Genomic and Biological Characterization of a Novel Proteus mirabilis Phage with Anti-Biofilm Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Sample Collection
2.2. Enrichment of Bacteriophages
2.3. Isolation and Purification of Bacteriophage
2.4. Concentration and Electron Microscopy Observation of Bacteriophage
2.5. Lytic Spectrum Profiling
2.6. Optimal Multiplicity of Infection (MOI) Determination
2.7. One-Step Growth Curve Analysis
2.8. Thermal Stability Assessment
2.9. pH Tolerance Evaluation
2.10. Bacteriophage-Mediated Anti-Biofilm Activity Analysis
2.11. Genomic DNA Extraction, Sequencing, and Bioinformatics Analysis
2.12. Data Analysis
3. Results
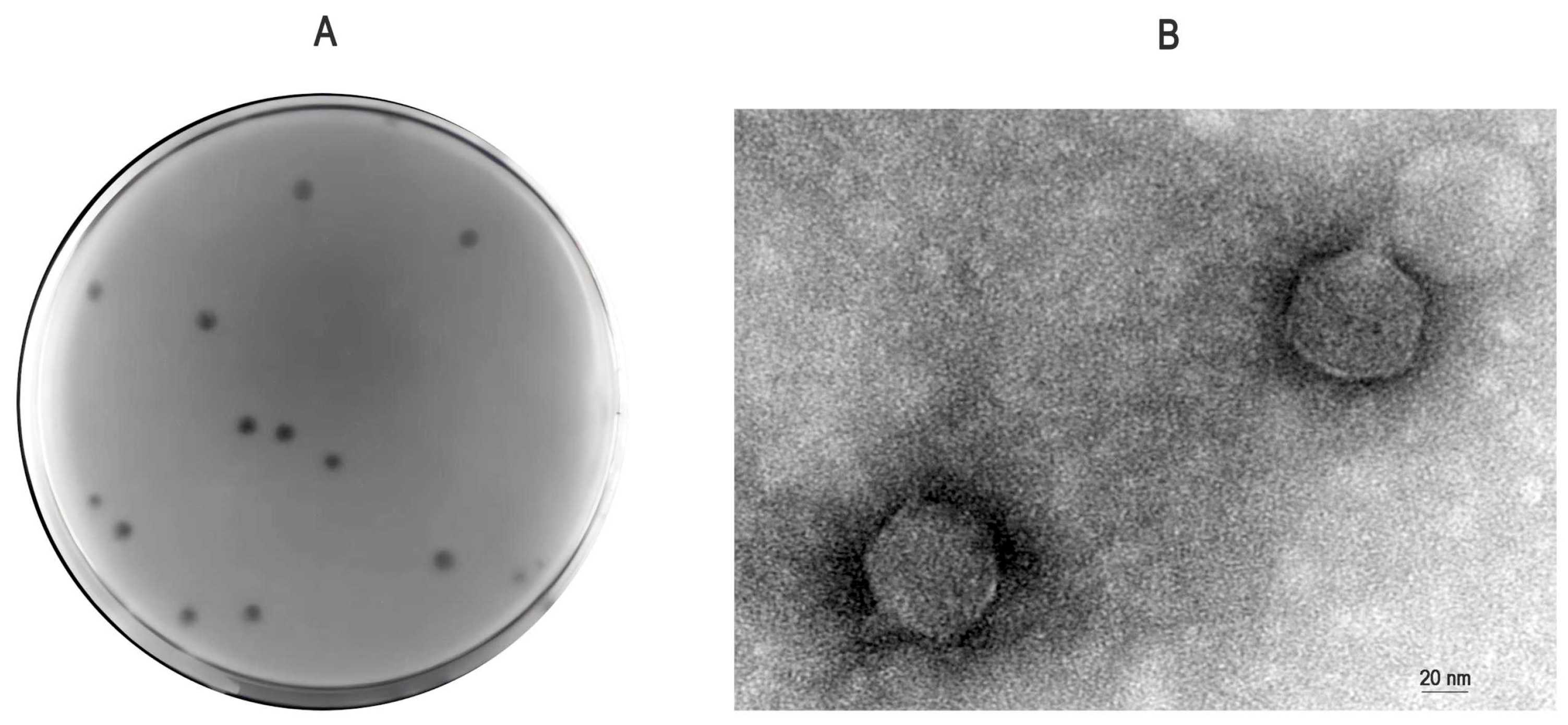
3.1. Isolation, Purification, and Electron Microscopy Morphology
3.2. Lytic Spectrum Determination
3.3. Optimal Multiplicity of Infection (MOI) and One-Step Growth Curve of P-07
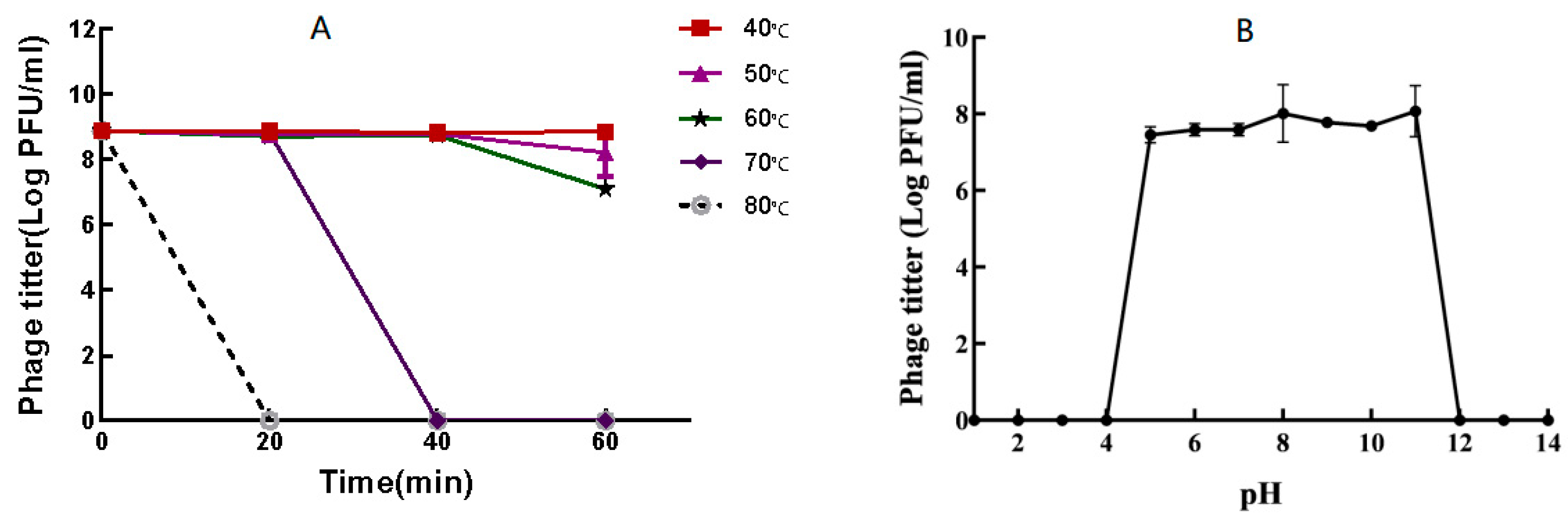
3.4. Thermal and pH Stability of Phage P-07
3.5. Biofilm Clearance Efficacy Against P. Mirabilis 07
3.6. Strain-Dependent Anti-Biofilm Activity
3.7. Differential Anti-Biofilm Efficacy Across Strains with Varying Biofilm-Forming Capacities
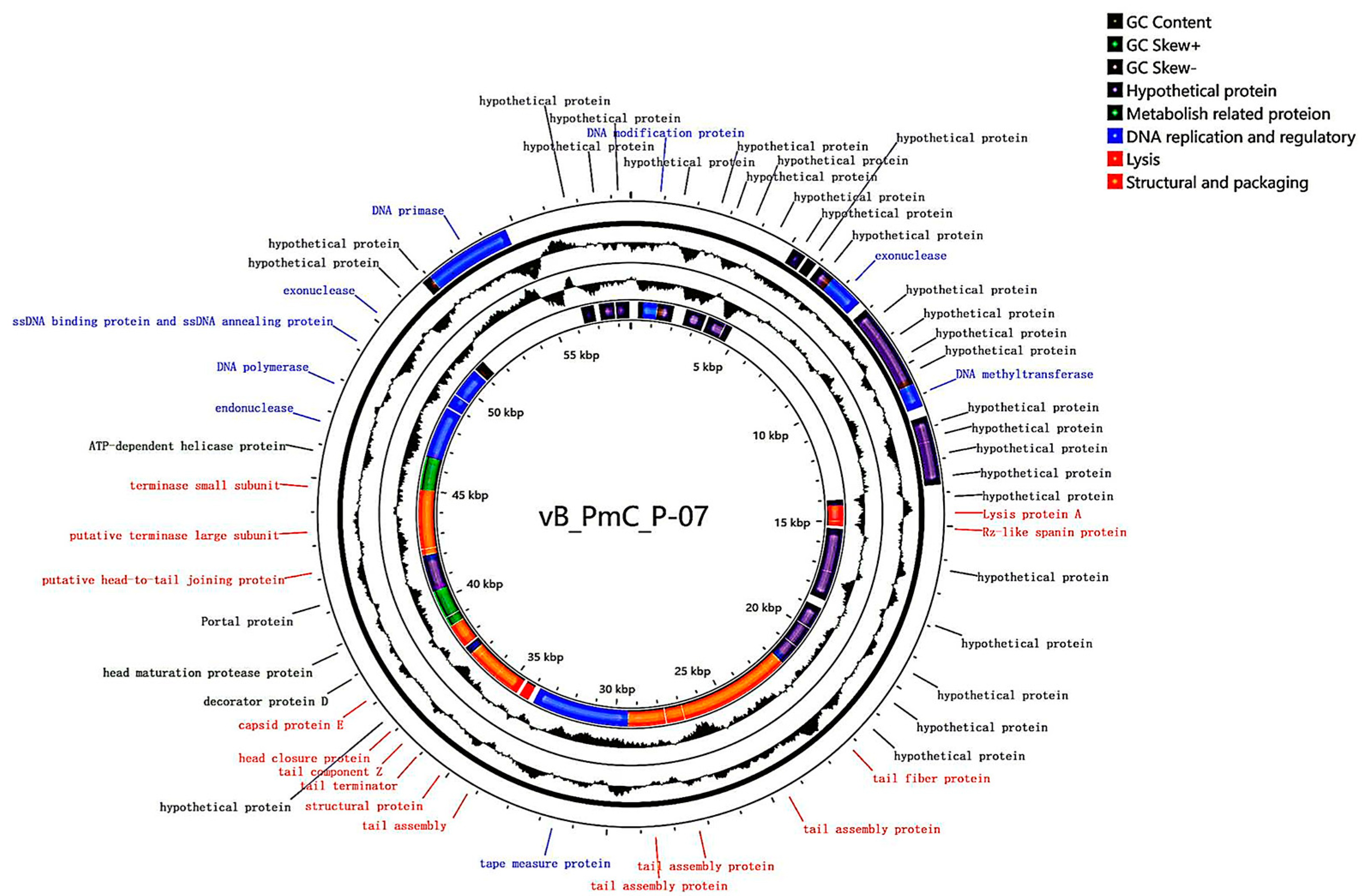
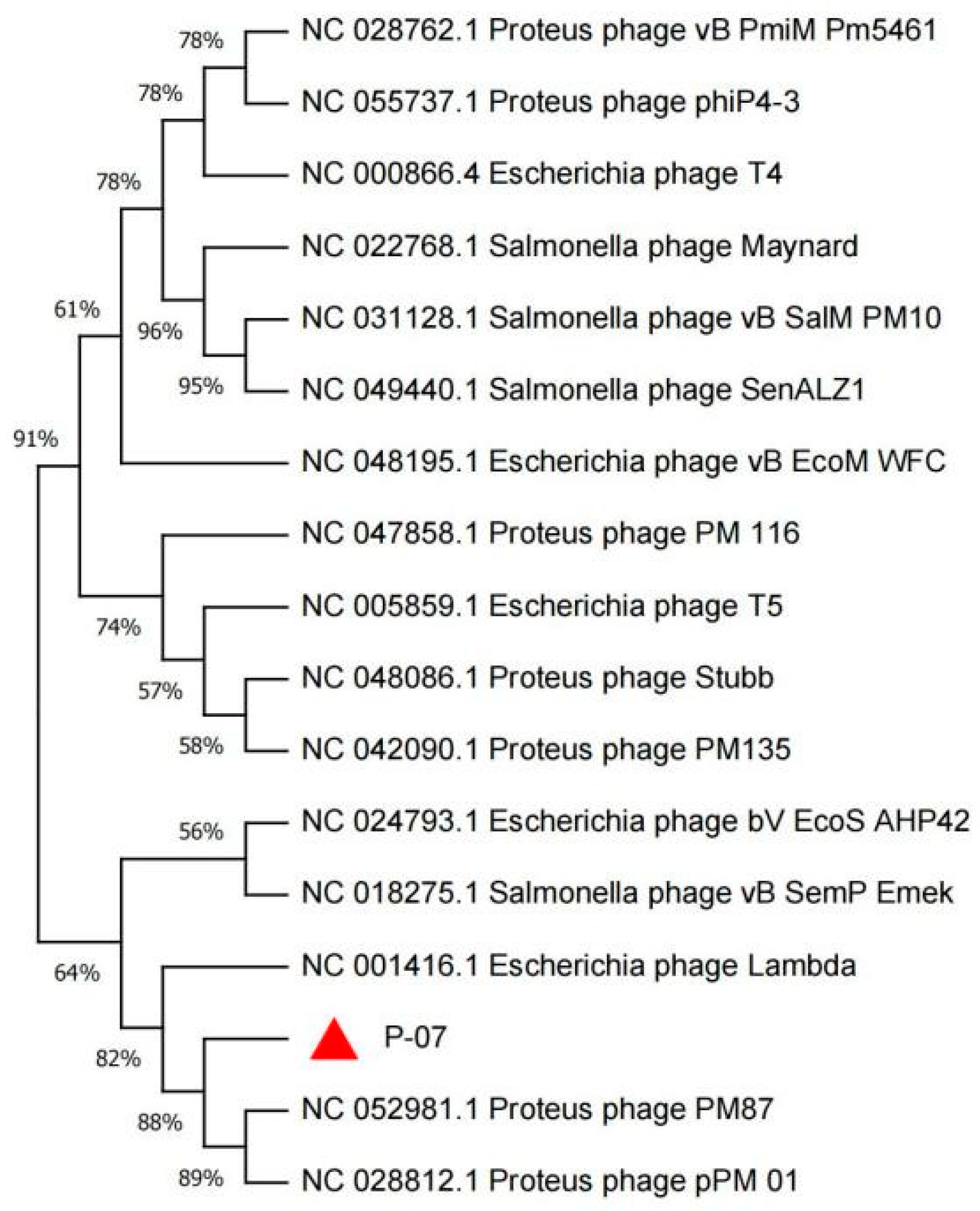
3.8. Whole-Genome Analysis of Phage P-07
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDR | Multidrug-Resistant |
| ESBL | Extended-Spectrum Beta-Lactamase |
| PFU | Plaque Forming Units |
| MOI | Multiplicity of Infection |
| CARD | Comprehensive Antibiotic Resistance Database |
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef]
- Colantonio, M.A.; Hartzell, M.; Shannon, B. Proteus mirabilis: An Unlikely Device-related Infection. J. Community Hosp. Intern. Med. Perspect. 2025, 15, 84–88. [Google Scholar] [CrossRef]
- Sun, Y.D.; Wen, S.S.; Zhao, L.L.; Xia, Q.Q.; Pan, Y.; Liu, H.H.; Wei, C.W.; Chen, H.Y.; Ge, J.W.; Wang, H.B. Association among biofilm formation, virulence gene expression, and antibiotic resistance in Proteus mirabilis isolates from diarrhetic animals in Northeast China. BMC Vet. Res. 2020, 16, 176. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Zhou, J.; Huang, H.; Wang, W.; Xiao, Y.; Tang, B.; Liu, H.; Xu, C.; Xiao, X. Genomic Investigation of Proteus mirabilis Isolates Recovered from Pig Farms in Zhejiang Province, China. Front. Microbiol. 2022, 13, 952982. [Google Scholar] [CrossRef] [PubMed]
- Keeratikunakorn, K.; Kaewchomphunuch, T.; Kaeoket, K.; Ngamwongsatit, N. Antimicrobial activity of cell free supernatants from probiotics inhibits against pathogenic bacteria isolated from fresh boar semen. Sci. Rep. 2023, 13, 5995. [Google Scholar] [CrossRef]
- Liu, X.L.; Wu, S.Y.; Yu, Z. Zoonotic Risks of Proteus mirabilis: Detection, Pathogenicity, and Antibiotic Resistance in Animals and Animal-Derived Foods. Microorganisms 2025, 13, 2060. [Google Scholar] [CrossRef] [PubMed]
- Chinnam, B.K.; Nelapati, S.; Tumati, S.R.; Bobbadi, S.; Chaitanya Peddada, V.; Bodempudi, B. Detection of β-Lactamase-Producing Proteus mirabilis Strains of Animal Origin in Andhra Pradesh, India and Their Genetic Diversity. J. Food Prot. 2021, 84, 1374–1379. [Google Scholar] [CrossRef]
- Alves, D.R.; Nzakizwanayo, J.; Dedi, C.; Olympiou, C.; Hanin, A.; Kot, W.; Hansen, L.; Lametschs, R.; Gahan, C.G.M.; Schellenberger, P.; et al. Genomic and Ecogenomic Characterization of Proteus mirabilis Bacteriophages. Front. Microbiol. 2019, 10, 1783. [Google Scholar] [CrossRef]
- Royer, S.; Morais, A.P.; da Fonseca Batistão, D.W. Phage therapy as strategy to face post-antibiotic era: A guide to beginners and experts. Arch. Microbiol. 2021, 203, 1271–1279. [Google Scholar] [CrossRef]
- Chen, J.; Han, J.; Yang, Z.; Zhou, W.; He, Y.; Chen, X.; Li, X.; Zou, T.; You, J. Bacteriophages as Potential Anti-Pathogenic Agents for Intestinal Health of Weaned Piglets in the Post-Antibiotic Era: An Updated Review. Animals 2025, 15, 1713. [Google Scholar] [CrossRef]
- Soffritti, I.; D’Accolti, M.; Bini, F.; Mazziga, E.; Volta, A.; Bisi, M.; Mazzacane, S.; De Cesare, A.; Indio, V.; Manfreda, G.; et al. Harnessing probiotics and bacteriophages to fight Salmonella and limit the use of antibiotics in broilers: A study in commercial conditions. Poult. Sci. 2025, 104, 105595. [Google Scholar] [CrossRef]
- Jamalludeen, N.; Johnson, R.P.; Friendship, R.T.; Kropinski, A.M.; Lingohr, E.J.; Gyles, C.L. Isolation and characterization of nine bacteriophages that lyse O149 enterotoxigenic Escherichia coli. Vet. Microbiol. 2007, 124, 47–57. [Google Scholar] [CrossRef]
- Chang, H.C.; Chen, C.R.; Lin, J.W.; Shen, G.H.; Chang, K.M.; Tseng, Y.H.; Weng, S.F. Isolation and characterization of novel giant Stenotrophomonas maltophilia phage phiSMA5. Appl. Environ. Microbiol. 2005, 71, 1387–1393. [Google Scholar] [CrossRef]
- Cai, R.P.; Wang, Z.J.; Wang, G.; Zhang, H.; Cheng, M.J.; Gu, Z.M.; Ji, Y.L.; Xi, H.Y.; Wang, X.W.; Xue, Y.B.; et al. Biological properties and genomics analysis of vB_KpnS_GH-K3, a Klebsiella phage with a putative depolymerase-like protein. Virus Genes 2019, 55, 696–706. [Google Scholar] [CrossRef]
- Wan, S.; Li, N.; Habib, S.; Zheng, P.; Li, Y.; Liang, Y.; Qu, Y. Biological Characteristics and Whole-Genome Analysis of a Porcine E. coli Phage. Vet. Sci. 2025, 12, 57. [Google Scholar] [CrossRef]
- Li, F.; Xing, S.; Fu, K.; Zhao, S.P.; Liu, J.F.; Tong, Y.G.; Zhou, L.J. Genomic and biological characterization of the Vibrio alginolyticus-infecting “Podoviridae” bacteriophage, vB_ValP_IME271. Virus Genes 2019, 55, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.; Zhou, H.; Yu, X.; Li, N.; Li, Y.; Qu, Y.; Li, Y.; Liang, Y.; Yan, D.; Yang, S.; et al. Biological characteristics and whole genome analysis of an ovine-derived multidrug-resistant Klebsiella pneumoniae phage. China Anim. Husb. Vet. Med. 2024, 51, 2047–2057. (In Chinese) [Google Scholar]
- Pajunen, M.; Kiljunen, S.; Skurnik, M. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 2000, 182, 5114–5120. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.Y. Safety Evaluation of a Phage Cocktail for Bovine Mastitis. Master’s Thesis, Shihezi University, Shihezi, China, 2022. (In Chinese). [Google Scholar]
- Shahin, K.; Bouzari, M.; Wang, R. Isolation, characterization and genomic analysis of a novel lytic bacteriophage vB_SsoS-ISF002 infecting Shigella sonnei and Shigella flexneri. J. Med. Microbiol. 2018, 67, 376–386. [Google Scholar] [CrossRef]
- Tochihara, T.; Sasaki, K.; Araki, O.; Morimoto, N.; Watanabe, K.; Hatada, Y.; Ito, S.; Ito, H.; Matsui, H. Site-directed mutagenesis establishes aspartic acids-227 and -342 as essential for enzyme activity in an isomalto-dextranase from Arthrobacter globiformis. Biotechnol. Lett. 2004, 26, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.D.; Sidhu, G.; Pot, I.; Brayer, G.D.; Withers, S.G.; McIntosh, L.P. Hydrogen bonding and catalysis: A novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 2000, 299, 255–279. [Google Scholar] [CrossRef] [PubMed]
- Esmael, A.; Abo-Elmaaty, S.A.; Khafaga, E.M.; Abdelrahman, S.; Hassan, M.G. Efficacy of three lytic bacteriophages for eradicating biofilms of multidrug-resistant Proteus mirabilis. Arch. Virol. 2021, 166, 3311–3322. [Google Scholar] [CrossRef]
- Pan, S.Y.; Cen, Q.B.; Wang, H.Y.; Kong, X.Y.; Pang, S.Y.; Gao, D.Y.; Song, J. Analysis of biological characteristics of a multidrug-resistant Proteus mirabilis phage and its antibacterial effect on chicken breast. Food Sci. 2025, 46, 89–97. [Google Scholar]
- Han, S.; Li, S.; Hu, G.; Lei, M.T.; Li, C.H.; Xiao, L.C.; Li, S.Q. Characterization and genomic analysis phage PmP19, a novel Proteus mirabilis phage with a broad host range. Arch. Virol. 2024, 169, 221. [Google Scholar] [CrossRef]
- Song, J.; Xu, R.Y.; Jin, Y.Q.; Chen, L.; Wang, Y.H.; Ruan, H.R.; Zheng, J.S.; Wu, R. Isolation, identification of canine-derived Proteus mirabilis phage and its effect on biofilm. Acta Vet. Zootech. Sin. 2020, 51, 2785–2793. [Google Scholar]
- Mah, T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- Mirzaei, A.; Wagemans, J.; Nasr, E.B.; Lavigne, R.; Moghim, S. A Phage Cocktail to Control Surface Colonization by Proteus mirabilis in Catheter-Associated Urinary Tract Infections. Microbiol. Spectr. 2022, 10, e0209222. [Google Scholar] [CrossRef]
- Soontarach, R.; Srimanote, P.; Voravuthikunchai, S.P.; Chusri, S. Antibacterial and Anti-Biofilm Efficacy of Endolysin LysAB1245 against a Panel of Important Pathogens. Pharmaceuticals 2024, 17, 155. [Google Scholar] [CrossRef]
- Baquero, D.P.; Liu, Y.; Wang, F.; Egelman, E.H.; Prangishvili, D.; Krupovic, M. Structure and assembly of archaeal viruses. Adv. Virus Res. 2020, 108, 127–164. [Google Scholar]


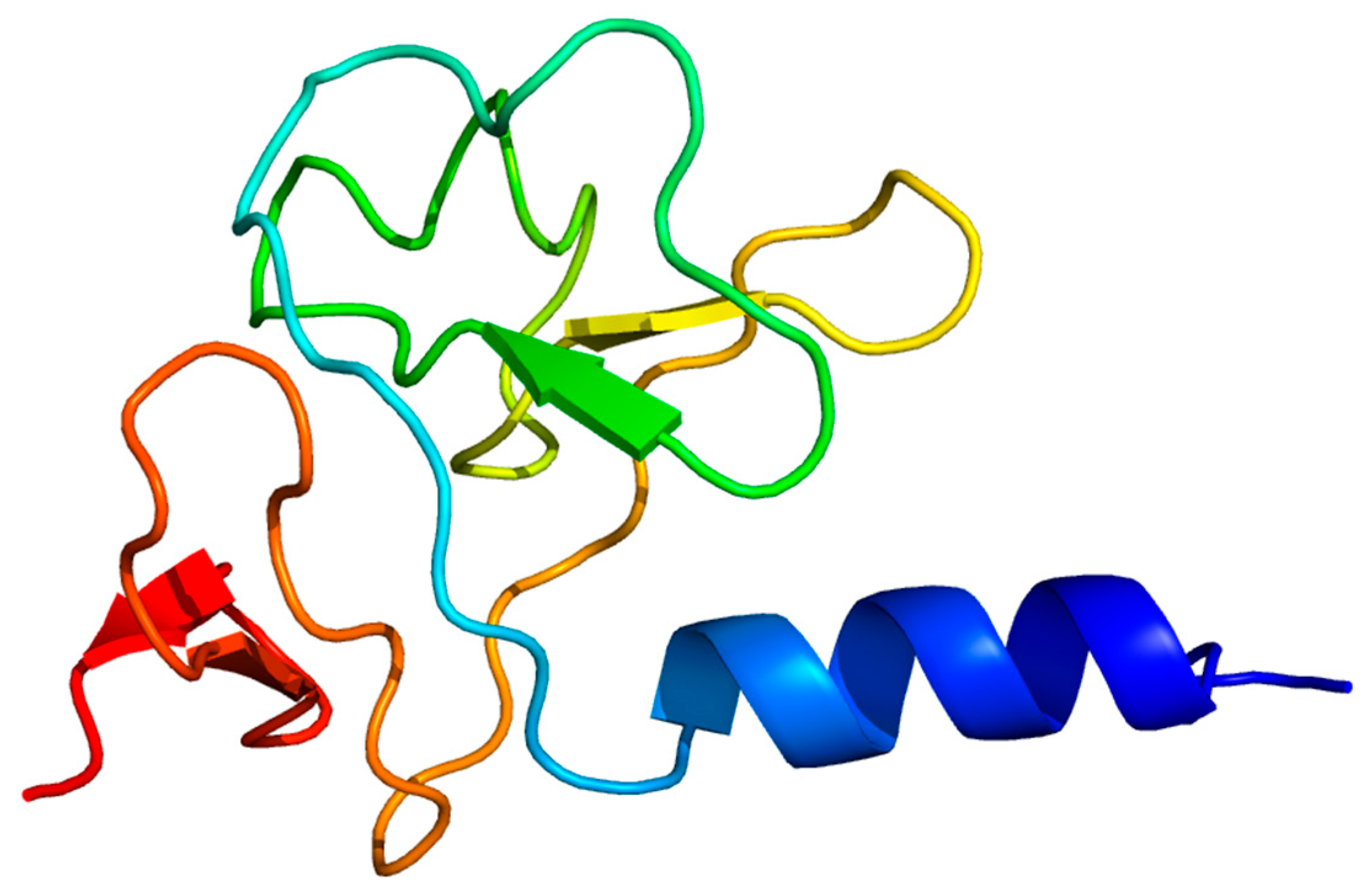
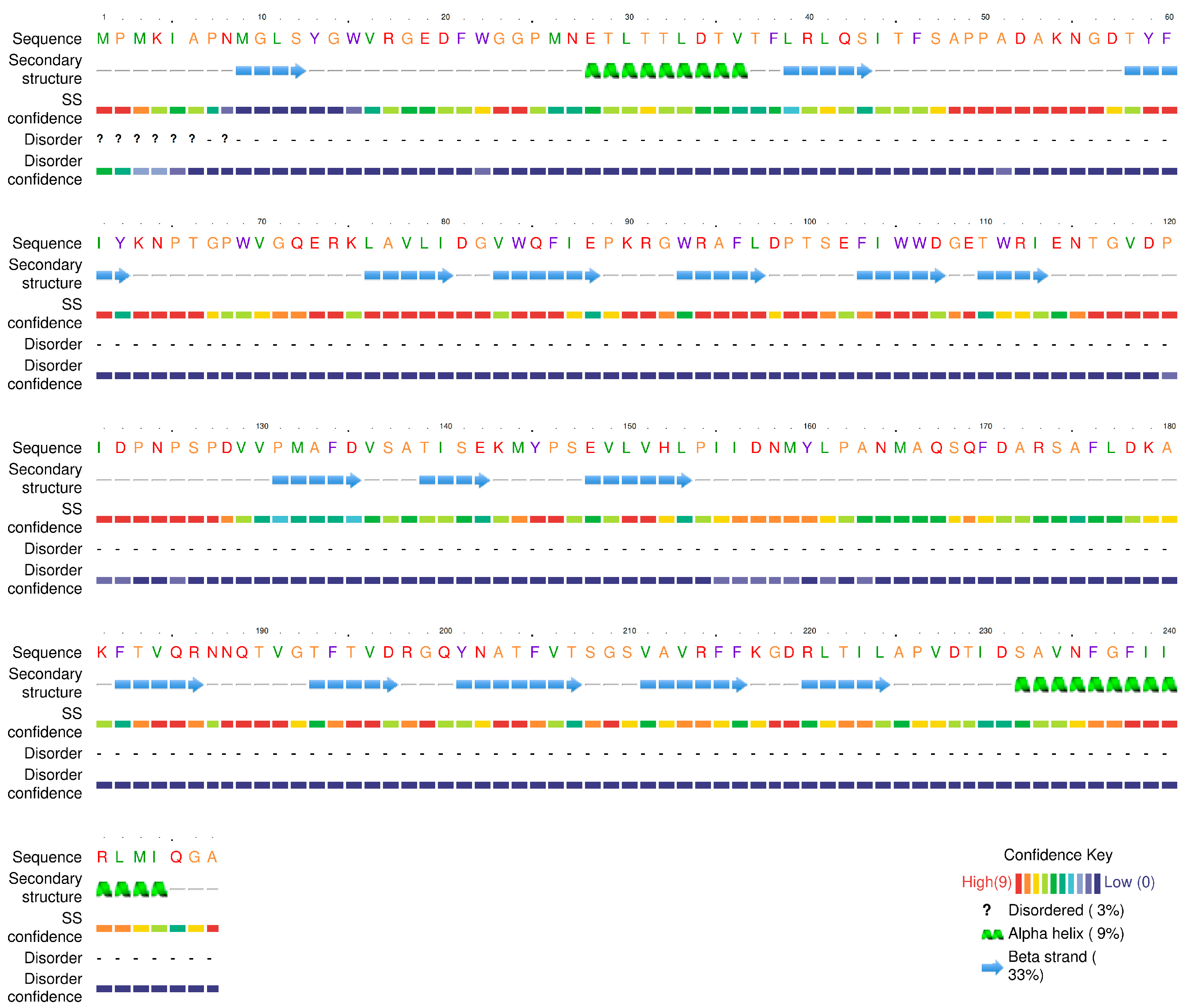
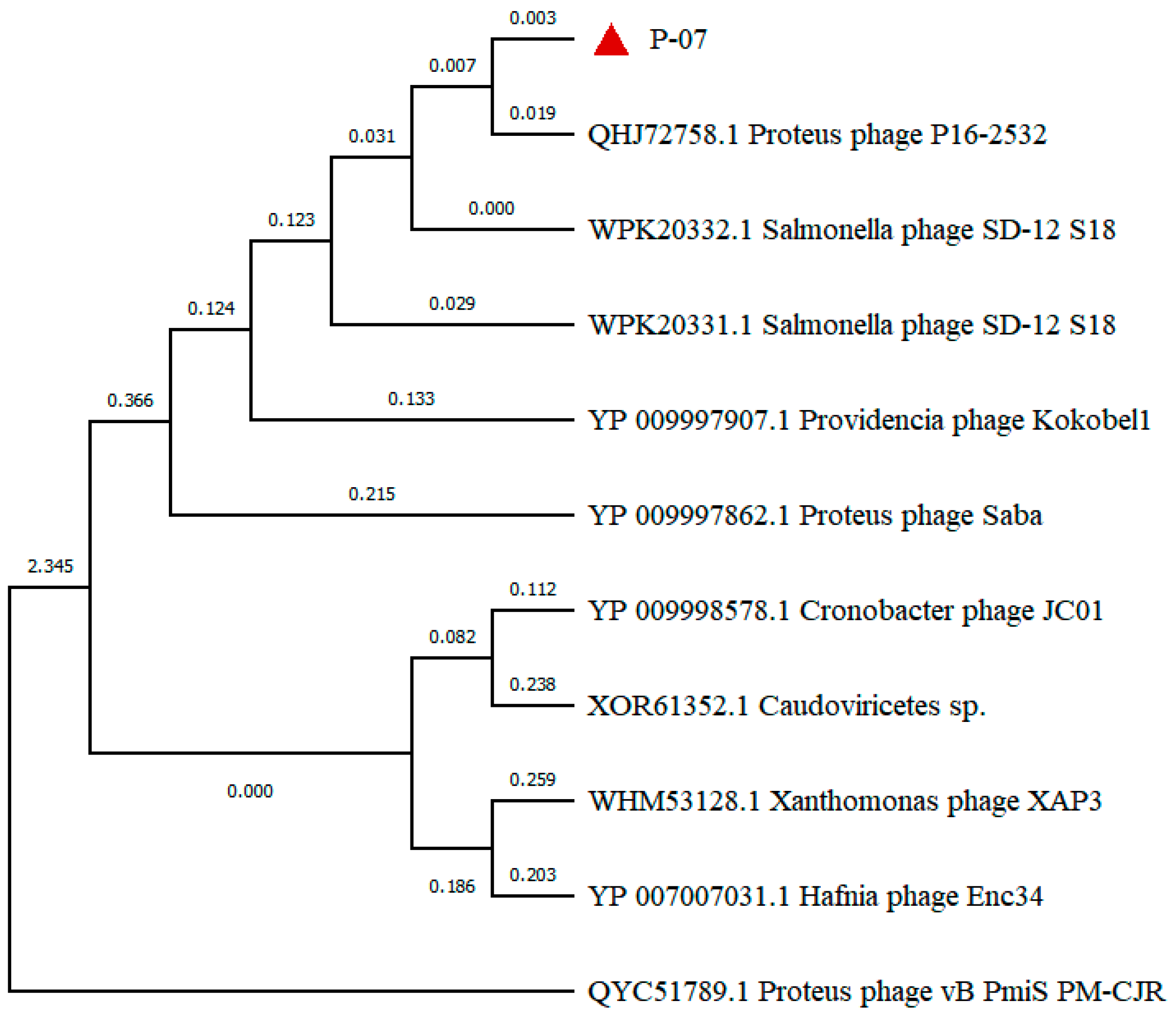
| ORF | START | STOP | Functional Module | Function | Identity/% | E Value | Accession Number |
|---|---|---|---|---|---|---|---|
| 46 | 40,452 | 39,175 | Metabolism-related protein | head maturation protease protein | 99.76% | 0 | YP_009199670.1 |
| 51 | 46,549 | 45,026 | ATP-dependent helicase protein | 99.80% | 0 | UGO49911.1 | |
| 2 | 1183 | 545 | DNA replication and regulatory | DNA modification | 99.53% | 2.00 × 10−153 | UGO51540.1 |
| 13 | 6597 | 7685 | exonuclease | 100.00% | 0 | YP_009997974.1 | |
| 18 | 10,605 | 11,342 | DNA methyltransferase | 99.59% | 0 | UGO36038.1 | |
| 37 | 33,756 | 29,425 | tape measure | 99.79% | 0 | UVD32353.1 | |
| 52 | 46,796 | 46,509 | endonuclease | 78.02% | 4.00 × 10−47 | YP_009997885.1 | |
| 53 | 48,829 | 46,796 | DNA polymerase | 99.11% | 0 | UNI72738.1 | |
| 54 | 49,499 | 48,894 | binding and annealing | 98.99% | 1.00 × 10−139 | YP_009998014.1 | |
| 55 | 50,859 | 49,531 | exonuclease | 97.74% | 0 | YP_010187223.1 | |
| 58 | 52,039 | 54,714 | DNA primase | 99.78% | 0 | UNI72672.1 | |
| 24 | 14,961 | 14,257 | Lysis | lysis protein A | 100.00% | 1.00 × 10−170 | UGO51513.1 |
| 25 | 15,286 | 14,972 | Structural and packaging | Rz-like spanin | 100.00% | 4.00 × 10−34 | YP_009199647.1 |
| 31 | 22,622 | 21,879 | tail fiber | 99.60% | 2.00 × 10−179 | YP_009199654.1 | |
| 32 | 26,443 | 22,622 | tail assembly | 99.29% | 0 | UGO48774.1 | |
| 33 | 26,645 | 26,436 | tail assembly | 81.54% | 5.00 × 10−31 | YP_009997909.1 | |
| 34 | 26,881 | 26,645 | tail assembly | 82.05% | 1.00 × 10−39 | YP_009997910.1 | |
| 35 | 27,709 | 26,894 | tail assembly | 82.29% | 5.00 × 10−171 | YP_009997911.1 | |
| 36 | 29,419 | 27,731 | tail assembly | 99.29% | 0 | YP_009997997.1 | |
| 38 | 34,449 | 33,997 | tail assembly | 100.00% | 8.00 × 10−102 | UGO51498.1 | |
| 39 | 35,775 | 34,588 | structural | 99.75% | 0 | UGO49923.1 | |
| 40 | 36,291 | 35,779 | tail terminator | 99.41% | 9.00 × 10−119 | YP_009199664.1 | |
| 41 | 36,898 | 36,281 | tail component Z | 99.51% | 1.00 × 10−144 | WOZ54555.1 | |
| 42 | 37,254 | 36,901 | head closure | 100.00% | 2.00 × 10−80 | UGO36011.1 | |
| 44 | 38,732 | 37,665 | capsid E | 99.72% | 0 | UGO49918.1 | |
| 45 | 39,161 | 38,745 | decorator protein D | 99.28% | 2.00 × 10−94 | YP_009199669.1 | |
| 48 | 42,367 | 42,092 | head-to-tail joining | 97.80% | 2.00 × 10−59 | UGO51620.1 | |
| 49 | 44,446 | 42,380 | terminase large subunit | 100.00% | 0 | UGO51619.1 | |
| 50 | 45,023 | 44,439 | terminase small subunit | 99.48% | 2.00 × 10−141 | UXY92233.1 | |
| 1 | 548 | 327 | Hypothetical protein | hypothetical protein | 98.63% | 2.00 × 10−46 | WP_196727143.1 |
| 3 | 1927 | 1187 | hypothetical protein | 99.59% | 7.00 × 10−177 | WOZ54581.1 | |
| 4 | 3403 | 2834 | hypothetical protein | 99.47% | 6.00 × 10−135 | WOZ54583.1 | |
| 5 | 3910 | 3509 | hypothetical protein | 100.00% | 2.00 × 10−94 | QHJ72730.1 | |
| 6 | 4079 | 3891 | hypothetical protein | 100.00% | 3.00 × 10−38 | UGO49958.1 | |
| 7 | 4311 | 4069 | hypothetical protein | 97.50% | 2.00 × 10−47 | EKV7661800.1 | |
| 8 | 4649 | 4323 | hypothetical protein | 100.00% | 5.00 × 10−72 | YP_009199627.1 | |
| 9 | 5119 | 5550 | hypothetical protein | 99.30% | 4.00 × 10−97 | WP_407818269.1 | |
| 10 | 5632 | 5943 | hypothetical protein | 100.00% | 9.00 × 10−68 | WP_318768141.1 | |
| 11 | 5949 | 6269 | hypothetical protein | 100.00% | 7.00 × 10−72 | WP_196727151.1 | |
| 12 | 6263 | 6610 | hypothetical protein | 100.00% | 3.00 × 10−75 | WP_196727152.1 | |
| 14 | 7955 | 8773 | hypothetical protein | 100.00% | 0 | WOZ54596.1 | |
| 15 | 8770 | 9537 | hypothetical protein | 99.61% | 0 | WOZ54597.1 | |
| 16 | 9534 | 9980 | hypothetical protein | 100.00% | 8.00 × 10−107 | YP_009998137.1 | |
| 17 | 9973 | 10,605 | hypothetical protein | 99.52% | 3.00 × 10−153 | WOZ54599.1 | |
| 19 | 11,644 | 12,177 | hypothetical protein | 100.00% | 4.00 × 10−129 | WP_196727159.1 | |
| 20 | 12,170 | 12,421 | hypothetical protein | 97.59% | 3.00 × 10−52 | UNI72704.1 | |
| 21 | 12,423 | 13,292 | hypothetical protein | 100.00% | 0 | WP_196727160.1 | |
| 22 | 13,289 | 13,753 | hypothetical protein | 97.40% | 1.00 × 10−108 | EKV7661814.1 | |
| 23 | 14,257 | 14,003 | hypothetical protein | 98.81% | 2.00 × 10−51 | YP_009199645.1 | |
| 26 | 17,282 | 15,297 | hypothetical protein | 96.52% | 0 | WP_318768180.1 | |
| 27 | 18,926 | 17,286 | hypothetical protein | 91.74% | 0 | WOZ54610.1 | |
| 28 | 19,952 | 18,936 | hypothetical protein | 99.41% | 0 | YP_009199651.1 | |
| 29 | 20,948 | 19,965 | hypothetical protein | 100.00% | 0 | YP_009199652.1 | |
| 30 | 21,879 | 20,959 | hypothetical protein | 99.35% | 0 | UNI72715.1 | |
| 43 | 37,601 | 37,254 | hypothetical protein | 98.26% | 3.00 × 10−73 | UXY92226.1 | |
| 56 | 51,402 | 50,920 | hypothetical protein | 96.88% | 1.00 × 10−103 | UGO51612.1 | |
| 57 | 51,755 | 52,039 | hypothetical protein | 79.79% | 2.00 × 10−48 | YP_009997934.1 | |
| 59 | 56,362 | 56,123 | hypothetical protein | 97.47% | 2.00 × 10−45 | UGO51678.1 | |
| 60 | 56,937 | 56,359 | hypothetical protein | 98.44% | 9.00 × 10−140 | YP_009199616.1 | |
| 61 | 57,146 | 56,934 | hypothetical protein | 98.57% | 4.00 × 10−41 | UGO49967.1 | |
| 62 | 57,848 | 57,159 | hypothetical protein | 97.38% | 9.00 × 10−164 | WOZ54577.1 | |
| 63 | 58,509 | 57,901 | hypothetical protein | 93.56% | 4.00 × 10−136 | UGO51542.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Li, N.; Wan, S.; Li, Y.; Li, Y.; Qu, Y. Genomic and Biological Characterization of a Novel Proteus mirabilis Phage with Anti-Biofilm Activity. Viruses 2025, 17, 1419. https://doi.org/10.3390/v17111419
Liang Y, Li N, Wan S, Li Y, Li Y, Qu Y. Genomic and Biological Characterization of a Novel Proteus mirabilis Phage with Anti-Biofilm Activity. Viruses. 2025; 17(11):1419. https://doi.org/10.3390/v17111419
Chicago/Turabian StyleLiang, Yan, Nana Li, Shenghui Wan, Yanfang Li, Yuwan Li, and Yonggang Qu. 2025. "Genomic and Biological Characterization of a Novel Proteus mirabilis Phage with Anti-Biofilm Activity" Viruses 17, no. 11: 1419. https://doi.org/10.3390/v17111419
APA StyleLiang, Y., Li, N., Wan, S., Li, Y., Li, Y., & Qu, Y. (2025). Genomic and Biological Characterization of a Novel Proteus mirabilis Phage with Anti-Biofilm Activity. Viruses, 17(11), 1419. https://doi.org/10.3390/v17111419







