Association Between Serum HBV DNA Levels and CCL-20, CD8a, CXCL-16, and GDF-15 in Patients with Chronic Hepatitis B
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Committee and Project Support Information
2.2. Study Population
2.3. Statistical Analysis
3. Results
4. Discussion
Limitations of Our Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khanam, A.; Chua, J.V.; Kottilil, S. Immunopathology of Chronic Hepatitis B Infection: Role of Innate and Adaptive Immune Response in Disease Progression. Int. J. Mol. Sci. 2021, 22, 5497. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Liu, X.; Tian, L.; Chen, Y. Cytokine-Mediated Immunopathogenesis of Hepatitis B Virus Infections. Clin. Rev. Allergy Immunol. 2016, 50, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Chisari, F.V.; Ferrari, C. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 1995, 13, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Hoare, M.; Davies, N.; Lopes, A.R.; Dunn, C.; Kennedy, P.T.; Alexander, G.; Finney, H.; Lawson, A.; Plunkett, F.J.; et al. Functional skewing of the global CD8 T cell population in chronic hepatitis B virus infection. J. Exp. Med. 2008, 205, 2111–2124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- CDC. Clinical Testing and Diagnosis for Hepatitis B. Available online: https://www.cdc.gov/hepatitis-b/hcp/diagnosis-testing/index.html (accessed on 1 September 2025).
- Park, J.J.; Wong, D.K.; Wahed, A.S.; Lee, W.M.; Feld, J.J.; Terrault, N.; Khalili, M.; Sterling, R.K.; Kowdley, K.V.; Bzowej, N.; et al. Hepatitis B virus-specific and global T-cell dysfunction in patients with chronic hepatitis B. Gastroenterology 2016, 150, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Pahuja, S.; Khare, P.; Kumar, A. Current Challenges and Future Perspectives of Diagnosis of Hepatitis B Virus. Diagnostics 2023, 13, 368. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, C.; Xue, R.; Wang, X.; Xiao, L.; Xian, J. High-sensitivity HBV DNA test for the diagnosis of occult HBV infection: Commonly used but not reliable. Front. Cell Infect. Microbiol. 2023, 13, 1186877. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mak, L.Y.; Seto, W.K.; Fung, J.; Yuen, M.F. New Biomarkers of Chronic Hepatitis B. Gut Liver 2019, 13, 589–595. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Poulsen, K.L.; Cajigas-Du Ross, C.K.; Chaney, J.K.; Nagy, L.E. Role of the chemokine system in liver fibrosis: A narrative review. Dig. Med. Res. 2022, 5, 30. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heydtmann, M.; Lalor, P.F.; Eksteen, J.A.; Hübscher, S.G.; Briskin, M.; Adams, D.H. CXC chemokine ligand 16 promotes integrin-mediated adhesion of liver-infiltrating lymphocytes to cholangiocytes and hepatocytes within the inflamed human liver. J. Immunol. 2005, 174, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Jin, Q.; Chen, H.; Wood, G.C.; Petrick, A.; Strodel, W.; Gabrielsen, J.; Benotti, P.; Mirshahi, T.; Carey, D.J.; et al. CCL20 is up-regulated in non-alcoholic fatty liver disease fibrosis and is produced by hepatic stellate cells in response to fatty acid loading. J. Transl. Med. 2018, 16, 108. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Affò, S.; Morales-Ibanez, O.; Rodrigo-Torres, D.; Altamirano, J.; Blaya, D.; Dapito, D.H.; Millán, C.; Coll, M.; Caviglia, J.M.; Arroyo, V.; et al. CCL20 mediates lipopolysaccharide induced liver injury and is a potential driver of inflammation and fibrosis in alcoholic hepatitis. Gut 2014, 63, 1782–1792. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, E.S.; Kim, S.H.; Kim, H.J.; Kim, K.H.; Lee, B.S.; Ku, B.J. Growth Differentiation Factor 15 Predicts Chronic Liver Disease Severity. Gut Liver 2017, 11, 276–282. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Allahmoradi, E.; Mohammadi, R.; Kheirandish, Z.P.; Alavian, S.M.; Heiat, M. The CD8+ T cell exhaustion mechanisms in chronic hepatitis B infection and immunotherapeutic strategies: A systematic review. Expert. Rev. Clin. Immunol. 2023, 19, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Fabio, M.; George, J.M.W.; Stephanie, R.; Gotto, J.; Behboudi, S.; Burroughs, A.K.; Dusheiko, G.M.; Williams, R.; Bertoletti, A. Tracking the Source of the Hepatitis B Virus–Specific CD8 T Cells during Lamivudine Treatment. J. Infect. Dis. 2003, 187, 679–682. [Google Scholar] [CrossRef]
- Calgin, M.K.; Cetinkol, Y. Decreased levels of serum zonulin and copeptin in chronic Hepatitis-B patients. Pak. J. Med. Sci. 2019, 35, 47–851. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, J.; Li, W.; Feng, L. Diagnostic accuracy for multiple categories: Statistical advice. Br. J. Psychiatry 2025. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Halim, M.H.; Abdulla, N.A.; Kamel, A.; El-Maksoud, N.A.; Ragab, H.M. Significance of growth differentiation factor 15 in chronic HCV patients. J. Genet. Eng. Biotechnol. 2017, 15, 403–407. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sometani, E.; Hikita, H.; Murai, K.; Toyoda, H.; Tanaka, S.; Oze, T.; Sung, J.; Shimoda, A.; Fukuoka, M.; Shigeno, S.; et al. High serum growth differentiation factor 15 is a risk factor for the occurrence of hepatocellular carcinoma in chronic hepatitis B patients treated with nucleos(t)ide analogs. Hepatol Res. 2024. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tang, D.; Xu, C.; Niu, Z.; Li, H.; Li, Y.; Zhang, P. Evaluation of Serum GDF15, AFP, and PIVKA-II as Diagnostic Markers for HBV-Associated Hepatocellular Carcinoma. Lab. Med. 2021, 52, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Hammerich, L.; Bangen, J.M.; Govaere, O.; Zimmermann, H.W.; Gassler, N.; Huss, S.; Liedtke, C.; Prinz, I.; Lira, S.A.; Luedde, T.; et al. Chemokine receptor CCR6-dependent accumulation of γδ T cells in injured liver restricts hepatic inflammation and fibrosis. Hepatology 2014, 59, 630–642. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Song, J.; Liu, L.; Wang, Z.; Xie, D.; Azami, N.L.B.; Lu, L.; Huang, Y.; Ye, W.; Zhang, Q.; Sun, M. CCL20 and CD8A as potential diagnostic biomarkers for HBV-induced liver fibrosis in chronic hepatitis B. Heliyon 2024, 10, e28329. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vachon, A.; Osiowy, C. Novel Biomarkers of Hepatitis B Virus and Their Use in Chronic Hepatitis B Patient Management. Viruses 2021, 13, 951. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, L.; Liao, F.; Yang, L.; Jiang, L.; Duan, L.; Wang, B.; Mu, D.; Chen, J.; Huang, Y.; Hu, Q.; et al. KLRG1-expressing CD8+ T cells are exhausted and polyfunctional in patients with chronic hepatitis B. PLoS ONE 2024, 19, e0303945. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wan, Y.; Mao, M.; Li, M.; Liu, J.; Tong, X.; Wang, J.; Li, J.; Yin, S.; Wu, C. Serum CXCL16: A new predictor of liver inflammation in patients with chronic hepatitis B. J. Viral Hepat. 2024, 31, 107–119. [Google Scholar] [CrossRef] [PubMed]

| Inclusion Criteria | Exclusion Criteria |
|---|---|
| 1. For at least six months, the diagnosis has been chronic hepatitis B with HBsAg positivity. | 1. Co-infection with hepatitis C (HCV), hepatitis D (HDV), or HIV. |
| 2. People who are already under regular medical supervision and have all their serum samples and test results on file. | 2. Any history of malignancy or current malignant disease. |
| 3. Patients who have not yet started antiviral therapy. | 3. Alcohol consumption >30 g/day for men and >20 g/day for women. |
| 4. For the control group, normal liver function and no evidence of chronic disease were observed in routine tests. | 4. Other chronic liver diseases include autoimmune hepatitis, non-alcoholic fatty liver disease (NAFLD), haemochromatosis, and Wilson’s disease. |
| 5. Acute infection, febrile illness, or systemic inflammation within the last month. | |
| 6. Presence of rheumatological or autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematosus, inflammatory bowel diseases, etc. | |
| 7. Type 1 or type 2 diabetes mellitus. | |
| 8. Chronic kidney disease (glomerular filtration rate <60 mL/min/1.73 m2). |
| Variable | Control (1) n= 30 1 | Mild Viral Load (2) n = 31 1 | Moderate Viral Load (3) n = 31 1 | Severe Viral Load (4) n = 34 1 | p2 Value |
|---|---|---|---|---|---|
| Age | 46.10 ± 7.43 | 51.39 ± 12.85 | 51.23 ± 16.65 | 50.47 ± 18.29 | 0.14 |
| Gender | 0.96 | ||||
| Female | 14 (46.67%) | 15 (48.39%) | 15 (48.39%) | 18 (52.94%) | |
| Male | 16 (53.33%) | 16 (51.61%) | 16 (51.61%) | 16 (47.06%) | |
| Hemoglobin (g/dL) | Normal range | 13.64 ± 2.41 | 13.26 ± 2.73 | 13.06 ± 3.07 | 0.68 |
| Leukocyte (×103/μl) | Normal range | 7.37 ± 1.85 | 6.82 ± 1.98 | 8.16 ± 5.04 | 0.28 |
| Platelet (×103/μl) | Normal range | 254.74 ± 65.87 | 250.58 ± 82.36 | 195.47 ± 86.68 | p (2–4): 0.009 p (3–4): 0.017 |
| Neutrophil (×103/μl) | Normal range | 4.57 ± 1.68 | 3.93 ± 1.56 | 4.40 ± 2.65 | 0.29 |
| Monocyte (×103/μl) | Normal range | 0.65 ± 0.33 | 0.58 ± 0.27 | 0.99 ± 1.29 | 0.17 |
| Lymphocyte (×103/μl) | Normal range | 1.97 ± 0.62 | 2.14 ± 0.83 | 2.63 ± 2.50 | 0.27 |
| MPV (femtoliter;fL) | Normal range | 10.38 ± 0.92 | 10.28 ± 0.89 | 10.30 ± 1.04 | 0.90 |
| PDW | Normal range | 12.27 ± 2.05 | 11.90 ± 1.70 | 12.15 ± 2.04 | 0.74 |
| ALP (IU/L) | Normal range | 89.77 ± 62.01 | 96.19 ± 46.92 | 111.18 ± 50.29 | 0.27 |
| GGT (IU/L) | Normal range | 20.00 (14.00–26.00) | 22.00 (15.00–38.00) | 44.00 (16.00–78.00) | p (2–4): 0.005 |
| LDH (IU/L) | Normal range | 242.48 ± 151.78 | 210.68 ± 97.09 | 247.12 ± 133.10 | 0.38 |
| AST (IU/L) | Normal range | 19.40 (14.00–28.10) | 20.30 (15.60–26.80) | 49.80 (29.20–75.00) | p (2–4): 0.025 p (3–4): 0.007 |
| ALT (IU/L) | Normal range | 19.90 (14.20–25.10) | 21.50 (14.40–33.70) | 65.70 (38.00–127.00) | 0.235 |
| AFP (ng/ml) | Normal range | 1.82 (1.25–2.46) | 2.37 (1.21–3.25) | 2.14 (1.34–4.36) | 0.28 |
| CD8a (ng/mL) | 14.91 (7.70–21.54) | 26.58 (12.73–55.32) | 21.52 (14.86–36.75) | 23.05 (10.36–36.35) | p (1–2): 0.004 |
| GDF-15 (pg/mL) | 317.11 (236.87–378.58) | 792.39 (556.81–1.496.25) | 821.30 (504.99–1.407.73) | 1.848.69 (1.094.06–2.721.25) | p (1–4): <0.001 p (2–4): 0.011 |
| CXCL-16 (pg/mL) | 7.31 (5.47–10.13) | 12.22 (9.38–14.23) | 9.17 (3.99–13.56) | 8.93 (4.37–14.33) | p (1–2): 0.009 |
| CCL-20 (ng/L) | 75.77 (29.78–113.93) | 87.46 (56.43–146.68) | 195.22 (63.16–481.16) | 440.00 (362.47–515.13) | p (1–3): <0.001 p (1–4): <0.001 p (2–3): <0.001 p (2–4): <0.001 p (3–4): 0.001 |
| Platelet/Lymphocyte | Normal range | 155.90 ± 120.34 | 141.44 ± 92.26 | 95.60 ± 58.13 | p (2–4): 0.028 |
| Neutrophil/Lymphocyte | Normal range | 2.76 ± 1.95 | 2.15 ± 1.25 | 2.40 ± 2.74 | 0.34 |
| Lymphocyte/Monocyte | Normal range | 3.64 ± 1.68 | 3.97 ± 1.52 | 3.66 ± 1.83 | 0.67 |
| Platelet/Neutrophil | Normal range | 63.51 ± 33.80 | 71.67 ± 33.53 | 55.16 ± 29.47 | 0.12 |
| MPV/Platelet | Normal range | 0.04 ± 0.01 | 0.05 ± 0.02 | 0.16 ± 0.51 | 0.39 |
| PDW/Platelet | Normal range | 0.05 ± 0.02 | 0.06 ± 0.03 | 0.18 ± 0.61 | 0.44 |
| PlateletxNeutrophil/Lymphocyte | Normal range | 526.73 (418.06–772.39) | 405.15 (277.50–763.51) | 380.52 (177.52–546.00) | 0.17 |
| NeutrophilxMonocyte/Lymphocyte | Normal range | 1.25(0.78–1.87) | 0.96(0.54–1.88) | 0.88(0.59–1.96) | 0.12 |
| AST/ALT | Normal range | 1.19 ± 0.60 | 1.02 ± 0.35 | 0.84 ± 0.45 | p (2–4): 0.011 |
| Age/Platelet | Normal range | 0.20 (0.15–0.27) | 0.20 (0.14–0.28) | 0.23 (0.12–0.41) | 0.20 |
| AST/Platelet | Normal range | 0.09 (0.06–0.11) | 0.09 (0.05–0.14) | 0.23 (0.11–0.72) | 0.13 |
| (AST/ALT)/Platelet | Normal range | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.02 ± 0.07 | 0.40 |
| GGT/Platelet | Normal range | 0.08 (0.06–0.13) | 0.10 (0.06–0.19) | 0.24 (0.07–0.54) | 0.10 |
| AST/GGT | Normal range | 1.02 (0.81–1.58) | 0.92 (0.64–1.34) | 1.37 (0.58–2.69) | 0.092 |
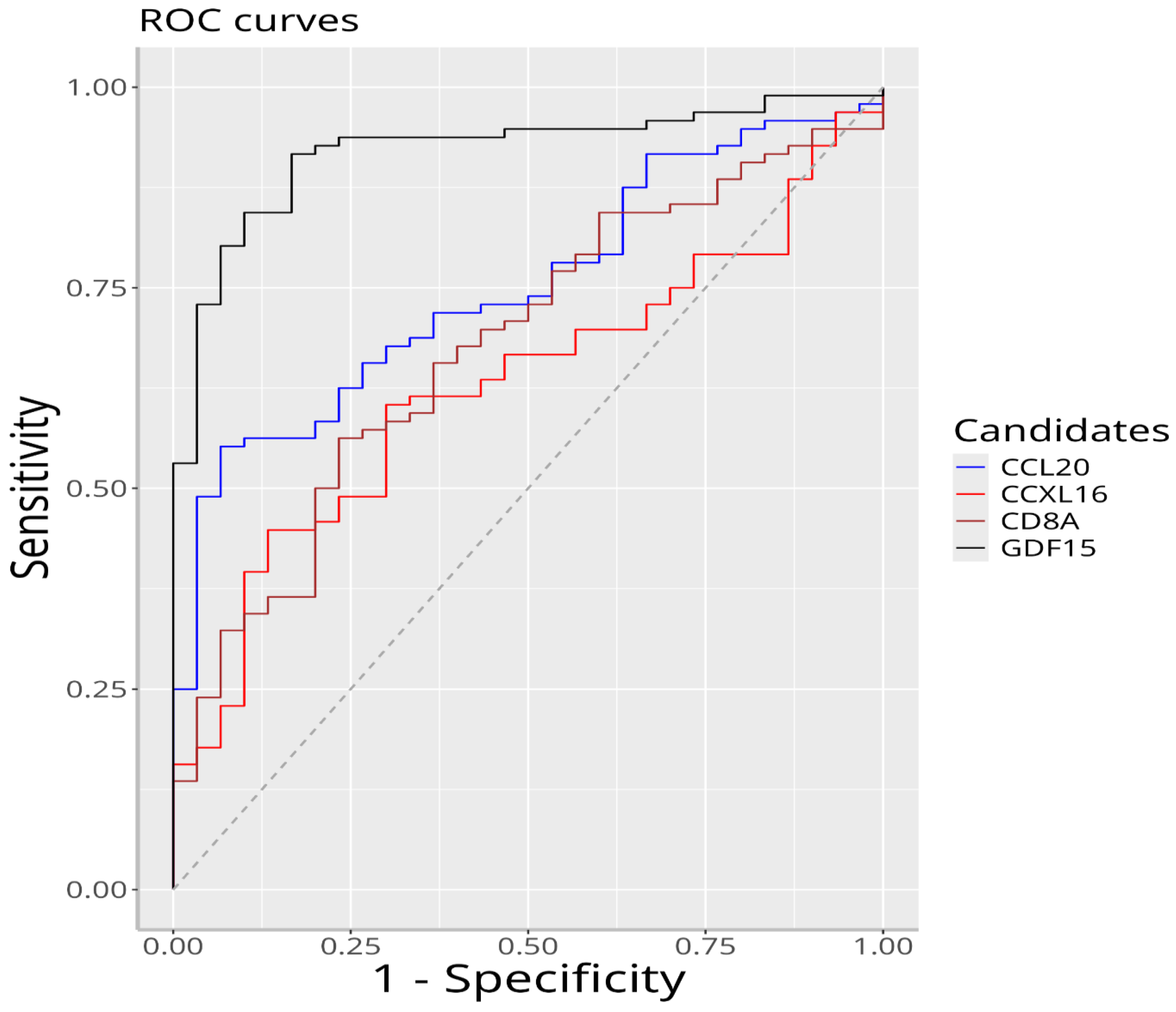
| Biomarker | Optimum Cutting Value | AUC | Sensitivity | Specificity |
|---|---|---|---|---|
| GDF-15 | 391.908 | 0.920 | 0.917 | 0.833 |
| CCL-20 | 183.946 | 0.751 | 0.552 | 0.933 |
| CD8a | 21.953 | 0.678 | 0.562 | 0.767 |
| CXCL-16 | 11.250 | 0.629 | 0.448 | 0.867 |
| Variable | (PlateletxNeutrophil)/Lymphocyte | Neutrophil × Monocyte/Lymphocyte | Neutrophil/Lymphocyte | AST/Platelet | GGT/Platelet |
|---|---|---|---|---|---|
| CCL-20 | −0.22 * | −0.02 | −0.2 * | 0.24 * | 0.18 |
| CD8a | 0.23 * | 0.23 * | 0.15 | −0.12 | 0.06 |
| GDF-15 | 0.01 | 0.19 | 0.09 | 0.31 ** | 0.23 * |
| CXCL-16 | 0.22 * | 0.41 ** | 0.26 ** | −0.01 | 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ezer, B.; Esen, H.S.; Ugrakli, S.; Iyisoy, M.S.; Ozdemir, M. Association Between Serum HBV DNA Levels and CCL-20, CD8a, CXCL-16, and GDF-15 in Patients with Chronic Hepatitis B. Viruses 2025, 17, 1352. https://doi.org/10.3390/v17101352
Ezer B, Esen HS, Ugrakli S, Iyisoy MS, Ozdemir M. Association Between Serum HBV DNA Levels and CCL-20, CD8a, CXCL-16, and GDF-15 in Patients with Chronic Hepatitis B. Viruses. 2025; 17(10):1352. https://doi.org/10.3390/v17101352
Chicago/Turabian StyleEzer, Burak, Hilal Sena Esen, Selin Ugrakli, Mehmet Sinan Iyisoy, and Mehmet Ozdemir. 2025. "Association Between Serum HBV DNA Levels and CCL-20, CD8a, CXCL-16, and GDF-15 in Patients with Chronic Hepatitis B" Viruses 17, no. 10: 1352. https://doi.org/10.3390/v17101352
APA StyleEzer, B., Esen, H. S., Ugrakli, S., Iyisoy, M. S., & Ozdemir, M. (2025). Association Between Serum HBV DNA Levels and CCL-20, CD8a, CXCL-16, and GDF-15 in Patients with Chronic Hepatitis B. Viruses, 17(10), 1352. https://doi.org/10.3390/v17101352







