the presence of recognizable hallmark genes - May miss minimal or highly degraded viruses lacking core proteins |
| 3 | Phigaro | Profile hidden Markov models | pVOGs | Prophage | 186 | - Slightly lower Jaccard index compared to PHASTER and VirSorter - Sensitivity depends on the selected detection mode |
| 4 | Phage Finder | HMM + BLAST + tRNAscan-SE + Aragorn + fasta33 + MUMMER | NCBI or WU BLASTP data, HMMSEARCH data, tRNAscan-SE data, Aragorn data, Phage_Finder information file, GenBank | Prophage | 368 | - Can skip “tandem” (piggy-back) prophages integrated into a single genomic site - Many putative prophage regions lack core HMM matches (large terminase, portal, major capsid), which may cause missed detections under strict mode settings |
| 5 | RdRp-scan | Hidden Markov Models | NCBI Riboviria’s proteins, PALMdb, data from public studies | Primarily eukaryotic RNA viruses (+ssRNA, -ssRNA, dsRNA), with partial coverage of prokaryotic RNA phages | 92 | - Short or fragmented ORFs (<200 aa) are often missed - Requires manual validation for some motifs and structural matches - Performance depends on database updates (must be regularly maintained) - May miss extremely divergent RdRps that fall outside of existing HMM or structural models |
| 6 | NeoRdRp2 | Hidden Markov Models | PALMdb, NCBI RNA Virus database, UniProtKB, GenBank, data from public studies | RNA viruses (primarily eukaryotic), including prokaryotic RNA phages such as Leviviricetes | 5 | - May split core RdRp motifs (A-C) during HMM construction - False positives in a small fraction (188/564k non-RdRp hits) - Extremely divergent RdRps may still escape detection |
| 7 | ViralRecall | Profile hidden Markov models | Data from public studies | nucleo-cytoplasmic large DNA viruses (NCLDV) | 80 | - Potential for false positives on short fragments - Time-consuming and not feasible for large datasets |
| No. | Tool | Methodology | Database Source | Viral Specialization | CI | Limitations |
|---|---|---|---|---|---|---|
| 1 | VirSorter 2 | ML (RF + hidden Markov models) | JGI Earth’s virome project, Xfams, NCBI RefSeq genomes, including archaea, bacteria, protists, fungi, and viruses | dsDNA phage, nucleo-cytoplasmic large DNA viruses (NCLDV), RNA viruses, ssDNA viruses, prophages | 1151 | - Performance drops on short contigs (<3 kb) - Some false positives with plasmids and eukaryotic mobile elements - High computational demand due to multi-classifier ensemble - RNA virus predictions require high-quality metatranscriptomic assemblies |
| 2 | VirFinder | ML (LR + k-mer) | RefSeq | Prokaryotic dsDNA viruses (bacteriophages and archaeal viruses) | 653 | - Lower performance for archaeal viruses |
| 3 | MARVEL | ML (RF) | RefSeq | dsDNA bacteriophages | 186 | - Dependent on upstream binning quality (chimeric bins cause errors) |
| 4 | ViraPipe | ML (RF)+ FFNN | NCBI GenBank, 19 different NGS experiments | Human DNA viruses | 56 | - Lower recall (few viral contigs detected at high precision) - Performance declines on very short or noisy contigs - Requires coding regions |
| 5 | VIBRANT | ML (NN) | RefSeq and Genbank, KEGG KoFam, Pfam (v32), and Virus Orthologous Groups (VOG) | Bacterial and archaeal dsDNA, ssDNA, and RNA viruses, capable of integrated provirus detection (Prophages) | 951 | - Lower recovery for very short (<1 kb) contigs - Depends on high-quality ORF prediction - Potential minor bias toward NCBI-trained proteins |
| 6 | PhiSpy | ML (RF) | Phantome server | Integrated bacterial dsDNA prophages | 561 | - Less effective for fragmented assemblies or metagenomic datasets - Misses very small or highly degraded prophages - Requires annotated ORFs for accurate detection. |
| 7 | DeepVirFinder | ML (CNN) | RefSeq | dsDNA viruses infecting bacteria and archaea | 594 | - Possible misclassification of eukaryotic contamination - Slightly less accurate for extremely short fragments (<150 bp) |
| 8 | PPR-Meta | ML (BiPathCNN) | RefSeq | Phages and plasmids (bacterial and archaeal MGEs, temperate and lytic) | 181 | - Slightly lower plasmid classification accuracy - Limited to prokaryotic MGEs (mobile genetic elements) |
| 9 | ViraMiner | ML (CNN) | Data from public studies | Human DNA viruses | 152 | - Lower recall for rare viral classes - Optimized for human-associated datasets - Less effective for environmental metagenomes |
| 10 | CHEER | ML(CNN) + k-mer | RefSeq | RNA viruses (human, animal, bacterial, archaeal, and environmental RNA viromes) | 65 | - Trained only on RefSeq viral genomes (may miss rare or novel taxa) |
| 11 | DeepMicroClass | ML (diPathCN) | NCBI Genome database, Kaiju, the PR2 database, MMETSP project, PLSDB, Virus–Host DB | Broad coverage of prokaryotic (including archaeal) and eukaryotic viruses—encompassing dsDNA, ssDNA, dsRNA, ssRNA viral genomes, plasmids, and prophages | 13 | - Possible misclassification between plasmids and prokaryotic chromosomes - Slightly reduced sensitivity for integrated proviruses - Not specialized for short reads (<500 bp) |
| 12 | VirDetect-AI | ML (CNN + RNN) | Virus Protein Database NCBI, | Eukaryotic DNA/RNA viruses | 2 | - Limited performance for short proteins (<300 aa) - Slightly reduced sensitivity for rare viral families (e.g., Orthoherpesviridae, Retroviridae) |
| 13 | Seeker | ML (LSTM) | RefSeq | Bacteriophages dsDNA (Caudovirales and unclassified tailed phages) | 131 | - Less accurate for very short sequences (<1 kbp) - May produce false positives for sequences with phage-like motifs but non-viral origin - Performance sensitive to sequence length distribution and GC-content biases |
| 14 | HVSeeker | ML (LSTM + ProteinBER) | NCBI, Integrated Microbial Genomes, Microbiomes–Viruses (IMGVR) | Bacteriophage dsDNA and bacterial host sequences | 1 | - RNN-based (LSTM) models can overfit long sequences (>1.5 kb) - Performance slightly lower for extremely short (<200 bp) fragments |
| 15 | Virtifier | ML (LSTM) | Refeq viral | Prokaryotic and eukaryotic DNA viruses | 48 | - Reduced accuracy when training/testing lengths mismatch |
| 16 | VirNucPro | ML (MLP) | Refeq viral | Broad viral DNA detection (both prokaryotic + archaeal and eukaryotic viruses), prophage detection | 1 | - Requires valid CDS regions for accurate analysis - Potential false positives from bacterial MGEs (mobile genetic elements) - Six-frame translation may introduce non-authentic protein products - Limited by standard codon usage assumptions |
| 17 | DETIRE | ML (CNN + BiLSTM) | Refeq viral | DNA viruses (bacteriophages + archaeal and eukaryotic) | 9 | - Accuracy drops slightly on very long contigs (>3 kbp) - The embedding model is optimized for 500 bp fragments |
| 18 | VirRep | ML (BERT + BiLSTM) | Published human gut virome catalogs, genomes of human gut prokaryotes | DNA viruses (prokaryotic and eukaryotic) | 4 | - Comprehensive evaluation across diverse environments is lacking - Requires biome-specific fine-tuning for non-gut or other environments |
| 19 | ViraLM | ML (DNABERT-2 + binary classifier) | NCBI RefSeq viral, data from public studies | DNA and RNA viruses from bacterial, archaeal, and eukaryotic hosts | 10 | - May lose long-range genomic context - Provides limited functional information - Faces computational challenges when processing lengthy input sequences |
| 20 | GCNFrame | ML (MLP) | NCBI and ICEberg | Prokaryotic dsDNA phages and bacterial host elements (ICEs), Prophages | 4 | - Sensitive to hyperparameter settings, which can affect model stability - Graph memory overhead due to inclusion of all possible k-mers (even those with zero counts) |
| 21 | PhaGCN | ML (CNN + GCN) | NCBI RefSeq | Bacteriophages (Caudovirales); validated families: Myoviridae, Siphoviridae, Podoviridae, Ackermannviridae, Herelleviridae, Demerecviridae, extensible to Rudiviridae, Inoviridae | 115 | - Performance decreases with shorter contigs (<4 kb) - Caudovirales-centric (covers ≈ 95.8% of known phages, but is not universal) |
| 22 | geNomad | ML (DNN + XGBoost) | GTDB, TOPAZ, PLSDB, RefSeq, IMG/VR version 3, Specialized sets of viruses (Nucleocytoviricota, Leviviridae, Asgard archaea viruses, etc.) | Broad-spectrum DNA viruses (prokaryotic + eukaryotic) and plasmids, capable of identifying novel MGEs, sequences of plasmids, proviruses, RNA, and giant viruses | 594 | - May not classify completely novel viral families lacking known markers - At lower taxonomic ranks, it shows limited family coverage (61.8%) |
| No. | Tool | Methodology | Database Source | Viral Specialization | CI | Limitations |
|---|---|---|---|---|---|---|
| 1 | Kraken2 | k-mer | Refeq viral | Broad DNA and RNA viruses, dsDNA bacteriophages and vertebrate DNA viruses, divergent RNA viruses | 6081 | - Slightly reduced specificity due to probabilistic hashing - Limited species-level resolution for highly similar taxa - Relies on the quality and completeness of the reference database |
| 2 | DisCVR | k-mer | NCBI taxonomy database | Human DNA and RNA viruses | 11 | - Lower sensitivity for low-coverage or highly fragmented samples |
| 3 | CLARK | k-mer | NCBI/Refeq viral | Bacterial, archaeal, and eukaryotic viruses are mainly DNA viruses, but RNA viruses can be detected if represented in the database | 775 | - Precision–sensitivity tradeoff depending on k-mer size |
| 4 | Vista | Pairwise alignment + k-mer profiles + ML | NCBI Viral Genomes Resource | Optimized for Caudoviricetes—tailed dsDNA bacteriophages infecting bacterial and archaeal hosts; eukaryotic viral families, spanning ssDNA, dsDNA, ssRNA, and dsRNA | 3 | - Bias toward taxa with many reference sequences - Difficulty handling singletons and segmented genomes - Requires frequent database updates to maintain accuracy |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galeeva, J.; Kuzmichenko, P.; Manolov, A.; Lukashev, A.; Ilina, E. Bioinformatics Tools and Approaches for Virus Discovery in Genomic Data: A Systematic Review. Viruses 2025, 17, 1538. https://doi.org/10.3390/v17121538
Galeeva J, Kuzmichenko P, Manolov A, Lukashev A, Ilina E. Bioinformatics Tools and Approaches for Virus Discovery in Genomic Data: A Systematic Review. Viruses. 2025; 17(12):1538. https://doi.org/10.3390/v17121538
Chicago/Turabian StyleGaleeva, Julia, Polina Kuzmichenko, Alexander Manolov, Alexander Lukashev, and Elena Ilina. 2025. "Bioinformatics Tools and Approaches for Virus Discovery in Genomic Data: A Systematic Review" Viruses 17, no. 12: 1538. https://doi.org/10.3390/v17121538
APA StyleGaleeva, J., Kuzmichenko, P., Manolov, A., Lukashev, A., & Ilina, E. (2025). Bioinformatics Tools and Approaches for Virus Discovery in Genomic Data: A Systematic Review. Viruses, 17(12), 1538. https://doi.org/10.3390/v17121538






