Metatranscriptomic Analysis of Oropharyngeal Samples Reveals Common Respiratory Viruses and a Potential Interspecies Transmitted Picobirnavirus in the Wayuu Population, La Guajira, Colombia
Abstract
1. Introduction
2. Materials and Methods
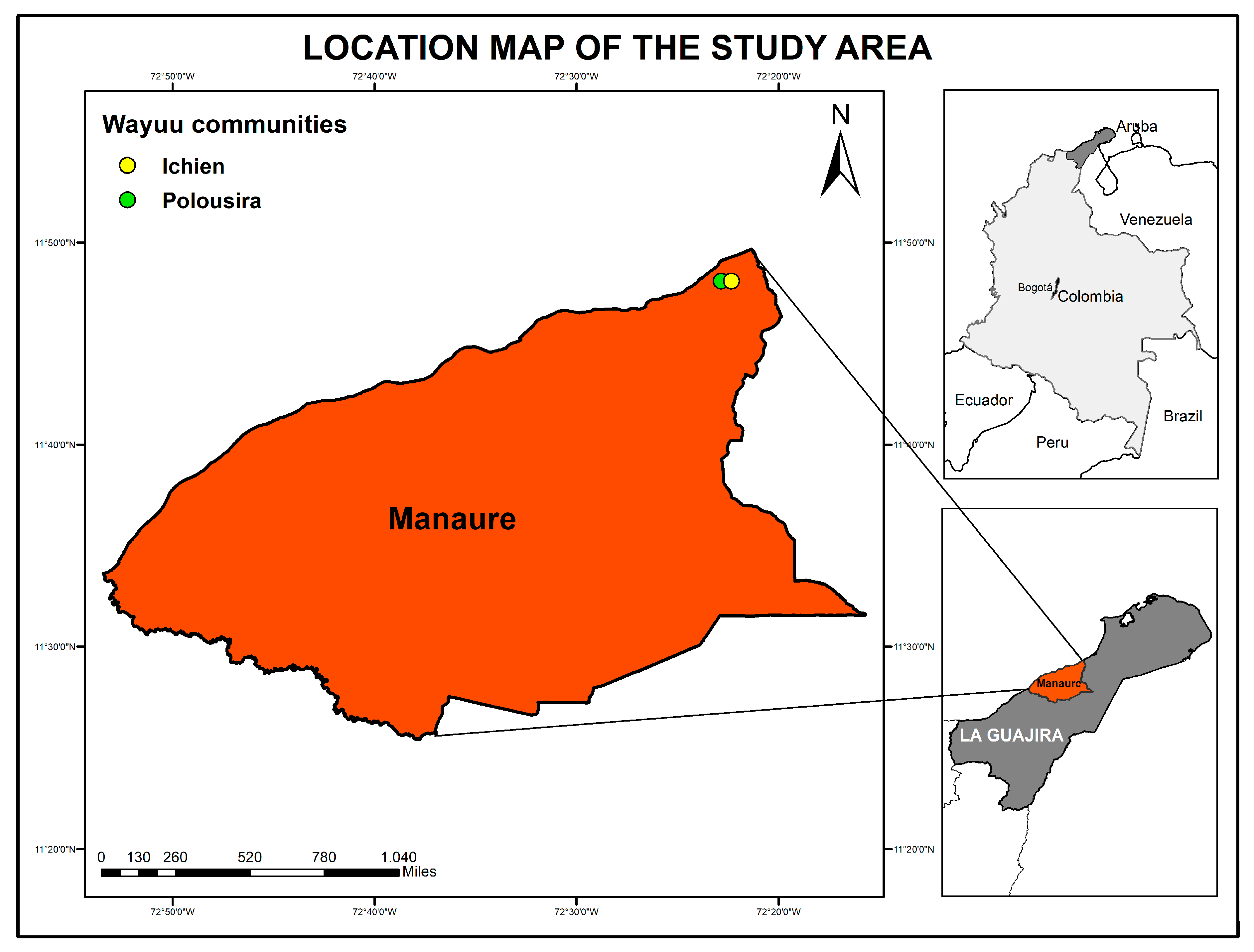
2.1. Participants, Samples and Study Area
2.2. RNA Purification and RT-qPCR for Virus Detection
2.3. Metatranscriptomic Sequencing
2.4. Bioinformatics Analysis
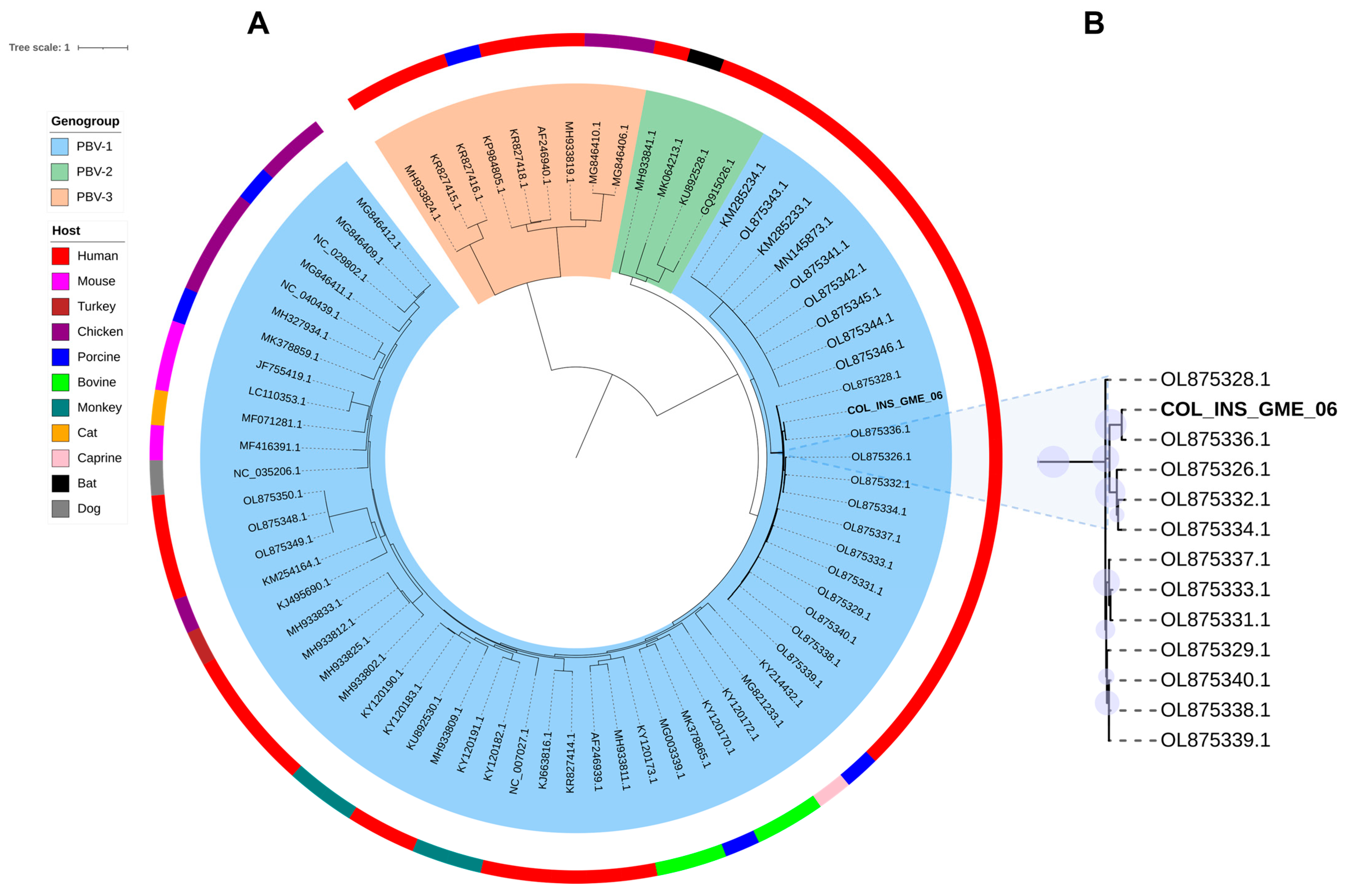
2.5. Phylogenetic Tree of RdRp from Orthopicobirnavirus Hominis
2.6. Ethical Considerations
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| The following abbreviations are used in this manuscript | . |
| MtNGS | Metatranscriptomic Next-Generation Sequencing |
| CZ-ID | Chan Zuckerberg ID |
| RT-qPCR | Reverse Transcription quantitative polymerase Chain Reaction |
References
- Fuentes, Y.V.; Ibáñez-Prada, E.D.; Serrano-Mayorga, C.C.; Pfizenmaier, C.G.; Cano, M.; Boada, N.; Rincon, P.; García-Gallo, E.; Duque, S.; Ocampo, A.F.; et al. Prevalence, incidence, and severity associated with viral respiratory tract infections in Colombian adults before the COVID-19 pandemic. J. Infect. Public Health 2022, 15, 1381–1387. [Google Scholar] [CrossRef]
- Yozwiak, N.L.; Skewes-Cox, P.; Stenglein, M.D.; Balmaseda, A.; Harris, E.; DeRisi, J.L. Virus Identification in Unknown Tropical Febrile Illness Cases Using Deep Sequencing. PLoS Negl. Trop. Dis. 2012, 6, e1485. [Google Scholar] [CrossRef]
- Di Bari, C.; Venkateswaran, N.; Fastl, C.; Gabriël, S.; Grace, D.; Havelaar, A.H.; Huntington, B.; Patterson, G.T.; Rushton, J.; Speybroeck, N.; et al. The global burden of neglected zoonotic diseases: Current state of evidence. One Health 2023, 17, 100595. [Google Scholar] [CrossRef] [PubMed]
- Silva-Ramos, C.R.; Rodriguez-Morales, A.J.; Hidalgo, M. Repercussions of the end of the armed conflict in Colombia and its influence on the emergence of zoonotic pathogens related to acute undifferentiated febrile illness: Future challenges to be addressed. Acta Trop. 2025, 267, 107680. [Google Scholar] [CrossRef]
- Hinds, A.; Suárez Aguilar, B.; Berrio, Y.D.; Ospina Galeano, D.; Gómez Vargas, J.H.; Ruiz, V.E.; Mignone, J. Consistency Between Administrative Health Records and Self-Reported Health Status and Health Care Use Among Indigenous Wayuu Health Insurance Enrollees: La Guajira, Colombia. Eval. Health Prof. 2024, 48, 222. [Google Scholar] [CrossRef]
- Instituto Nacional de Salud (INS). Infección Respiratoria Aguda—IRA Comportamiento Epidemiológico; Instituto Nacional de Salud (INS): Bogotá, Colombia, 2024. [Google Scholar]
- Krieger, J.; Higgins, D.L. Housing and Health: Time Again for Public Health Action. Am. J. Public Health 2011, 92, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Parsania, M.; Poopak, B.; Pouriayevali, M.H.; Haghighi, S.; Amirkhani, A.; Nateghian, A. Detection of Human Metapneumovirus and Respiratory Syncytial Virus by Real-Time Polymerase Chain Reaction Among Hospitalized Young Children in Iran. Jundishapur J. Microbiol. 2016, 9, 32974. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Weinberg, G.A.; Schnabel, K.C.; Erdman, D.D.; Prill, M.M.; Iwane, M.K.; Shelley, L.M.; Whitaker, B.L.; Szilagyi, P.G.; Hall, C.B. Field evaluation of TaqMan Array Card (TAC) for the simultaneous detection of multiple respiratory viruses in children with acute respiratory infection. J. Clin. Virol. 2013, 57, 254–260. [Google Scholar] [CrossRef]
- Lu, X.; Erdman, D.D. Quantitative real-time PCR assays for detection and type-specific identification of the endemic species C human adenoviruses. J. Virol. Methods 2016, 237, 174–178. [Google Scholar] [CrossRef]
- Aamir, U.B.; Alam, M.M.; Sadia, H.; Zaidi, S.S.Z.; Kazi, B.M. Correction: Molecular Characterization of Circulating Respiratory Syncytial Virus (RSV) Genotypes in Gilgit Baltistan Province of Pakistan during 2011–2012 Winter Season. PLoS ONE 2015, 10, e0145599. [Google Scholar] [CrossRef]
- Gunson, R.N.; Collins, T.C.; Carman, W.F. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions. J. Clin. Virol. 2005, 33, 341–344. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Yip, C.C.Y.; Huang, Y.; Yuen, K.Y. More and More Coronaviruses: Human Coronavirus HKU1. Viruses 2009, 1, 57–71. [Google Scholar] [CrossRef]
- Dare, R.K.; Fry, A.M.; Chittaganpitch, M.; Sawanpanyalert, P.; Olsen, S.J.; Erdman, D.D. Human Coronavirus Infections in Rural Thailand: A Comprehensive Study Using Real-Time Reverse-Transcription Polymerase Chain Reaction Assays. J. Infect. Dis. 2007, 196, 1321–1328. [Google Scholar] [CrossRef]
- World Health Organization. Information Forthe Molecular Detectionof Influenza Viruses. Available online: https://cdn.who.int/media/docs/default-source/influenza/molecular-detention-of-influenza-viruses/protocols_influenza_virus_detection_2024.pdf?sfvrsn=df7d268a_8 (accessed on 10 January 2025).
- Berg, M.G.; Forberg, K.; Perez, L.J.; Luk, K.C.; Meyer, T.V.; Cloherty, G.A. Emergence of a Distinct Picobirnavirus Genotype Circulating in Patients Hospitalized with Acute Respiratory Illness. Viruses 2021, 13, 2534. [Google Scholar] [CrossRef]
- Álvarez-Díaz, D.A.; Quintero, P.A.; Peláez-Carvajal, D.; Ajami, N.J.; Usme-Ciro, J.A. Novel pan-serotype control RNA for dengue virus typing through real-time reverse transcription-polymerase chain reaction. J. Virol. Methods 2019, 271, 113677. [Google Scholar] [CrossRef]
- Álvarez-Díaz, D.A.; Valencia-Álvarez, E.; Rivera, J.A.; Rengifo, A.C.; Usme-Ciro, J.A.; Peláez-Carvajal, D.; Lozano-Jiménez, Y.Y.; Torres-Fernández, O. An updated RT-qPCR assay for the simultaneous detection and quantification of chikungunya, dengue and zika viruses. Infect. Genet. Evol. 2021, 93, 104967. [Google Scholar] [CrossRef] [PubMed]
- Chan Zuckerberg ID—Detect & Track Infectious Diseases. Available online: https://czid.org/ (accessed on 20 August 2025).
- Knox, M.A.; Gedye, K.R.; Hayman, D.T.S. The Challenges of Analysing Highly Diverse Picobirnavirus Sequence Data. Viruses 2018, 10, 685. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.J.; Cloherty, G.A.; Berg, M.G. Understanding the genetic diversity of picobirnavirus: A classification update based on phylogenetic and pairwise sequence comparison approaches. Viruses 2021, 13, 1476. [Google Scholar] [CrossRef]
- Perez, L.J.; Cloherty, G.A.; Berg, M.G. Parallel evolution of picobirnaviruses from distinct ancestral origins. Microbiol. Spectr. 2023, 11, e02693-23. [Google Scholar] [CrossRef]
- Alhamid, G.; Tombuloglu, H.; Rabaan, A.A.; Al-Suhaimi, E. SARS-CoV-2 detection methods: A comprehensive review. Saudi J. Biol. Sci. 2022, 29, 103465. [Google Scholar] [CrossRef]
- Altan, E.; Dib, J.C.; Gulloso, A.R.; Juandigua, D.E.; Deng, X.; Bruhn, R.; Hildebrand, K.; Freiden, P.; Yamamoto, J.; Schultz-Cherry, S.; et al. Effect of Geographic Isolation on the Nasal Virome of Indigenous Children. J. Virol. 2019, 93, 17. [Google Scholar] [CrossRef]
- Rajagopala, S.V.; Bakhoum, N.G.; Pakala, S.B.; Shilts, M.H.; Rosas-Salazar, C.; Mai, A.; Boone, H.H.; McHenry, R.; Yooseph, S.; Halasa, N.; et al. Metatranscriptomics to characterize respiratory virome, microbiome, and host response directly from clinical samples. Cell Rep. Methods 2021, 1, 100091. [Google Scholar] [CrossRef] [PubMed]
- Martyn, C.; Hayes, B.M.; Lauko, D.; Midthun, E.; Castaneda, G.; Bosco-Lauth, A.; Salkeld, D.J.; Kistler, A.; Pollard, K.S.; Chou, S. Metatranscriptomic investigation of single Ixodes pacificus ticks reveals diverse microbes, viruses, and novel mRNA-like endogenous viral elements. mSystems 2024, 9, e00321-24. [Google Scholar] [CrossRef] [PubMed]
- Shamim, U.; Yadav, A.; Maurya, R.; Devi, P.; Kumari, P.; Kanika; Khare, K.; Tarai, B.; Pandey, R. Functional metagenomics highlights varied infection states with dynamics of pathogens and antibiotic resistance in lower respiratory tract infections. Heliyon 2024, 10, e38380. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Kalantar, K.L.; Glascock, A.L.; Chu, V.T.; Guerrero, E.S.; Bernick, N.; Butcher, X.; Ewing, K.; Fahsbender, E.; Holmes, O.; et al. Simultaneous detection of pathogens and antimicrobial resistance genes with the open source, cloud-based, CZ ID pipeline. Genome Med. 2024, 17, 46. [Google Scholar] [CrossRef]
- Meng, Y.; Xiao, L.; Chen, W.; Zhao, F.; Zhao, X. An efficient metatranscriptomic approach for capturing RNA virome and its application to SARS-CoV-2. J. Genet. Genomics. 2021, 48, 860. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, G.; Yang, H.; Li, H.; Guo, Y.; Liu, H.; Xu, X.; Zhang, C. Estimating the prevalence of six common respiratory viral infections in Zhangzhou, China using nasopharyngeal swabs in adults and throat swabs in Children. Sci. Rep. 2025, 15, 487. [Google Scholar] [CrossRef]
- NS: Infección Respiratoria Aguda Colombia. 2025. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2025_Boletin_epidemiologico_semana_16.pdf (accessed on 30 September 2025).
- Jo, K.J.; Choi, S.-H.; Oh, C.E.; Kim, H.; Choi, B.S.; Jo, D.S.; Park, S.E. Epidemiology and Clinical Characteristics of Human Coronaviruses-Associated Infections in Children: A Multi-Center Study. Front. Pediatr. 2022, 10, 877759. [Google Scholar] [CrossRef]
- van der Hoek, L.; Sure, K.; Ihorst, G.; Stang, A.; Pyrc, K.; Jebbink, M.F.; Petersen, G.; Forster, J.; Berkhout, B.; Überla, K. Croup Is Associated with the Novel Coronavirus NL63. PLoS Med. 2005, 2, e240. [Google Scholar] [CrossRef]
- Lesmes-Rodríguez, L.C.; Castillo, L.N.P.; Jaramillo-Hernández, D.A. Seroprevalencia de HCoV-NL63 y HCoV-HKU1 y su relación con las características clínicas de pacientes con COVID-19 de Villavicencio, Colombia. Biomédica 2024, 44, 340–354. [Google Scholar] [CrossRef] [PubMed]
- INS: Los Virus Respiratorios Como La Influenza, Rinovirus, Enterovirus, El Sincitial Respiratorio, Entre Otros, Son Virus Respiratorios Estacionales Ya Conocidos. Available online: https://www.ins.gov.co/Noticias/Paginas/INS-explica-la-circulaci%C3%B3n-de-virus-respiratorios-en-el-pa%C3%ADs.aspx (accessed on 18 August 2025).
- Sadiq, S.; Holmes, E.C.; Mahar, J.E. Genomic and phylogenetic features of the Picobirnaviridae suggest microbial rather than animal hosts. Virus Evol. 2024, 10, 33. [Google Scholar] [CrossRef]
- Roux, S.; Matthijnssens, J.; Dutilh, B.E. Metagenomics in Virology. In Encyclopedia of Virology, 4th ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 1–5, pp. 133–140. Available online: https://www.sciencedirect.com/science/article/abs/pii/B9780128096338209576?via%3Dihub (accessed on 21 August 2025).
- Mirzaei, M.K.; Xue, J.; Costa, R.; Ru, J.; Schulz, S.; Taranu, Z.E.; Deng, L. Challenges of Studying the Human Virome—Relevant Emerging Technologies. Trends Microbiol. 2021, 29, 171–181. [Google Scholar] [CrossRef]
- Liu, S.; Niu, R.; Wang, X.; Cui, J.; Cui, M.; Zhou, H.; Li, J.; Holmes, E.C.; Shi, W.; Li, C. Meta-transcriptomic sequencing reveals divergent RNA viruses in geckos. Virus Res. 2025, 354, 199551. [Google Scholar] [CrossRef]
- Cummings, M.J.; Tokarz, R.; Bakamutumaho, B.; Kayiwa, J.; Byaruhanga, T.; Owor, N.; Namagambo, B.; Wolf, A.; Mathema, B.; Lutwama, J.J.; et al. Precision Surveillance for Viral Respiratory Pathogens: Virome Capture Sequencing for the Detection and Genomic Characterization of Severe Acute Respiratory Infection in Uganda. Clin. Infect. Dis. 2019, 68, 1118–1125. [Google Scholar] [CrossRef]
- Smits, S.L.; van Leeuwen, M.; Schapendonk, C.M.; Schürch, A.C.; Bodewes, R.; Haagmans, B.L.; Osterhaus, A.D. Picobirnaviruses in the human respiratory tract. Emerg. Infect. Dis. 2012, 18, 1539–1540. [Google Scholar] [CrossRef] [PubMed]
- Wakuda, M.; Pongsuwanna, Y.; Taniguchi, K. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J. Virol. Methods 2005, 126, 165–169. [Google Scholar] [CrossRef] [PubMed]

| Virus Name | Common/Formerly Name | Family/Genus | Genome Type |
|---|---|---|---|
| Betacoronavirus pandemicum | SARS-CoV-2 | Coronaviridae/Betacoronavirus | ssRNA (+) |
| Alphainfluenzavirus influenzae | Influenza A | Orthomyxoviridae/Alphainfluenzavirus | ssRNA (−), segmented |
| Betainfluenzavirus influenzae | Influenza B | Orthomyxoviridae/Betainfluenzavirus | ssRNA (−), segmented |
| Alphacoronavirus amsterdamense | HCoV-NL63 | Coronaviridae/Alphacoronavirus | ssRNA (+) |
| Betacoronavirus hongkongense | HCoV-HKU1 | Coronaviridae/Betacoronavirus | ssRNA (+) |
| Alphacoronavirus chicagoense | HCoV-229E | Coronaviridae/Alphacoronavirus | ssRNA (+) |
| Orthopneumovirus hominis | Respiratory Syncytial Virus (RSV) | Pneumoviridae/Orthopneumovirus | ssRNA (−) |
| Metapneumovirus | Human Metapneumovirus | Pneumoviridae/Metapneumovirus | ssRNA (−) |
| Adenovirus | Human adenovirus | Adenoviridae/Mastadenovirus | dsDNA |
| Enterovirus betacoxsackie | Coxsackievirus (Enterovirus B) | Picornaviridae/Enterovirus | ssRNA (+) |
| Enterovirus alpharhino | Rhinovirus A | Picornaviridae/Enterovirus | ssRNA (+) |
| Orthopicobirnavirus hominis | Human Picobirnavirus | Picobirnaviridae/Orthopicobirnavirus | dsRNA, segmented |
| Category | Description | Number | Percentage |
|---|---|---|---|
| Gender | Male | 27 | 39.1 |
| Female | 42 | 60.9 | |
| Total | 69 | 100 | |
| Age | <5 years | 23 | 33.3 |
| 5–14 years | 27 | 39.1 | |
| 15–29 years | 10 | 14.5 | |
| 30–59 years | 8 | 11.6 | |
| ≥60 years | 1 | 1.5 | |
| Total | 69 | 100 | |
| Symptoms * | Fever | 36 | 52.2 |
| Cold/Flu-like symptoms | 30 | 43.5 | |
| Diarrhea | 22 | 31.9 | |
| Cough | 13 | 18.8 | |
| Stomach pain | 4 | 5.8 | |
| Headache | 3 | 4.3 | |
| Sore throat | 3 | 4.3 | |
| Vomiting | 2 | 2.9 | |
| Nausea | 1 | 1.4 | |
| Ear pain | 1 | 1.4 | |
| Skin rash/spots | 1 | 1.4 | |
| Kidney pain | 1 | 1.4 | |
| Loss of appetite | 1 | 1.4 | |
| None/No symptoms | 8 | 11.6 |
| Sample Name | Type of Sample | RT-qPCR | Metatranscriptomic | ||
|---|---|---|---|---|---|
| Ct | Virus | Reads Per Million | Virus | ||
| COL_INS_GME_01 | nasopharyngeal swab | - | Negative | - | Negative |
| COL_INS_GME_02 | nasopharyngeal swab | - | Negative | - | Negative |
| COL_INS_GME_03 | nasopharyngeal swab | 34.14 | Alphacoronavirus amsterdamense | 31.3 | Alphacoronavirus amsterdamense |
| COL_INS_GME_04 | nasopharyngeal swab | 30.53 | Enterovirus alpharino | 129.4 | Enterovirus alpharino |
| COL_INS_GME_05 | nasopharyngeal swab | 29.64 | Enterovirus alpharino | - | Negative |
| COL_INS_GME_06 | nasopharyngeal swab | 29.74 | Influenza A/B | 21.91 | Betainfluenzavirus influenzae |
| 70.48 | Orthopicobirnavirus hominis | ||||
| COL_INS_GME_07 | nasopharyngeal swab | 28.85 | Enterovirus alpharino | 75.24 | Enterovirus alpharino |
| COL_INS_GME_08 | nasopharyngeal swab | 25.64 | Enterovirus sp. | 878 | Enterovirus betacoxsackie |
| COL_INS_GME_09 | nasopharyngeal swab | - | Negative | - | Negative |
| COL_INS_GME_10 | nasopharyngeal swab | - | Negative | - | Negative |
| COL_INS_GME_11 | nasopharyngeal swab | - | Negative | - | Negative |
| COL_INS_GME_12 | nasopharyngeal swab | - | Negative | - | Negative |
| COL_INS_GME_13 | nasopharyngeal swab | - | Negative | - | Negative |
| COL_INS_GME_14 | Serum | - | Negative | - | Negative |
| COL_INS_GME_15 | Serum | - | Negative | - | Negative |
| COL_INS_GME_16 | Serum | - | Negative | - | Negative |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De arco-Rodríguez, B.E.; Pérez-Lozada, J.T.; Laiton-Donato, K.; Peláez-Carvajal, D.; Puerto-Castro, G.M.; Álvarez-Díaz, D.A. Metatranscriptomic Analysis of Oropharyngeal Samples Reveals Common Respiratory Viruses and a Potential Interspecies Transmitted Picobirnavirus in the Wayuu Population, La Guajira, Colombia. Viruses 2025, 17, 1397. https://doi.org/10.3390/v17101397
De arco-Rodríguez BE, Pérez-Lozada JT, Laiton-Donato K, Peláez-Carvajal D, Puerto-Castro GM, Álvarez-Díaz DA. Metatranscriptomic Analysis of Oropharyngeal Samples Reveals Common Respiratory Viruses and a Potential Interspecies Transmitted Picobirnavirus in the Wayuu Population, La Guajira, Colombia. Viruses. 2025; 17(10):1397. https://doi.org/10.3390/v17101397
Chicago/Turabian StyleDe arco-Rodríguez, Beatriz Elena, Jhindy Tatiana Pérez-Lozada, Katherine Laiton-Donato, Dioselina Peláez-Carvajal, Gloria Mercedes Puerto-Castro, and Diego Alejandro Álvarez-Díaz. 2025. "Metatranscriptomic Analysis of Oropharyngeal Samples Reveals Common Respiratory Viruses and a Potential Interspecies Transmitted Picobirnavirus in the Wayuu Population, La Guajira, Colombia" Viruses 17, no. 10: 1397. https://doi.org/10.3390/v17101397
APA StyleDe arco-Rodríguez, B. E., Pérez-Lozada, J. T., Laiton-Donato, K., Peláez-Carvajal, D., Puerto-Castro, G. M., & Álvarez-Díaz, D. A. (2025). Metatranscriptomic Analysis of Oropharyngeal Samples Reveals Common Respiratory Viruses and a Potential Interspecies Transmitted Picobirnavirus in the Wayuu Population, La Guajira, Colombia. Viruses, 17(10), 1397. https://doi.org/10.3390/v17101397








