Abstract
Background: The HPV16 E2 gene plays a crucial role in viral replication and oncogene regulation. This study aimed to assess the genetic variability of the E2 gene and to identify immunogenic epitopes of the E2 protein. Methods: The E2 gene was amplified and sequenced. T-cell epitope prediction and evaluation were performed using IEDB, NetMHCpan v4.0, NetMHCIIpan v4.1, VaxiJen, ToxNet, and pLM4Alg. Results: Phylogenetic analysis of 47 E2 sequences demonstrated co-circulation of the D (n = 4) and A (n = 43) HPV16 lineages in Cajamarca. Twenty-eight Single Nucleotide Polymorphism (SNPs) were identified in E2, 21 of which were nonsynonymous. Seventeen variations were associated with positive Papanicolaou (Pap) test results. Epitope prediction identified 2 MHC class I and 27 MHC class II epitopes classified as potentially antigenic, non-toxic, and non-allergenic, with an overall global population coverage across both MHC classes of 99.78%. Conclusions: The A HPV16 lineage predominated among the women studied. The identified SNPs indicate substantial variability in the E2 gene and a relationship with endocervical lesions. In total, 29 E2-derived T-cell epitopes with immunogenic potential were identified.
1. Introduction
Human papillomavirus (HPV) is a global health concern because of its association with the development of endocervical lesions and cervical cancer (CC). It is estimated that HPV is linked to more than 600,000 new cases of CC each year, and, unfortunately, more than half of those affected die from this malignancy [1]. In Peru, the prevalence of HPV infection is alarmingly high, affecting a substantial proportion of the female population. In 2020, 4270 cases of CC were reported, leading to 2288 deaths, making it the second leading cause of mortality among women in the country [2,3,4].
To date, more than 200 HPV genotypes have been identified, with high-risk types closely associated with endocervical lesions and CC. Among high-risk HPVs, genotype 16 (HPV16) is highly oncogenic, and its persistent infection is strongly associated with the onset of endocervical lesions and the development of cervical cancer [5]. Notably, this genotype is among the most prevalent worldwide and in Peru [6].
Genomic investigations underscore the importance of studying HPV genetic variability—particularly SNPs—many of which have been associated with increased risk of endocervical lesions and CC [7]. Several of these variants map to the E2 gene, which encodes the HPV E2 protein. E2 plays a key role in viral replication and in regulating transcription of the oncogenes E6 and E7 [8]. Given its expression at multiple stages of infection and critical functional (enzymatic) properties for the viral cycle, E2 stands out as a promising target for therapeutic vaccine development.
Computational epitope prediction—short amino-acid sequences within viral proteins—has become a rational and valuable approach to initiate the design of new therapeutic vaccines capable of stimulating effective immune responses, with encouraging results [9,10].
In this context, the present study aims to predict T-cell epitopes in the HPV16 E2 protein while considering genetic variation within the E2 region observed in our local setting, and to explore their association with endocervical lesions in Peruvian women. This approach will provide valuable insights into HPV biology and enable the proposal of in silico vaccine candidates.
2. Materials and Methods
2.1. Cervical Sample Collection
A total of 1433 endocervical samples were collected from asymptomatic women who voluntarily attended outpatient gynecological checkups at the Hospital Regional Docente de Cajamarca during 2021–2024, within the framework of regional epidemiological surveillance. Samples were obtained by endocervical swabbing following standard procedures and preserved in a PBS-based transport medium. Clinical–epidemiological data were abstracted from each patient’s medical record. All participants provided written informed consent prior to enrollment.
2.2. DNA Extraction and HPV Detection by PCR
Genetic material was extracted from 200 μL of each specimen using the Magbead Viral DNA/RNA Kit (CWBIO, Taizhou, China) according to the manufacturer’s instructions. Detection and genotyping of HPV16 were performed by conventional PCR using the extracted DNA as template. DNA amplification employed Taq DNA polymerase (Kappa Biosystem, Wilmington, MA, USA), specific primers, and cycling conditions previously described in the literature [11].
2.3. Amplification and Sequencing of the HPV16 E2 Gene
The HPV16 E2 gene was amplified from DNA of samples positive for genotype 16. Conventional PCR was performed using Taq DNA polymerase (Kappa Biosystem, Wilmington, MA, USA) with primers flanking the E2 gene (1185 bp): HPV16-E2F 5′-CAGATTAAGTTTGCACGAGGACGAG-3′ and HPV16-E2R 5′-CAAAGCAAAAAGCACGCCAGTAA-3′. PCR conditions were: 95 °C for 5 min; 55 cycles of 95 °C for 30 s, 57 °C for 45 s, and 72 °C for 1 min 30 s; followed by 72 °C for 10 min, then hold at 10 °C. Amplicons were resolved by 2% agarose gel electrophoresis (Agarose Type I, Calbiochem, Merck, Darmstadt, Germany) and stained with SYBR™ Safe DNA Gel Stain at a final concentration of 5 μg/mL. Positive PCR products were purified using the SpinPrep™ Gel DNA Kit (Merck) and stored at −80 °C for downstream analyses.
All purified PCR amplicons were sequenced using Oxford Nanopore Technology with the Ligation Sequencing Kit v14 (SQK-LSK114, Oxford Nanopore Technologies, Oxford, UK), following the manufacturer’s instructions. The DNA input was normalized to 200 fmol prior to library preparation. Sequencing was carried out on a MinION device equipped with a Flow Cell (R10.4.1). The resulting reads were processed in Epi2me using the Amplicon workflow (wf-amplicons v1.1.4) to generate consensus sequences.
2.4. Identification of Variants in HPV16 E2 and Phylogenetic Analysis
Sequences were aligned to the HPV16 reference (GenBank: NC_001526.3) using ClustalW within MEGA v12.0. Phylogenetic trees for the E2 region were constructed by the neighbor-joining method in MEGA 12.0. Tree reliability was assessed by 1000-replicate bootstrap analysis. Study sequences were compared against representative HPV16 lineage variants retrieved from GenBank (28 sequences): HQ644283.1 (A1), HQ644268.1 (A1), HQ644280.1 (A1), HQ644282.1 (A1), AF536179.1 (A2), HQ644236.1 (A3), HQ644248.1 (A4), HQ644251.1 (A4), AF534061.1 (A4), HQ644235.1 (A4), HQ644240.1 (B1), HQ644290.1 (B1), HQ644238.1 (B1), HQ644298.1 (B3), HQ644237.1 (C), HQ644239.1 (C), HQ644249.1 (C), HQ644250.1 (C), AF472509.1 (C), HQ644257.1 (D1), HQ644279.1 (D2), HQ644281.1 (D2), HQ644263.1 (D2), HQ644277.1 (D2), HQ644247.1 (D3), HQ644253.1 (D3), HQ644255.1 (D3), AF402678.1 (D3).
The E2 protein was modeled to visualize amino acid substitutions derived from SNP analysis. Each of the three functional domains (transactivation, hinge, and DNA-binding) was independently modeled using the AlphaFold Server, employing a template-guided approach. The structural quality of the models was assessed through Ramachandran plot analysis, and the domains were assembled into a complete structure using UCSF Chimera version 1.19.0.
2.5. Statistical Analysis
Frequencies of HPV16 E2 variants were computed by automated counting in Python. Associations between E2 variants and a positive Papanicolaou (Pap) test were assessed with Fisher’s exact test. Statistical analyses were performed using SciPy v1.16.0 in Python v3.12.7, with p < 0.05 considered statistically significant.
2.6. T-Cell Epitope Prediction and Selection
Epitope prediction was conducted from the consensus E2 sequence derived from the phylogenetic analyses using the IEDB server. Cytotoxic T-lymphocyte (CTL) epitopes were predicted with NetMHCpan v4.0, assuming lengths of 8–11 amino acids, restricted to MHC class I supertypes selected by IEDB and those most frequent in South America [9,12]. Helper T-cell epitopes were predicted with NetMHCIIpan v4.1, assuming lengths of 11–18 amino acids, targeted to HLA-DR, HLA-DQ, and HLA-DP alleles selected by IEDB and the most prevalent in South America [9,12].
Predicted epitopes were evaluated for sequence conservation against a panel of 520 E2 sequences retrieved from UniProtKB to confirm their presence across known E2 proteins. Subsequently, antigenicity, toxicity, and allergenicity were assessed using VaxiJen [13], ToxinPred [14], and pLM4Alg [15], respectively. Finally, epitopes meeting criteria for immunogenic potential, non-toxicity, and non-allergenicity were evaluated for population coverage using the IEDB Population Coverage tool.
3. Results
3.1. Epidemiological Characteristics of the Analyzed Samples
We analyzed 1433 endocervical swab samples from asymptomatic women by PCR to detect human papillomavirus (HPV). Of these, 395 tested positives for HPV, yielding a prevalence of 27.56%. HPV16 was identified in 76 samples, corresponding to a prevalence of 5.3% in the study population (Figure 1).
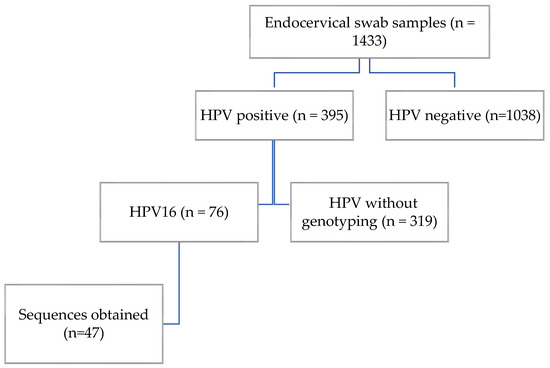
Figure 1.
Participant flow diagram.
3.2. Analysis of Variation in the HPV16 E2 Gene
Out of 76 HPV16-positive samples, only 47 were successfully amplified and sequenced. The remaining samples showed low DNA quality or insufficient amplification of the E2 gene region, possibly due to degradation or low viral load in the clinical specimens. Across these 47 E2 sequences, we identified 28 distinct single nucleotide polymorphisms (SNPs), distributed across the transactivation domain (8 SNPs), the hinge region (12 SNPs), and the DNA-binding domain (8 SNPs). Most SNPs were in the hinge region of the E2 protein, with fewer in the DNA-binding and transactivation domains (Figure 2 and Table 1). The most frequent variation was C3410T, present in 45 samples (95.7%) which is present in lineages A and D. The next most frequent was C3684A, detected in 5 samples (10.6%) which is present in lineage D. Several variations showed a frequency of 8.5%, including C2860A, C3159A, G3182A, T3224A, G3249A, A3362G, C3377G, G3449A, C3516A, T3517C, A3538C, T3566G, T3694A, T3706C, G3778T, C3787A, and T3805G. Variations C3161T and G3416A were observed in 3 samples each (6.4%). Finally, T3118C, A3181C, T3384C, T3387C, C3551G, T3664C, and G3767A each occurred in a single sample (2.1% of sequences).
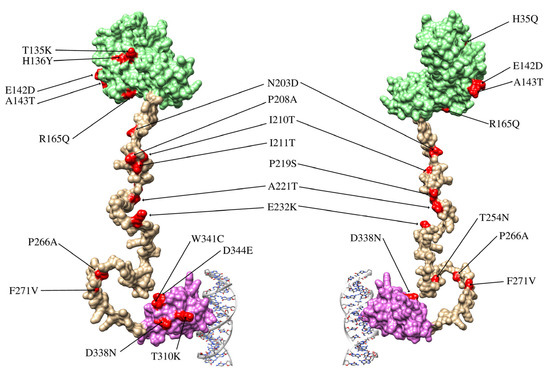
Figure 2.
Structure of the E2 protein and locations of the variations identified in this study. The E2 protein comprises three domains: the transactivation domain (light green), the hinge domain (pale gold), and the DNA-binding domain (lilac). The hinge domain lacks a well-defined three-dimensional structure, as its main function is to connect the functional domains of the protein. The figure shows the front and back views of the E2 protein in complex with a DNA fragment.

Table 1.
Variations identified in HPV16 (European sublineage A1, GenBank: NC_001526.3) and their relationship with Pap smear results. p values obtained from Fisher’s exact test (FET) < 0.05, indicating statistical significance, are shown in bold.
3.3. Phylogenetic Analysis of the HPV16 E2 Gene
Phylogenetic analysis of the 47 nucleotide sequences showed that four clustered within lineage D (Asian American), three of which were derived from Papanicolaou (Pap) test-positive samples. The remaining 43 sequences clustered within lineage A (European). No samples from lineages B (African-1), C (African-2), or A4 (Asian) were detected. These findings indicate a predominance of the A lineage in the study population (Figure 3).
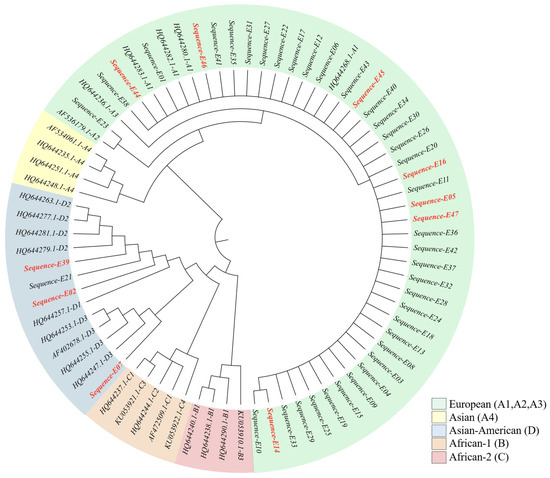
Figure 3.
Phylogenetic tree based on the HPV16 E2 gene sequence. Sequences corresponding to patients with a positive Pap smear are shown in red.
3.4. Association Between Variations and Endocervical Lesions
Associations between the presence of each variation and Pap test result were assessed using Fisher’s exact test for small samples. Seventeen variations showed statistically significant associations (Table 1). Specifically, C2860A, C3159A, G3182A, T3224A, G3249A, A3362G, C3377G, G3449A, C3516A, T3517C, A3538C, T3566G, T3694A, T3706C, G3778T, C3787A, and T3805G yielded p < 0.05, suggesting a possible association with a positive Pap test result. In contrast, T3118C, T3384C, C3551G, G3767A, C3161T, A3181C, T3387C, C3410T, G3416A, T3664C, and C3684A showed no significant association (p ≥ 0.05). Finally, Fisher’s exact test applied to lineage versus Pap result revealed a significant association between the D lineage and a positive Pap test (Table 2).

Table 2.
Association between HPV16 lineages and Pap smear results (Fisher’s exact test, p = 0.026).
3.5. T-Cell Epitope Prediction: Characteristics and Population Coverage
CD4+ T-cell epitope prediction was performed with the IEDB platform using NetMHCIIpan 4.1 EL, evaluating peptide processing and binding against a panel of 29 MHC class II alleles representative of global and South American genetic diversity.
From all peptides analyzed, 108 were selected for showing binding to ≥25% of the tested MHC-II alleles (Figure 4).
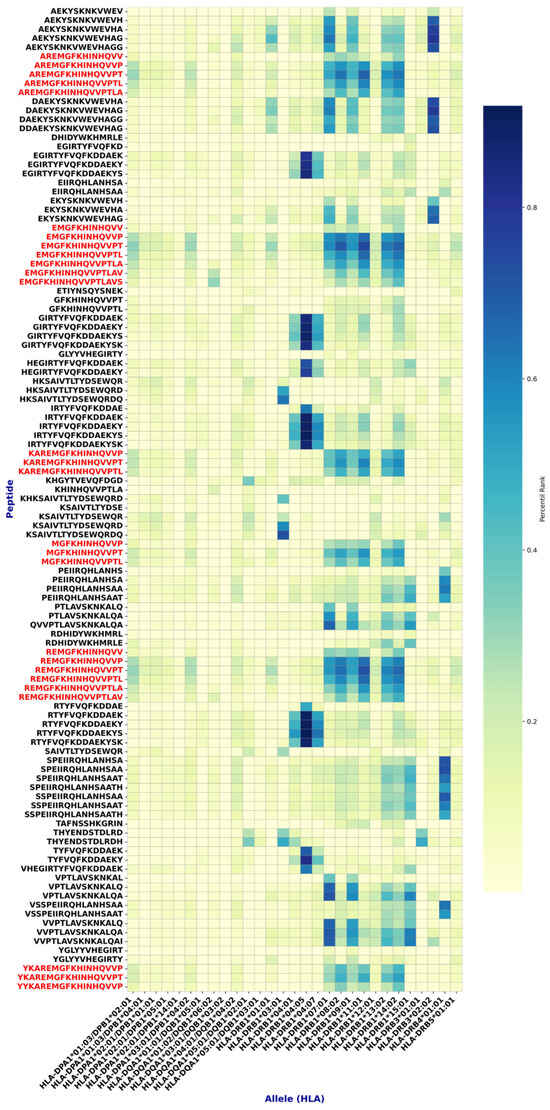
Figure 4.
Epitopes found with high affinity for MHC class II. Highly conserved epitopes with a good safety profile are shown in red. Note that these epitopes have a common core: FKHINHQVV.
For CD8+ T-cell epitopes, NetMHCpan 4.1 EL was applied against a panel of 33 MHC class I alleles. Nine peptides were selected for binding to >25% of the tested alleles (Figure 4).
Subsequent analyses identified multiple epitopes with >70% sequence conservation, meeting the criterion to be considered conserved. Among them, 2 MHC-I and 27 MHC-II epitopes (Figure 4 and Figure 5, and Table 3) demonstrated, by in silico analyses, a favorable safety profile (non-toxic and non-allergenic) and immunogenic potential. Notably, the MHC class II epitopes shared a common core motif (FKHINHQVV) across these sequences (Figure 4).
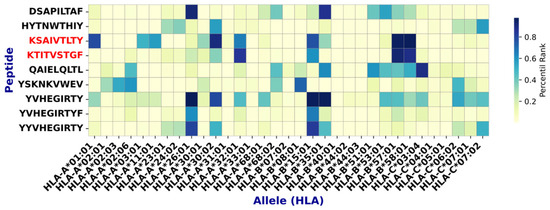
Figure 5.
Epitopes found with high affinity for MHC class I. Epitopes with high conservation and good safety profiles are shown in red.

Table 3.
Characterization of epitopes with high immunological potential.
Overall, the selected epitopes showed broad combined global population coverage, averaging 99.78%, indicating strong potential to elicit immune responses across diverse populations. Regionally, high coverages were observed in Europe (99.99%), North America (100.00%), and South Asia (99.85%). In contrast, Central America (91.17%) and Southwest Africa (91.20%) exhibited lower, though still substantial, combined coverage—reflecting differences in HLA allele distributions across populations. By class, MHC-I epitopes achieved a mean global coverage of 89.80%, with notable values in Europe (92.44%) and East Asia (90.11%); MHC-II epitopes reached a mean of 97.87%, exceeding 99% in East Africa, South America, and Europe (Figure 6).
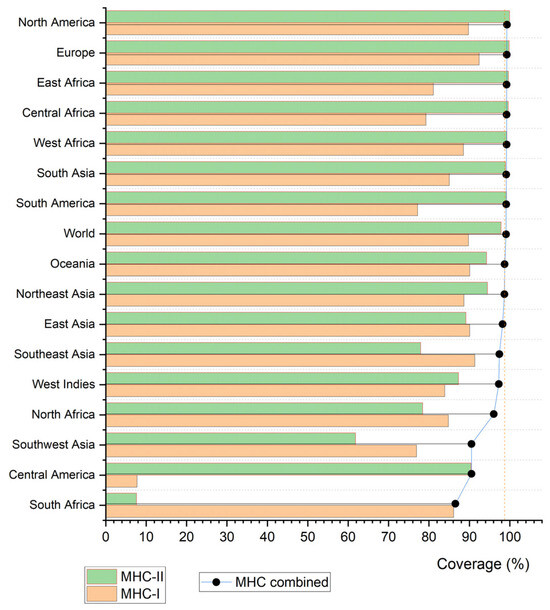
Figure 6.
Bar graph illustrating the population coverage of the evaluated epitopes.
4. Discussion
HPV prevalence varies globally—from 6.6% in Europe to 22.9% in Africa—and also regionally (Asia 8.3%, North America 13.8%, South America 14.3%, Central America 20.5%) [22], suggesting the influence of geographic, socioeconomic, and cultural factors. Genotype distribution is likewise heterogeneous, with HPV16 among the most prevalent high-risk types. In Peru, overall HPV and HPV16 prevalences are variable: Iwasaki et al. (2014) reported 34.49% (any HPV) and 10.77% (HPV16) [23], whereas del Valle-Mendoza et al. (2021) found 19.08% (any HPV) and 3.24% (HPV16) in Cajamarca [6]. In the present study, we observed an HPV prevalence of 27.56% and an HPV16 prevalence of 5.3% in clinically healthy women—values consistent with del Valle-Mendoza but lower than those of Iwasaki—potentially indicating a lower prevalence of HPV, particularly HPV16, in Cajamarca.
HPV16 is classified into several lineages and sublineages based on genetic variation. Four major lineages are currently recognized, each subdivided into sublineages [24]. Lineage A includes sublineages A1, A2, A3 (European), and A4 (Asian). Lineage B comprises B1 (African-1, Afr1a) and B2 (African-1, Afr1b). Lineage C corresponds to African-2 (Afr2a). Lineage D includes D1 (North American, NA1), D2 (Asian-American, AA2), and D3 (Asian-American, AA1) [25,26]. Several studies have shown that lineage D, particularly the Asian-American (AA) variants, display higher oncogenic potential compared with European variants, possibly due to mutations in viral regulatory genes such as E2 that can alter viral transcription and replication control [16,27,28,29].
Earlier studies (2004, 2007) by Tornesello et al. and Sichero et al. reported that non-European HPV16 variants—such as AA and AFR1—confer a higher risk of cervical intraepithelial lesions than European (EUR) variants [17,27]. Similarly, Schiffman et al. (2010) noted that AFR1, AFR2, and AA variants were more prevalent than EUR and As variants and were associated with greater viral persistence and carcinogenic potential [28]. More recent work by Dai (2018) and Wang (2021) found that Asian and Asian-American HPV16 variants were associated with cervical cancer in Chinese cohorts, suggesting that non-European variants may carry a higher risk of progression to lesions and cancer [16,29]. In our study, most amino acid variations significantly associated with positive Pap results were characteristic of lineage D. This suggests that the statistical significance observed for positive Pap results mainly reflects the contribution of lineage D variants. Given that E2 regulates the transcription of the E6 and E7 oncogenes, alterations found within lineage D may affect this regulation, potentially enhancing viral persistence and the carcinogenic process [16,28,29].
We identified 17 variations significantly associated with endocervical lesions by Fisher’s exact test, in line with previous findings. However, five variations (A3181C, T3384C, T3387C, T3664C, and C3684A) did not show significant associations, contrary to other reports [16,29,30]. Regarding their distribution, we observed greater diversity in the hinge (bridge) domain of E2, consistent with prior studies and suggesting greater tolerance for genetic variation due to the lack of enzymatic functions. The C3410T variation stood out for its high frequency (95.7%), suggesting it is an established variation in circulating HPV16 within the analyzed population. The higher variability observed within the hinge domain may indicate adaptive flexibility, whereas lineage D–specific substitutions in functional regions of E2 could compromise its regulatory role, leading to enhanced oncogene expression and an increased risk of cervical lesion development.
Functionally, the HPV16 E2 protein is pivotal for viral genome replication and repression of the oncogenes E6 and E7, which are implicated in carcinogenesis. Variations in E2 may alter its function and influence viral behavior. In the DNA-binding domain, variations such as G3778T (W341C), T3694A (T313T), and C3684A (T310K) could enhance conformational stability, potentially affecting E2′s repressive capacity on E6/E7 [25,29,31,32]. In the transactivation domain, C2860A (H35Q) may alter E2–E1 interactions and thereby impact viral replication, although its effect remains unclear [31]. Other variations such as T135K and H136Y have been linked to increased HPV16 persistence, although the underlying mechanisms are not fully understood [32]. As for the hinge region, its role in the progression of endocervical lesions has not been extensively studied in HPV16, but in other types (HPV11 and HPV2) it is relevant for nuclear localization, DNA binding, and transactivation activity [33,34], suggesting that variations in this region could modulate E2 function.
T-cell epitope-based vaccines have shown promise in treating viral infections and cancers. For HPV, a vaccine aimed at inducing a robust T-cell response against E2 could complement current L1-based prophylactic vaccines—particularly for individuals already infected or with precancerous lesions [35,36]. Selecting peptides with diverse MHC-binding specificities maximizes population coverage by accounting for MHC polymorphism and allele frequency differences across populations, thereby avoiding ethnic bias. This strategy ensures that epitopes can be presented by a broad array of MHC alleles, which is critical for developing widely applicable vaccines. Accordingly, we identified 2 promising MHC-I epitopes and 27 MHC-II epitopes, the latter exhibiting high similarity to one another. The epitopes also showed a high degree of conservation, supporting their functional relevance and potential for universal HPV vaccine design [37].
We predicted peptides of 8–11 amino acids for MHC-I and 11–18 amino acids for MHC-II, recognizing that MHC-I typically binds shorter epitopes (8–11 aa), whereas MHC-II accommodates a broader length range (~13–25 aa). Notably, in MHC-II, only a central core of ~9 amino acids directly interact with the binding groove [38]. This length flexibility allows identification of a wider variety of candidate epitopes—particularly important for pathogens with high genetic variability such as HPV.
The epitopes identified here display a high degree of conservation, underscoring their likely functional importance in immune recognition. This is especially relevant given that HPV16 variants can harbor amino-acid substitutions that may alter adaptive immune responses. For example, variations W341C and D344E in the EUR variant lie within the epitope HKSAIVTLTYDSEWQRDQ [39]. Likewise, E142D and A143T, detected in this study, map to regions corresponding to epitopes EEASVTVVEGQVDYY and YICEEASVTV [39,40], suggesting a potential impact on antigen presentation. Similarly, R145Q may affect the structure or function of the YVHEGIRTY epitope, diminishing immune recognition [41]. In future work, we propose investigating the functional impact of the variations identified within known epitopes to validate their effects on antigen presentation and immune responses.
Taken together, the observed variations in HPV16 E2 may favor immune-evasion mechanisms, facilitating viral persistence in the host. These findings highlight the need to incorporate HPV16 genetic diversity into therapeutic and vaccine strategies to achieve effective immune responses against diverse viral variants.
5. Conclusions
We identified an HPV16 prevalence of 5.3% among asymptomatic women in the Cajamarca region. Analysis of the E2 gene revealed substantial genetic diversity, with 28 single-nucleotide polymorphisms (SNPs) distributed across the gene, 17 of which were significantly associated with positive Papanicolaou (Pap) test results. In silico epitope prediction yielded 29 candidates with high predicted MHC affinity that were immunogenic, non-toxic, non-allergenic, and highly conserved across the HPV16 variants analyzed. These epitopes demonstrated broad population coverage, underscoring their potential utility for developing more inclusive and effective vaccine strategies. Taken together, our findings highlight the importance of incorporating HPV16 genetic variability into the design of future diagnostic and prevention tools.
Author Contributions
Conceptualization, E.B.-V.d.V., M.A.A.-L. and J.d.V.-M.; methodology, E.B.-V.d.V., M.A.A.-L., Y.T.-C., J.d.V.-M., W.S.-C. and D.A.-L.; validation, M.A.A.-L., Y.T.-C., J.d.V.-M., L.B.-G., D.A.-L. and R.A.-O.; formal analysis, E.B.-V.d.V., M.A.A.-L., Y.T.-C., J.d.V.-M. and D.A.-L.; investigation, E.B.-V.d.V., M.A.A.-L., Y.T.-C., L.B.-G., D.A.-L., D.D.-H.,W.S.-C., R.A.-O., A.C.-T. and J.d.V.-M.; resources, J.d.V.-M. and M.A.A.-L.; data curation, E.B.-V.d.V., M.A.A.-L., Y.T.-C., L.B.-G., D.A.-L. and J.d.V.-M.; writing—original draft preparation, E.B.-V.d.V., M.A.A.-L., Y.T.-C., L.B.-G., D.A.-L., D.D.-H., W.S.-C., R.A.-O., A.C.-T. and J.d.V.-M.; writing—review and editing, E.B.-V.d.V., M.A.A.-L., Y.T.-C., L.B.-G., D.A.-L., D.D.-H., W.S.-C., R.A.-O., A.C.-T. and J.d.V.-M.; supervision, J.d.V.-M., W.S.-C. and M.A.A.-L.; project administration, J.d.V.-M. and M.A.A.-L.; funding acquisition, J.d.V.-M. and M.A.A.-L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with a grant from CONCYTEC through the PROCIENCIA program within the framework of the “E041-2023-02” competition, according to contract or agreement PE501083623-2023 and by Dirección de Investigación-Universidad Peruana de Ciencias Aplicadas (2025 C-020-2025), Lima-Peru.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Approval of the Cajamarca Regional Teaching Hospital (LETTER No. 002-2024-GR.CAJ-DRS/HRDC-DCEI) dated 18 January 2024.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The nucleotide sequences generated in this study have been deposited in the NCBI GenBank database under accession numbers PX439089–PX439135. Dataset presented in the study are openly available in: https://doi.org/10.6084/m9.figshare.30267019 (accessed on 2 October 2025).
Acknowledgments
We thank the staff of the health network from Hospital Regional Docente de Cajamarca.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Kombe Kombe, A.J.; Li, B.; Zahid, A.; Mengist, H.M.; Bounda, G.-A.; Zhou, Y.; Jin, T. Epidemiology and Burden of Human Papillomavirus and Related Diseases, Molecular Pathogenesis, and Vaccine Evaluation. Front. Public Health 2021, 8, 552028. [Google Scholar] [CrossRef] [PubMed]
- Silva-Caso, W.; Olivera-Irazábal, M.; León-Álvarez, P.; Del Valle, L.J.; Díaz-Estacio, S.; Vargas, M.; Ruiz, J.; Bermúdez-García, A.; del Valle Mendoz, J. Identification of human papillomavirus as a preventive strategy for cervical cancer in asymptomatic women in the Peruvian Andes. Asian Pac. J. Trop. Med. 2014, 7, S121–S126. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Poniewierza, P.; Panek, G. Cervical Cancer Prophylaxis—State-of-the-Art and Perspectives. Healthcare 2022, 10, 1325. [Google Scholar] [CrossRef]
- del Valle-Mendoza, J.; Becerra-Goicochea, L.; Aguilar-Luis, M.A.; Pinillos-Vilca, L.; Carrillo-Ng, H.; Silva-Caso, W.; Palomares-Reyes, C.; Taco-Masias, A.-A.; Aquino-Ortega, R.; Tinco-Valdez, C.; et al. Genotype-specific prevalence of human papillomavirus infection in asymptomatic Peruvian women: A community-based study. BMC Res. Notes 2021, 14, 172. [Google Scholar] [CrossRef]
- Hirose, Y.; Yamaguchi-Naka, M.; Onuki, M.; Tenjimbayashi, Y.; Tasaka, N.; Satoh, T.; Tanaka, K.; Iwata, T.; Sekizawa, A.; Matsumoto, K.; et al. High Levels of Within-Host Variations of Human Papillomavirus 16 E1/E2 Genes in Invasive Cervical Cancer. Front. Microbiol. 2020, 11, 596334. [Google Scholar] [CrossRef] [PubMed]
- Anayannis, N.V.; Schlecht, N.F.; Ben-Dayan, M.; Smith, R.V.; Belbin, T.J.; Ow, T.J.; Blakaj, D.M.; Burk, R.D.; Leonard, S.M.; Woodman, C.B.; et al. Association of an intact E2 gene with higher HPV viral load, higher viral oncogene expression, and improved clinical outcome in HPV16 positive head and neck squamous cell carcinoma. PLoS ONE 2018, 13, e0191581. [Google Scholar] [CrossRef]
- Requena, D.; Médico, A.; Chacón, R.D.; Ramírez, M.; Marín-Sánchez, O. Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country. Front. Immunol. 2020, 11, 2008. [Google Scholar] [CrossRef]
- Zhu, L.; Cui, X.; Yan, Z.; Tao, Y.; Shi, L.; Zhang, X.; Yao, Y.; Shi, L. Design and evaluation of a multi-epitope DNA vaccine against HPV16. Hum. Vaccines Immunother. 2024, 20, 2352908. [Google Scholar] [CrossRef]
- Carrillo-Ng, H.; Becerra-Goicochea, L.; Tarazona-Castro, Y.; Pinillos-Vilca, L.; del Valle, L.J.; Aguilar-Luis, M.A.; Tinco-Valdez, C.; Silva-Caso, W.; Martins-Luna, J.; Peña-Tuesta, I.; et al. Variations in cervico-vaginal microbiota among HPV-positive and HPV-negative asymptomatic women in Peru. BMC Res. Notes 2021, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P.; et al. IEDB-AR: Immune epitope database—Analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Rathore, A.S.; Arora, A.; Choudhury, S.; Tijare, P.; Raghava, G.P.S. ToxinPred 3.0: An improved method for predicting the toxicity of peptides. Comput. Biol. Med. 2023, 179, 108926. [Google Scholar] [CrossRef]
- Du, Z.; Xu, Y.; Liu, C.; Li, Y. pLM4Alg: Protein Language Model-Based Predictors for Allergenic Proteins and Peptides. J. Agric. Food Chem. 2024, 72, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Yao, Y.; Yan, Z.; Zhou, Z.; Shi, L.; Wang, X.; Sun, L.; Zhang, R.; Yao, Y. The association of human papillomavirus type 16 E2 variations with cervical cancer in a Han Chinese population. Infect. Genet. Evol. 2018, 64, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, F.; Fu, S.; Zhang, C.; Zhe, X.; Li, H.; Li, D.; Shao, R.; Pan, Z. Analysis of genetic variation in human papillomavirus type 16 E1 and E2 in women with cervical infection in Xinjiang, China. BMC Med. Genom. 2021, 14, 268. [Google Scholar] [CrossRef]
- Cheung, J.L.; Cheung, T.H.; Yu, M.Y.; Chan, P.K. Virological characteristics of cervical cancers carrying pure episomal form of HPV16 genome. Gynecol. Oncol. 2013, 131, 374–379. [Google Scholar] [CrossRef]
- Smith, B.; Chen, Z.; Reimers, L.; van Doorslaer, K.; Schiffman, M.; Desalle, R.; Herrero, R.; Yu, K.; Wacholder, S.; Wang, T.; et al. Sequence imputation of HPV16 genomes for genetic association studies. PLoS ONE 2011, 6, e21375. [Google Scholar] [CrossRef]
- Zu, Y.; Ou, Z.; Wu, D.; Liu, W.; Liu, L.; Wu, D.; Zhao, Y.; Ren, P.; Zhang, Y.; Li, W.; et al. Genetic characteristics of human papillomavirus type 16, 18, 52 and 58 in southern China. Genomics 2021, 113, 3895–3906. [Google Scholar] [CrossRef]
- Brown, C.; Kowalczyk, A.M.; Taylor, E.R.; Morgan, I.M.; Gaston, K. P53 represses human papillomavirus type 16 DNA replication via the viral E2 protein. Virol. J. 2008, 5, 5. [Google Scholar] [CrossRef]
- Crow, J.M. HPV: The global burden. Nature 2012, 488, S2–S3. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, R.; Galvez-Philpott, F.; Arias-Stella, J.; Arias-Stella, J. Prevalence of high-risk human papillomavirus by cobas 4800 HPV test in urban Peru. Braz. J. Infect. Dis. 2014, 18, 469–472. [Google Scholar] [CrossRef]
- Cornet, I.; Gheit, T.; Iannacone, M.R.; Vignat, J.; Sylla, B.S.; Del Mistro, A.; Franceschi, S.; Tommasino, M.; Clifford, G.M. HPV16 genetic variation and the development of cervical cancer worldwide. Br. J. Cancer 2013, 108, 240–244. [Google Scholar] [CrossRef]
- Kahla, S.; Hammami, S.; Kochbati, L.; Chanoufi, M.B.; Oueslati, R. HPV16 E2 variants correlated with radiotherapy treatment and biological significance in cervical cell carcinoma. Infect. Genet. Evol. 2018, 65, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wu, E.; Ma, L.; Zhang, G.; Shi, Y.; Huang, J.; Zha, X. Lineage distribution and E2 sequence variation of high-risk human papillomavirus types isolated from patients with cervical cancer in Sichuan province, China. Arch. Virol. 2015, 160, 2845–2855. [Google Scholar] [CrossRef]
- Sichero, L.; Ferreira, S.; Trottier, H.; Duarte-Franco, E.; Ferenczy, A.; Franco, E.L.; Villa, L.L. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int. J. Cancer 2007, 120, 1763–1768. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Duraturo, M.L.; Salatiello, I.; Buonaguro, L.; Losito, S.; Botti, G.; Stellato, G.; Greggi, S.; Piccoli, R.; Pilotti, S.; et al. Analysis of human papillomavirus type-16 variants in Italian women with cervical intraepithelial neoplasia and cervical cancer. J. Med. Virol. 2004, 74, 117–126. [Google Scholar] [CrossRef]
- Schiffman, M.; Rodriguez, A.C.; Chen, Z.; Wacholder, S.; Herrero, R.; Hildesheim, A.; Desalle, R.; Befano, B.; Yu, K.; Safaeian, M.; et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010, 70, 3159–3169. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.T.; Müller, M. Next generation prophylactic human papillomavirus vaccines. Lancet Oncol. 2015, 16, e217–e225. [Google Scholar] [CrossRef]
- van der Weele, P.; Meijer, C.J.L.M.; King, A.J. Whole-Genome Sequencing and Variant Analysis of Human Papillomavirus 16 Infections. J. Virol. 2017, 91, e00844-17. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, H.; Yang, B.; Geffre, C.P.; Zhang, A.; Zhou, A.; Cao, H.; Wang, J.; Zhang, Z.; Zheng, W. Variants of human papillomavirus type 16 predispose toward persistent infection. Int. J. Clin. Exp. Pathol. 2015, 8, 8453–8459. [Google Scholar]
- Gao, C.; Pan, M.-M.; Lei, Y.-J.; Tian, L.-Q.; Jiang, H.-Y.; Li, X.-L.; Shi, Q.; Tian, C.; Yuan, Y.K.; Fan, G.X.; et al. A point mutation in the DNA-binding domain of HPV-2 E2 protein increases its DNA-binding capacity and reverses its transcriptional regulatory activity on the viral early promoter. BMC Mol. Biol. 2012, 13, 5. [Google Scholar] [CrossRef]
- Zou, N.; Lin, B.Y.; Duan, F.; Lee, K.-Y.; Jin, G.; Guan, R.; Yao, G.; Lefkowitz, E.J.; Broker, T.R.; Chow, L.T. The Hinge of the Human Papillomavirus Type 11 E2 Protein Contains Major Determinants for Nuclear Localization and Nuclear Matrix Association. J. Virol. 2000, 74, 3761–3770. [Google Scholar] [CrossRef] [PubMed]
- Melief, C.J.M.; van der Burg, S.H. Immunotherapy of established (pre)malignant disease by synthetic long peptide vaccines. Nat. Rev. Cancer 2008, 8, 351–360. [Google Scholar] [CrossRef]
- Trimble, C.L.; Morrow, M.P.; Kraynyak, K.A.; Shen, X.; Dallas, M.; Yan, J.; Edwards, L.; Parker, R.L.; Denny, L.; Giffear, M.; et al. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: A randomised, double-blind, placebo-controlled phase 2b trial. Lancet 2015, 386, 2078–2088. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.-H.; Sidney, J.; Li, W.; Fusseder, N.; Sette, A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinform. 2007, 8, 361. [Google Scholar] [CrossRef]
- Rizarullah; Aditama, R.; Giri-Rachman, E.A.; Hertadi, R. Designing a Novel Multiepitope Vaccine from the Human Papilloma Virus E1 and E2 Proteins for Indonesia with Immunoinformatics and Molecular Dynamics Approaches. ACS Omega 2024, 9, 16547–16562. [Google Scholar] [CrossRef]
- Kristensen, N.P.; Dionisio, E.; Bentzen, A.K.; Tamhane, T.; Kemming, J.S.; Nos, G.; Voss, L.F.; Hansen, U.K.; Lauer, G.M.; Hadrup, S.R. Simultaneous analysis of pMHC binding and reactivity unveils virus-specific CD8 T cell immunity to a concise epitope set. Sci. Adv. 2024, 10, eadm8951. [Google Scholar] [CrossRef]
- Stellato, G.; Paavonen, J.; Nieminen, P.; Hibma, M.; Vilja, P.; Lehtinen, M. Diagnostic phase antibody response to the human papillomavirus type 16 E2 protein is associated with successful treatment of genital HPV lesions with systemic interferon alpha-2b. Clin. Diagn. Virol. 1997, 7, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, K.H.; Neller, M.A.; Srihari, S.; Crooks, P.; Lekieffre, L.; Aftab, B.T.; Liu, H.; Smith, C.; Kenny, L.; Porceddu, S.; et al. Profiling HPV-16-specific T cell responses reveals broad antigen reactivities in oropharyngeal cancer patients. J. Exp. Med. 2020, 217, e20200389. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).