A Highly Sensitive BRET-Based Reporter for Live-Cell Detection of HIV-1 Protease Activity and Inhibitor Screening
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids. Mammalian Expression Plasmids
2.2. Protease Inhibitors. Saquinavir Mesylate
2.3. Cell Culture
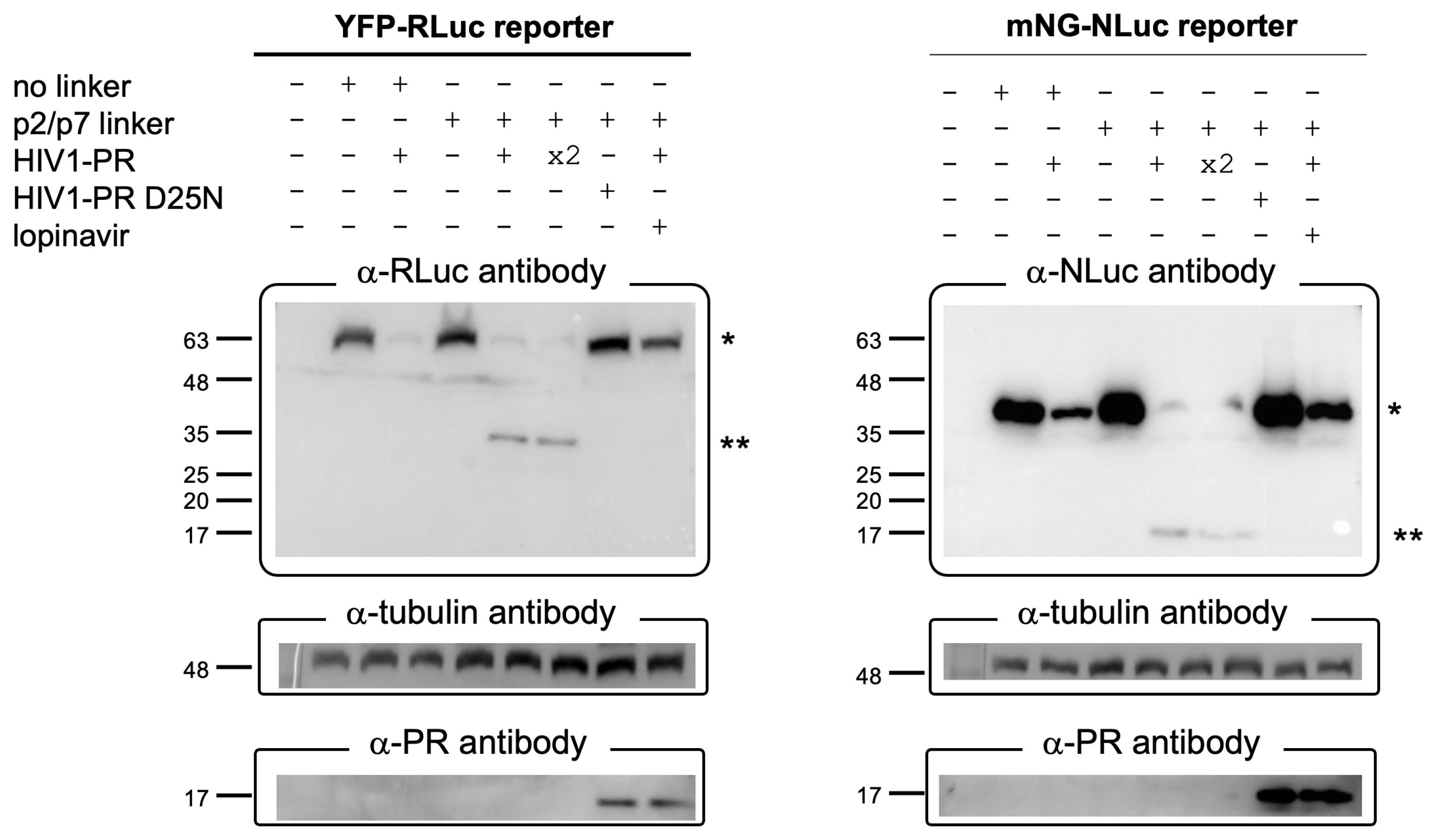
2.4. Western Blotting Analysis
2.5. Spectral Measurements
2.6. Bioluminescence Resonance Energy Transfer (BRET) Assays
2.7. PR Inhibitors Screening Using mNG-p2/p7-NLuc
2.8. Statistical Analysis
3. Results
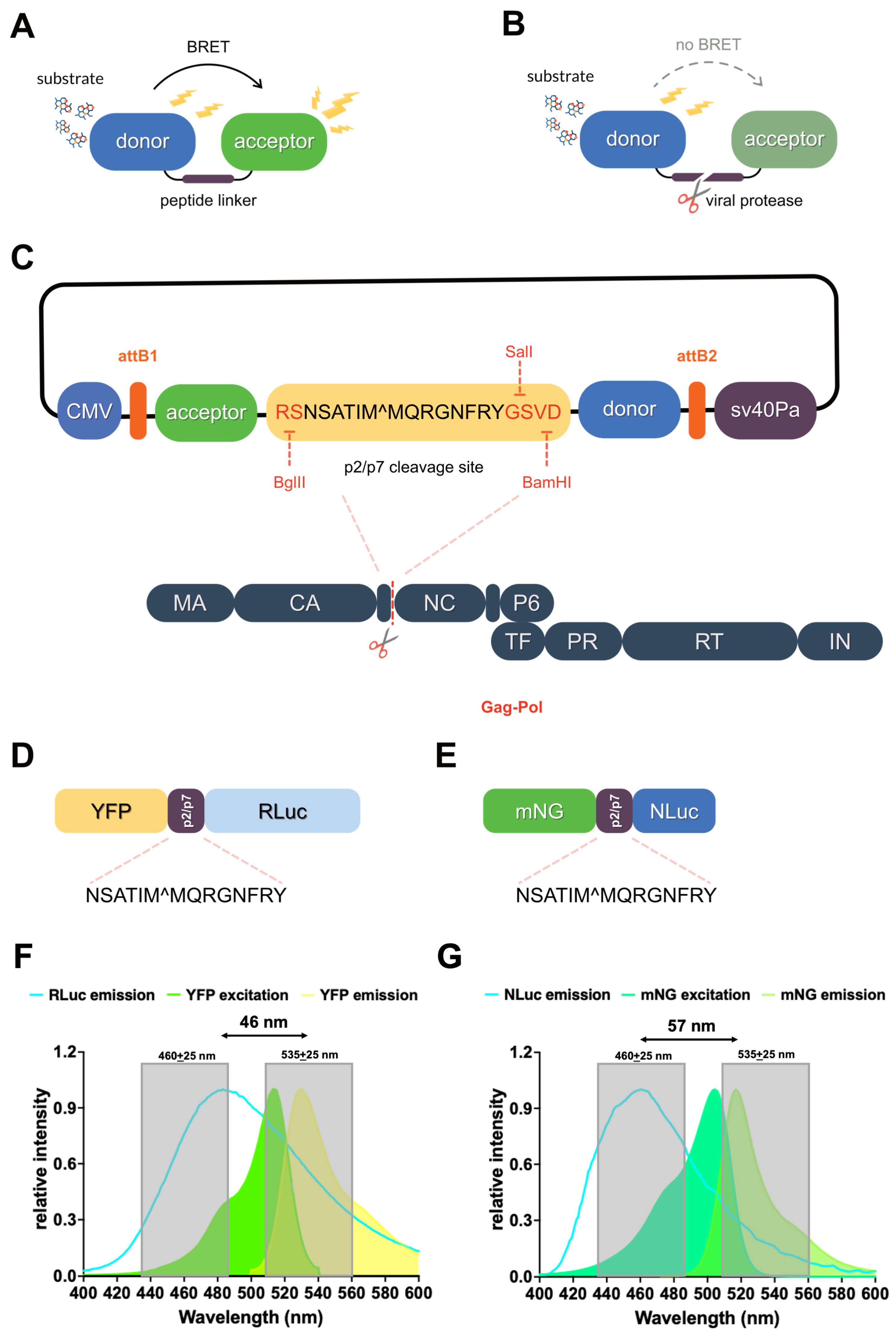
3.1. Design of BRET-Based HIV-1 PR Reporters
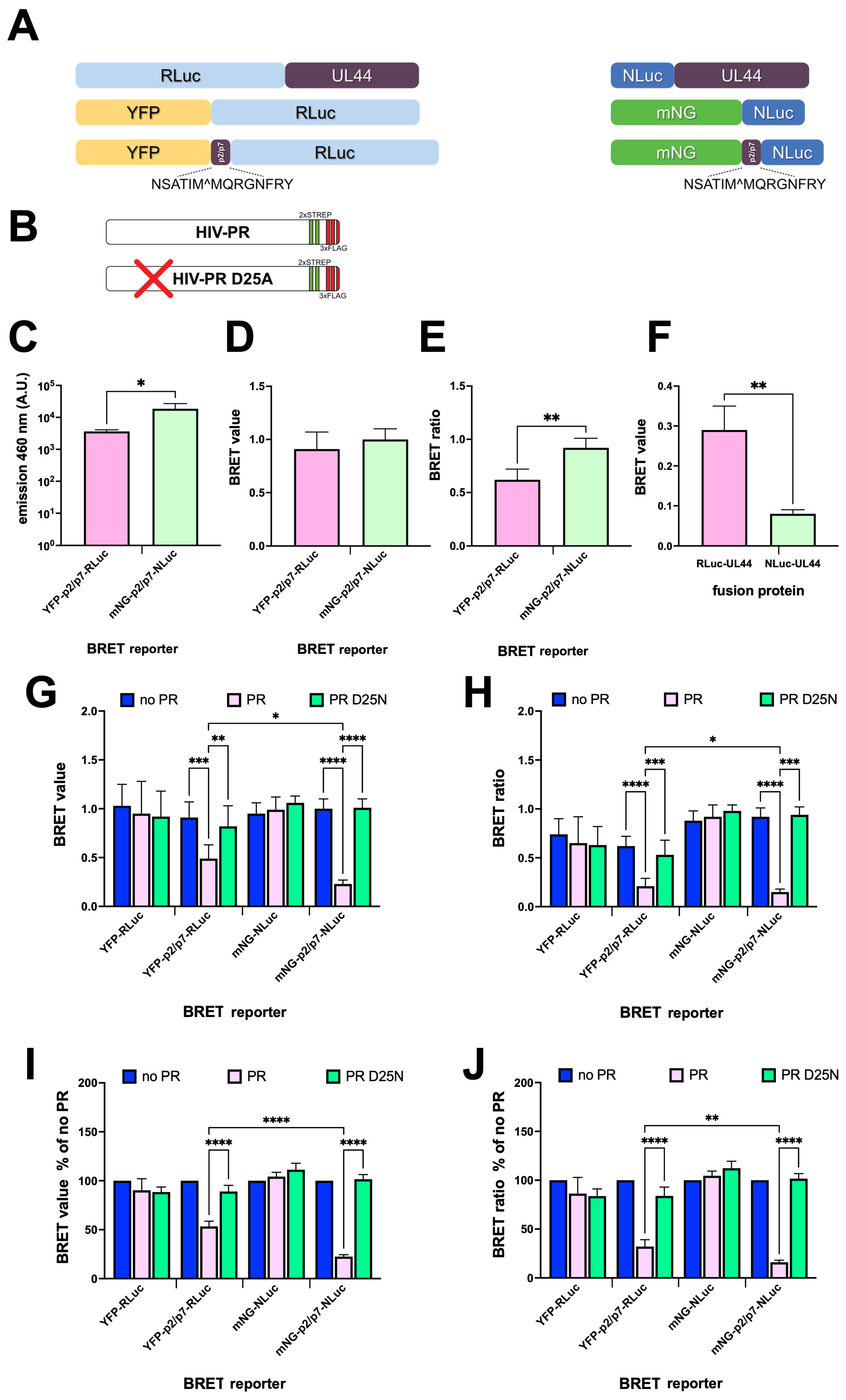
3.2. Validation of BRET-Based HIV-1 PR Reporters in a Cellular Context
3.3. Spectral Properties of the YFP-p2/p7-RLuc and mNG-p2/p7-NLuc BRET Reporters
3.4. The BRET Reporters Are Cleaved by HIV-1 PR
3.5. mNG-p2/p7-NLuc Detects HIV-1 PR Activity in a Dose Dependent Manner
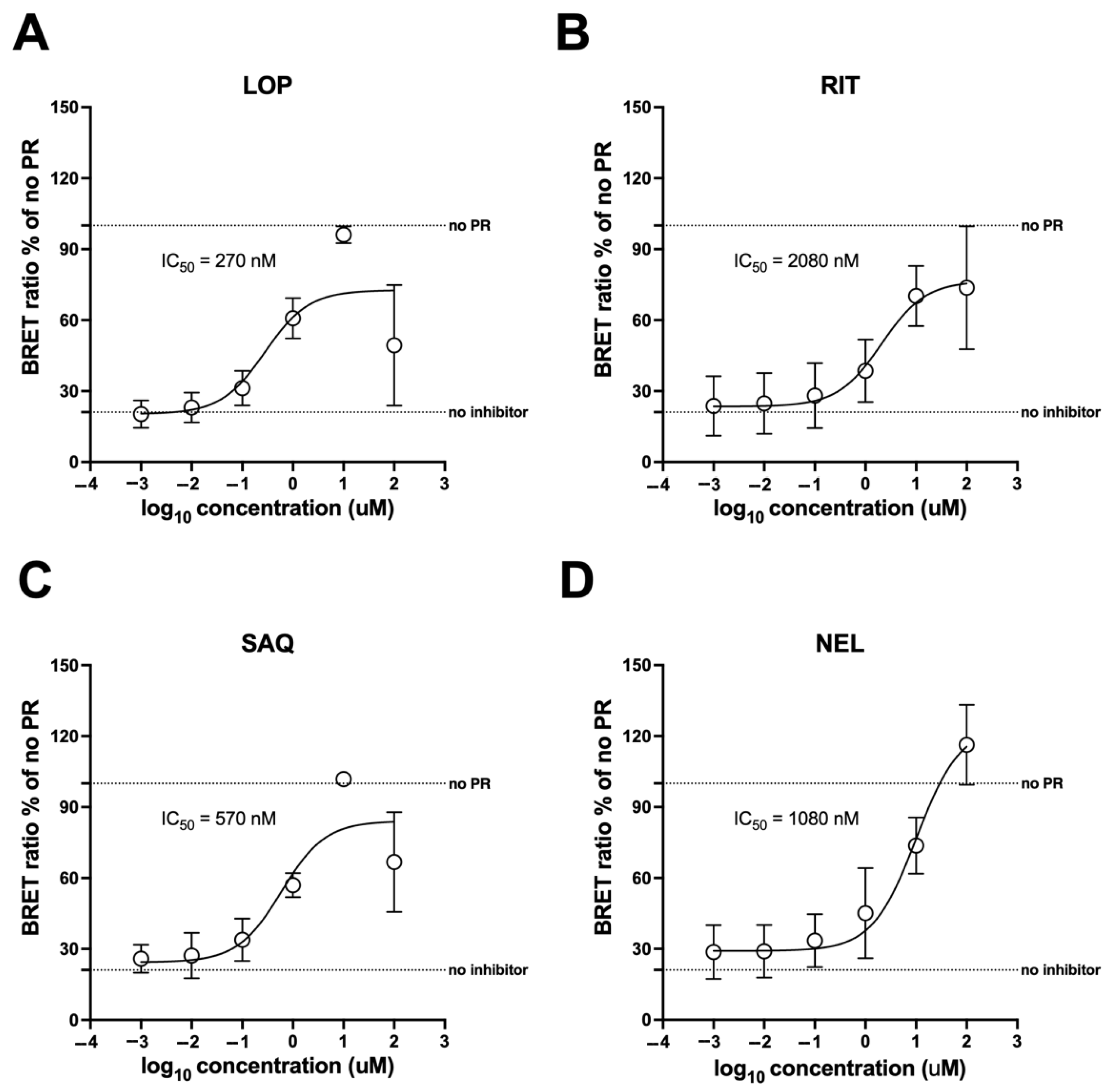
3.6. mNG-p2/p7-NLuc as a Tool for Antiviral Drug Discovery
3.7. The mNG-p2/p7-NLuc Reporter Allows Measuring the Potency of HIV-1 PR Inhibitors in Living Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BRET | Bioluminescent resonance energy transfer |
| RLuc | Renilla luciferase |
| NLuc | Nanoluc luciferase |
| h-CTZ | h-coelenterazine |
| mNG | mNeonGreen |
| HIV-1 | Human Immunodeficiency Virus 1 |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | Fetal bovine serum |
| IC50 | Half-maximal inhibitory concentration |
References
- Borges, P.H.O.; Ferreira, S.B.; Silva, F.P., Jr. Recent Advances on Targeting Proteases for Antiviral Development. Viruses 2024, 16, 366. [Google Scholar] [CrossRef]
- Centazzo, M.; Manganaro, L.; Alvisi, G. Cellular Targets of HIV-1 Protease: Just the Tip of the Iceberg? Viruses 2023, 15, 712. [Google Scholar] [CrossRef]
- De Clercq, E. The design of drugs for HIV and HCV. Nat. Rev. Drug Discov. 2007, 6, 1001–1018. [Google Scholar] [CrossRef] [PubMed]
- Manns, M.P.; von Hahn, T. Novel therapies for hepatitis C—One pill fits all? Nat. Rev. Drug Discov. 2013, 12, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Ohsaki, Y.; Sasaki, T.; Umekage, Y.; Yanada, H.; Ishikawa, M.; Yoshida, R. Real-World Treatment Outcomes in the First and Subsequent Coronavirus Disease 2019 (COVID-19) Hospital Clusters. Cureus 2025, 17, e78981. [Google Scholar] [CrossRef]
- Hong, S.J.; Resnick, S.J.; Iketani, S.; Cha, J.W.; Albert, B.A.; Fazekas, C.T.; Chang, C.W.; Liu, H.; Dagan, S.; Abagyan, M.R.; et al. A multiplex method for rapidly identifying viral protease inhibitors. Mol. Syst. Biol. 2025, 21, 158–172. [Google Scholar] [CrossRef]
- Ihssen, J.; Faccio, G.; Yao, C.; Sirec, T.; Spitz, U. Fluorogenic in vitro activity assay for the main protease M(pro) from SARS-CoV-2 and its adaptation to the identification of inhibitors. STAR Protoc. 2021, 2, 100793. [Google Scholar] [CrossRef]
- Arias-Arias, J.L.; MacPherson, D.J.; Hill, M.E.; Hardy, J.A.; Mora-Rodriguez, R. A fluorescence-activatable reporter of flavivirus NS2B-NS3 protease activity enables live imaging of infection in single cells and viral plaques. J. Biol. Chem. 2020, 295, 2212–2226. [Google Scholar] [CrossRef]
- Pahmeier, F.; Neufeldt, C.J.; Cerikan, B.; Prasad, V.; Pape, C.; Laketa, V.; Ruggieri, A.; Bartenschlager, R.; Cortese, M. A Versatile Reporter System To Monitor Virus-Infected Cells and Its Application to Dengue Virus and SARS-CoV-2. J. Virol. 2021, 95, e01715-20. [Google Scholar] [CrossRef]
- van der Linden, L.; Ulferts, R.; Nabuurs, S.B.; Kusov, Y.; Liu, H.; George, S.; Lacroix, C.; Goris, N.; Lefebvre, D.; Lanke, K.H.; et al. Application of a cell-based protease assay for testing inhibitors of picornavirus 3C proteases. Antivir. Res. 2014, 103, 17–24. [Google Scholar] [CrossRef]
- Bacart, J.; Corbel, C.; Jockers, R.; Bach, S.; Couturier, C. The BRET technology and its application to screening assays. Biotechnol. J. 2008, 3, 311–324. [Google Scholar] [CrossRef]
- Hu, K.; Clement, J.F.; Abrahamyan, L.; Strebel, K.; Bouvier, M.; Kleiman, L.; Mouland, A.J. A human immunodeficiency virus type 1 protease biosensor assay using bioluminescence resonance energy transfer. J. Virol. Methods 2005, 128, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- England, C.G.; Ehlerding, E.B.; Cai, W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjug. Chem. 2016, 27, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Inouye, S.; Sato, J.I.; Sahara-Miura, Y.; Tomabechi, Y.; Sumida, Y.; Sekine, S.I.; Shirouzu, M.; Hosoya, T. Reverse mutants of the catalytic 19 kDa mutant protein (nanoKAZ/nanoLuc) from Oplophorus luciferase with coelenterazine as preferred substrate. PLoS ONE 2022, 17, e0272992. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lambert, G.G.; Chammas, A.; Ni, Y.; Cranfill, P.J.; Baird, M.A.; Sell, B.R.; Allen, J.R.; Day, R.N.; Israelsson, M.; et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 2013, 10, 407–409. [Google Scholar] [CrossRef]
- den Hamer, A.; Dierickx, P.; Arts, R.; de Vries, J.; Brunsveld, L.; Merkx, M. Bright Bioluminescent BRET Sensor Proteins for Measuring Intracellular Caspase Activity. ACS Sens. 2017, 2, 729–734. [Google Scholar] [CrossRef]
- Di Antonio, V.; Palu, G.; Alvisi, G. Live-Cell Analysis of Human Cytomegalovirus DNA Polymerase Holoenzyme Assembly by Resonance Energy Transfer Methods. Microorganisms 2021, 9, 928. [Google Scholar] [CrossRef]
- Messa, L.; Celegato, M.; Bertagnin, C.; Mercorelli, B.; Alvisi, G.; Banks, L.; Palu, G.; Loregian, A. The Dimeric Form of HPV16 E6 Is Crucial to Drive YAP/TAZ Upregulation through the Targeting of hScrib. Cancers 2021, 13, 4083. [Google Scholar] [CrossRef]
- Trevisan, M.; Di Antonio, V.; Radeghieri, A.; Palu, G.; Ghildyal, R.; Alvisi, G. Molecular Requirements for Self-Interaction of the Respiratory Syncytial Virus Matrix Protein in Living Mammalian Cells. Viruses 2018, 10, 109. [Google Scholar] [CrossRef]
- Scaturro, P.; Trist, I.M.; Paul, D.; Kumar, A.; Acosta, E.G.; Byrd, C.M.; Jordan, R.; Brancale, A.; Bartenschlager, R. Characterization of the mode of action of a potent dengue virus capsid inhibitor. J. Virol. 2014, 88, 11540–11555. [Google Scholar] [CrossRef]
- Smith, K.M.; Di Antonio, V.; Bellucci, L.; Thomas, D.R.; Caporuscio, F.; Ciccarese, F.; Ghassabian, H.; Wagstaff, K.M.; Forwood, J.K.; Jans, D.A.; et al. Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting. Biochim. Biophys. Acta 2018, 1865, 1114–1129. [Google Scholar] [CrossRef]
- Petersen, G.F.; Pavan, S.; Ariawan, D.; Tietz, O.; Nematollahzadeh, S.; Sarker, S.; Forwood, J.K.; Alvisi, G. Nuclear trafficking of Anelloviridae capsid protein ORF1 reflects modular evolution of subcellular targeting signals. Virus Evol. 2025, 11, veaf069. [Google Scholar] [CrossRef]
- Sinigalia, E.; Alvisi, G.; Mercorelli, B.; Coen, D.M.; Pari, G.S.; Jans, D.A.; Ripalti, A.; Palu, G.; Loregian, A. Role of homodimerization of human cytomegalovirus DNA polymerase accessory protein UL44 in origin-dependent DNA replication in cells. J. Virol. 2008, 82, 12574–12579. [Google Scholar] [CrossRef] [PubMed]
- Jager, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV-human protein complexes. Nature 2011, 481, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Cross, E.M.; Akbari, N.; Ghassabian, H.; Hoad, M.; Pavan, S.; Ariawan, D.; Donnelly, C.M.; Lavezzo, E.; Petersen, G.F.; Forwood, J.K.; et al. A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals. Protein Sci. 2024, 33, e4876. [Google Scholar] [CrossRef] [PubMed]
- Elbadawy, H.M.; Mohammed Abdul, M.I.; Aljuhani, N.; Vitiello, A.; Ciccarese, F.; Shaker, M.A.; Eltahir, H.M.; Palu, G.; Di Antonio, V.; Ghassabian, H.; et al. Generation of Combinatorial Lentiviral Vectors Expressing Multiple Anti-Hepatitis C Virus shRNAs and Their Validation on a Novel HCV Replicon Double Reporter Cell Line. Viruses 2020, 12, 1044. [Google Scholar] [CrossRef]
- Sinigalia, E.; Alvisi, G.; Segre, C.V.; Mercorelli, B.; Muratore, G.; Winkler, M.; Hsiao, H.H.; Urlaub, H.; Ripalti, A.; Chiocca, S.; et al. The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner. PLoS ONE 2012, 7, e49630. [Google Scholar] [CrossRef]
- Ghassabian, H.; Falchi, F.; Timmoneri, M.; Mercorelli, B.; Loregian, A.; Palu, G.; Alvisi, G. Divide et impera: An In Silico Screening Targeting HCMV ppUL44 Processivity Factor Homodimerization Identifies Small Molecules Inhibiting Viral Replication. Viruses 2021, 13, 941. [Google Scholar] [CrossRef]
- Alvisi, G.; Roth, D.M.; Camozzi, D.; Pari, G.S.; Loregian, A.; Ripalti, A.; Jans, D.A. The flexible loop of the human cytomegalovirus DNA polymerase processivity factor ppUL44 is required for efficient DNA binding and replication in cells. J. Virol. 2009, 83, 9567–9576. [Google Scholar] [CrossRef]
- Perales, C.; Carrasco, L.; Ventoso, I. Cleavage of eIF4G by HIV-1 protease: Effects on translation. FEBS Lett. 2003, 533, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Alvisi, G.; Manaresi, E.; Cross, E.M.; Hoad, M.; Akbari, N.; Pavan, S.; Ariawan, D.; Bua, G.; Petersen, G.F.; Forwood, J.; et al. Importin alpha/beta-dependent nuclear transport of human parvovirus B19 nonstructural protein 1 is essential for viral replication. Antivir. Res. 2023, 213, 105588. [Google Scholar] [CrossRef] [PubMed]
- Alvisi, G.; Manaresi, E.; Pavan, S.; Jans, D.A.; Wagstaff, K.M.; Gallinella, G. Avermectins Inhibit Replication of Parvovirus B19 by Disrupting the Interaction Between Importin alpha and Non-Structural Protein 1. Viruses 2025, 17, 220. [Google Scholar] [CrossRef] [PubMed]
- Ravlo, E.; Ianevski, A.; Schjolberg, J.O.; Solvang, V.; Dumaru, R.; Lysvand, H.; Hankinson, J.; Vaha-Koskela, M.; Vainionpaa, S.; Varhe, A.; et al. Synergistic combination of orally available safe-in-man pleconaril, AG7404, and mindeudesivir inhibits enterovirus infections in human cell and organoid cultures. Cell. Mol. Life Sci. 2025, 82, 57. [Google Scholar] [CrossRef]
- Tripathi, A.; Chauhan, S.; Khasa, R. A Comprehensive Review of the Development and Therapeutic Use of Antivirals in Flavivirus Infection. Viruses 2025, 17, 74. [Google Scholar] [CrossRef]
- Hilton, B.J.; Wolkowicz, R. An assay to monitor HIV-1 protease activity for the identification of novel inhibitors in T-cells. PLoS ONE 2010, 5, e10940. [Google Scholar] [CrossRef]
- Corbel, C.; Wang, Q.; Bousserouel, H.; Hamdi, A.; Zhang, B.; Lozach, O.; Ferandin, Y.; Tan, V.B.; Gueritte, F.; Colas, P.; et al. First BRET-based screening assay performed in budding yeast leads to the discovery of CDK5/p25 interaction inhibitors. Biotechnol. J. 2011, 6, 860–870. [Google Scholar] [CrossRef]
- Drinovec, L.; Kubale, V.; Nøhr Larsen, J.; Vrecl, M. Mathematical models for quantitative assessment of bioluminescence resonance energy transfer: Application to seven transmembrane receptors oligomerization. Front. Endocrinol. 2012, 3, 104. [Google Scholar] [CrossRef]
- Mo, X.L.; Fu, H. BRET: NanoLuc-Based Bioluminescence Resonance Energy Transfer Platform to Monitor Protein-Protein Interactions in Live Cells. Methods Mol. Biol. 2016, 1439, 263–271. [Google Scholar] [CrossRef]
- Gaber, R.; Majerle, A.; Jerala, R.; Bencina, M. Noninvasive high-throughput single-cell analysis of HIV protease activity using ratiometric flow cytometry. Sensors 2013, 13, 16330–16346. [Google Scholar] [CrossRef]
- Lv, Z.; Chu, Y.; Wang, Y. HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV AIDS Auckl. 2015, 7, 95–104. [Google Scholar] [CrossRef]
- Majerova, T.; Konvalinka, J. Viral proteases as therapeutic targets. Mol. Asp. Med. 2022, 88, 101159. [Google Scholar] [CrossRef]
- Ali, R.; Ramadurai, S.; Barry, F.; Nasheuer, H.P. Optimizing fluorescent protein expression for quantitative fluorescence microscopy and spectroscopy using herpes simplex thymidine kinase promoter sequences. FEBS Open Bio 2018, 8, 1043–1060. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Centazzo, M.; Djossou, A.V.-V.; Pavan, S.; Alvisi, G. A Highly Sensitive BRET-Based Reporter for Live-Cell Detection of HIV-1 Protease Activity and Inhibitor Screening. Viruses 2025, 17, 1391. https://doi.org/10.3390/v17101391
Centazzo M, Djossou AV-V, Pavan S, Alvisi G. A Highly Sensitive BRET-Based Reporter for Live-Cell Detection of HIV-1 Protease Activity and Inhibitor Screening. Viruses. 2025; 17(10):1391. https://doi.org/10.3390/v17101391
Chicago/Turabian StyleCentazzo, Matteo, Atalie Verra-Victoria Djossou, Silvia Pavan, and Gualtiero Alvisi. 2025. "A Highly Sensitive BRET-Based Reporter for Live-Cell Detection of HIV-1 Protease Activity and Inhibitor Screening" Viruses 17, no. 10: 1391. https://doi.org/10.3390/v17101391
APA StyleCentazzo, M., Djossou, A. V.-V., Pavan, S., & Alvisi, G. (2025). A Highly Sensitive BRET-Based Reporter for Live-Cell Detection of HIV-1 Protease Activity and Inhibitor Screening. Viruses, 17(10), 1391. https://doi.org/10.3390/v17101391






