Identification and Full-Genome Characterisation of Genomoviruses in Cassava Leaves Infected with Cassava Mosaic Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Initial Sequencing, De Novo Assembly and Taxonomic Classification
2.3. Sequencing of Full Genomovirus Sequences
2.4. Bioinformatic Analyses
2.5. Genome Annotation and Motif Scan
2.6. Phylogenetic Analysis
3. Results
3.1. Recovery and Sequencing of Full Genomovirus Genomes
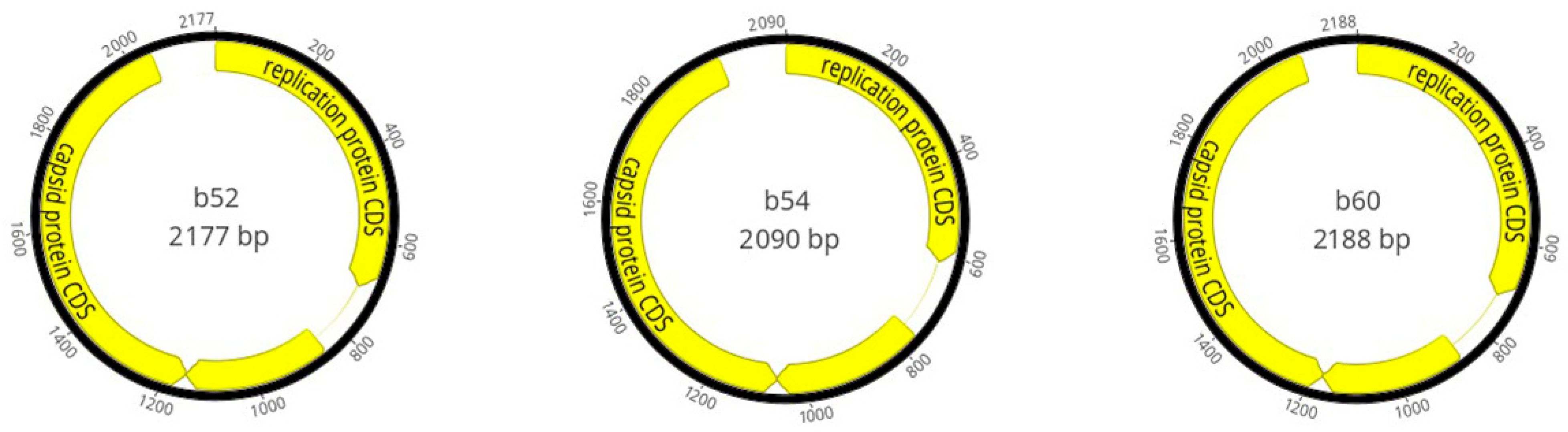
3.2. Genome Characterisation
3.3. Conserved Motifs
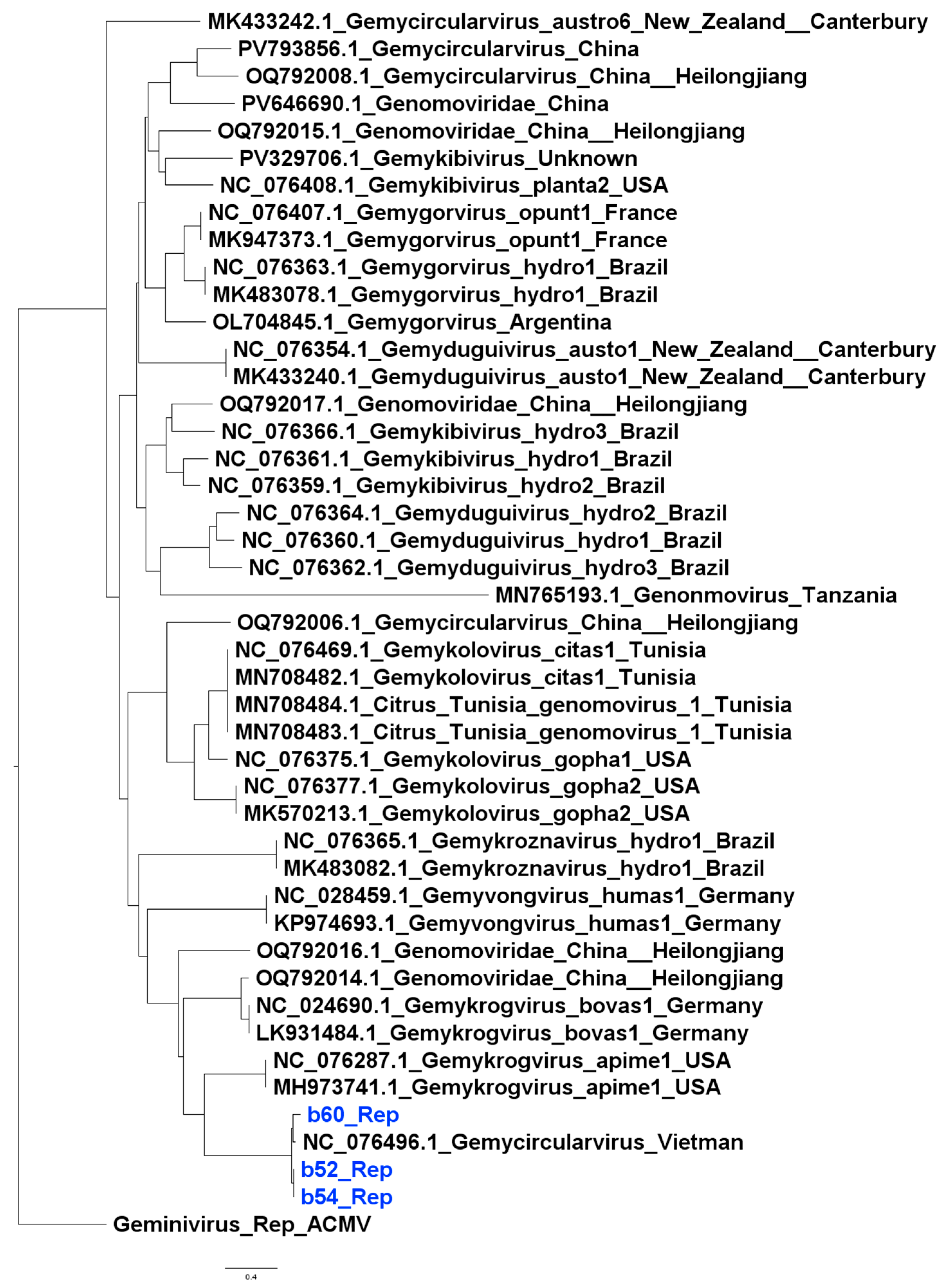
3.4. Phylogenetic Inference
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varsani, A.; Krupovic, M. Family Genomoviridae: 2021 Taxonomy Update. Arch. Virol. 2021, 166, 2911–2926. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Ghabrial, S.A.; Jiang, D.; Varsani, A. Genomoviridae: A New Family of Widespread Single-Stranded DNA Viruses. Arch. Virol. 2016, 161, 2633–2643. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, B.; Fu, Y.; Jiang, D.; Ghabrial, S.A.; Li, G.; Peng, Y.; Xie, J.; Cheng, J.; Huang, J.; et al. A Geminivirus-Related DNA Mycovirus That Confers Hypovirulence to a Plant Pathogenic Fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 8387–8392. [Google Scholar] [CrossRef]
- Li, P.; Wang, S.; Zhang, L.; Qiu, D.; Zhou, X.; Guo, L. A Tripartite SsDNA Mycovirus from a Plant Pathogenic Fungus Is Infectious as Cloned DNA and Purified Virions. Sci. Adv. 2020, 6, eaay9634. [Google Scholar] [CrossRef]
- Eni, A.O.; Efekemo, O.P.; Onile-Ere, O.A.; Pita, J.S. Survey Dataset on the Epidemiological Assessment of Cassava Mosaic Disease in South West and North Central Regions of Nigeria Reveals Predominance of Single Viral Infection. Data Br. 2021, 38, 107282. [Google Scholar] [CrossRef]
- Eni, A.O.; Efekemo, O.P.; Onile-ere, O.A.; Pita, J.S. South West and North Central Nigeria: Assessment of Cassava Mosaic Disease and Field Status of African Cassava Mosaic Virus and East African Cassava Mosaic Virus. Ann. Appl. Biol. 2020, 178, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.; Doyle, J. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- ONT Ligation Sequencing Amplicons—Native Barcoding Kit 96 V14 (SQK-NBD114.96). Available online: https://store.nanoporetech.com/native-barcoding-kit-96-v14.html (accessed on 24 June 2025).
- NanoporeTech Dorado 2024. Available online: https://github.com/nanoporetech/dorado (accessed on 13 October 2025).
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H. Minimap2 2024. Available online: https://github.com/lh3/minimap2 (accessed on 8 October 2025).
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Bickhart, D.M.; Behsaz, B.; Gurevich, A.; Rayko, M.; Shin, S.B.; Kuhn, K.; Yuan, J.; Polevikov, E.; Smith, T.P.L.; et al. MetaFlye: Scalable Long-Read Metagenome Assembly Using Repeat Graphs. Nat. Methods 2020, 17, 1103–1110. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k -Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Šikić, M. Time- and Memory-Efficient Genome Assembly with Raven. Nat. Comput. Sci. 2021, 1, 332–336. [Google Scholar] [CrossRef]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and Accurate de Novo Genome Assembly from Long Uncorrected Reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and Processing Long-Read Sequencing Data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Pavan, R.R.; Sullivan, M.B.; Tisza, M. CRESSENT: A Bioinformatic Toolkit to Explore and Improve SsDNA Virus Annotation 2025. bioRxiv 2025. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Varsani, A.; Krupovic, M. Sequence-Based Taxonomic Framework for the Classification of Uncultured Single-Stranded DNA Viruses of the Family Genomoviridae. Virus Evol. 2017, 3, vew037. [Google Scholar] [CrossRef]
- Dai, Z.; Wang, H.; Xu, J.; Lu, X.; Ni, P.; Yang, S.; Shen, Q.; Wang, X.; Li, W.; Wang, X.; et al. Unveiling the Virome of Wild Birds: Exploring CRESS-DNA Viral Dark Matter. Genome Biol. Evol. 2024, 16, evae206. [Google Scholar] [CrossRef]
- Fontenele, R.S.; Roumagnac, P.; Richet, C.; Kraberger, S.; Stainton, D.; Aleamotu’a, M.; Filloux, D.; Bernardo, P.; Harkins, G.W.; McCarthy, J.; et al. Diverse Genomoviruses Representing Twenty-Nine Species Identified Associated with Plants. Arch. Virol. 2020, 165, 2891–2901. [Google Scholar] [CrossRef]
- Nakasu, E.Y.T.; Melo, F.L.; Michereff-Filho, M.; Nagata, T.; Ribeiro, B.M.; Ribeiro, S.G.; Lacorte, C.; Inoue-Nagata, A.K. Discovery of Two Small Circular SsDNA Viruses Associated with the Whitefly Bemisia Tabaci. Arch. Virol. 2017, 162, 2835–2838. [Google Scholar] [CrossRef]
- Hao, F.; Wu, M.; Li, G. Characterization of a Novel Genomovirus in the Phytopathogenic Fungus Botrytis Cinerea. Virology 2021, 553, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, C.J.; Marzano, S.-Y.L. Characterization of Transcriptional Responses to Genomovirus Infection of the White Mold Fungus, Sclerotinia Sclerotiorum. Viruses 2022, 14, 1892. [Google Scholar] [CrossRef] [PubMed]
- Orton, J.P.; Morales, M.; Fontenele, R.S.; Schmidlin, K.; Kraberger, S.; Leavitt, D.J.; Webster, T.H.; Wilson, M.A.; Kusumi, K.; Dolby, G.A.; et al. Virus Discovery in Desert Tortoise Fecal Samples: Novel Circular Single-Stranded DNA Viruses. Viruses 2020, 12, 143. [Google Scholar] [CrossRef] [PubMed]

| Isolate | Year/Location | No Reads | Coverage | Min Base Coverage | Genome Length (bp) | GC (%) | BLASTn Search Results | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Best Hit | % Coverage | E Value | Identify | |||||||
| b52 | 2015/Benue State | 2557 | 779.81× | 228x | 2177 | 47.50 | MN765193.1 | 100% | 0.0 | 95.37% |
| b54 | 2015/Oyo State | 601 | 162.17× | 120x | 2090 | 47.61 | NC_076496.1 | 95% | 0.0 | 84.81% |
| b60 | 2017/Ondo State | 2970 | 1015.83× | 23x | 2188 | 47.76 | NC_076496.1 | 57% | 0.0 | 93.78% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onile-ere, O.; John, O.; Sonowo, O.; Name, P.E.; Tibiri, E.B.; Tiendrébéogo, F.; Pita, J.; Oranusi, S.; Eni, A.O. Identification and Full-Genome Characterisation of Genomoviruses in Cassava Leaves Infected with Cassava Mosaic Disease. Viruses 2025, 17, 1418. https://doi.org/10.3390/v17111418
Onile-ere O, John O, Sonowo O, Name PE, Tibiri EB, Tiendrébéogo F, Pita J, Oranusi S, Eni AO. Identification and Full-Genome Characterisation of Genomoviruses in Cassava Leaves Infected with Cassava Mosaic Disease. Viruses. 2025; 17(11):1418. https://doi.org/10.3390/v17111418
Chicago/Turabian StyleOnile-ere, Olabode, Oluwagboadurami John, Oreoluwa Sonowo, Pakyendou Estel Name, Ezechiel Bionimian Tibiri, Fidèle Tiendrébéogo, Justin Pita, Solomon Oranusi, and Angela O. Eni. 2025. "Identification and Full-Genome Characterisation of Genomoviruses in Cassava Leaves Infected with Cassava Mosaic Disease" Viruses 17, no. 11: 1418. https://doi.org/10.3390/v17111418
APA StyleOnile-ere, O., John, O., Sonowo, O., Name, P. E., Tibiri, E. B., Tiendrébéogo, F., Pita, J., Oranusi, S., & Eni, A. O. (2025). Identification and Full-Genome Characterisation of Genomoviruses in Cassava Leaves Infected with Cassava Mosaic Disease. Viruses, 17(11), 1418. https://doi.org/10.3390/v17111418








