Metagenomics Study of the Commercial Tomato Virome Focused on Virus Species of Epidemiological Interest
Abstract
1. Introduction
2. Materials and Methods
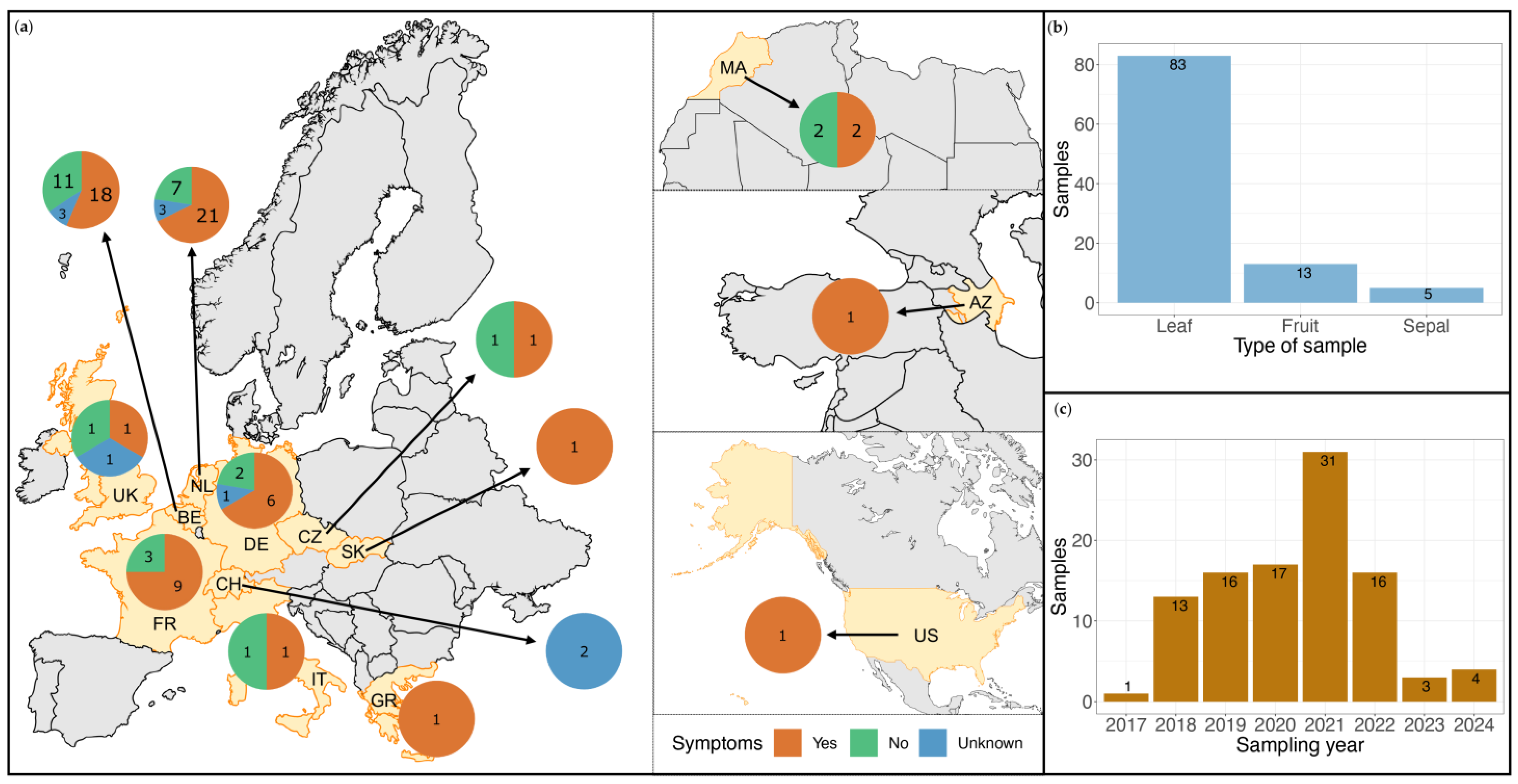
2.1. Sample Selection
2.2. Molecular Viral Detection
2.3. Sample Processing and HTS
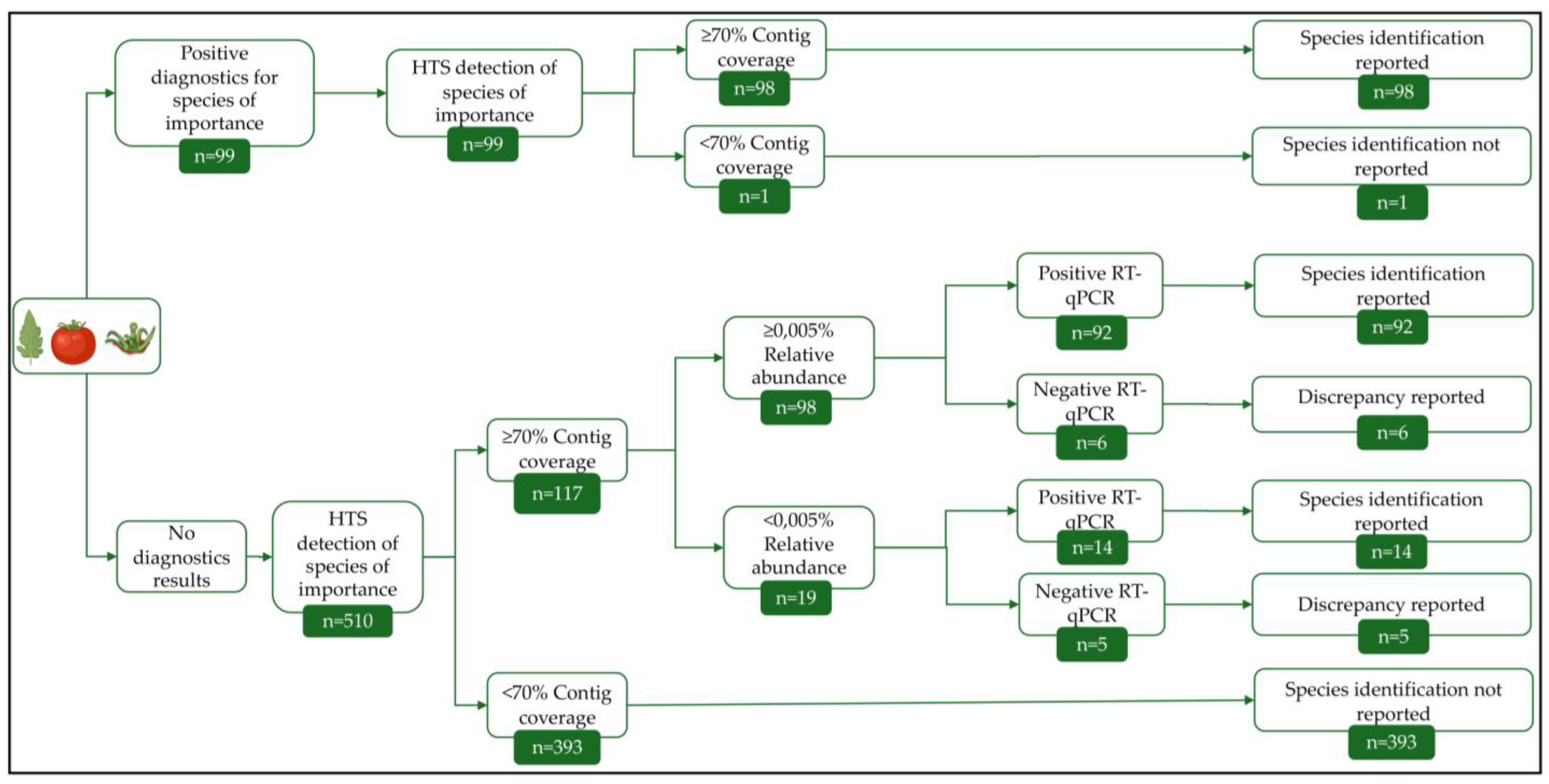
2.4. Bioinformatics Analysis
2.5. Phylogenetic Analysis
3. Results
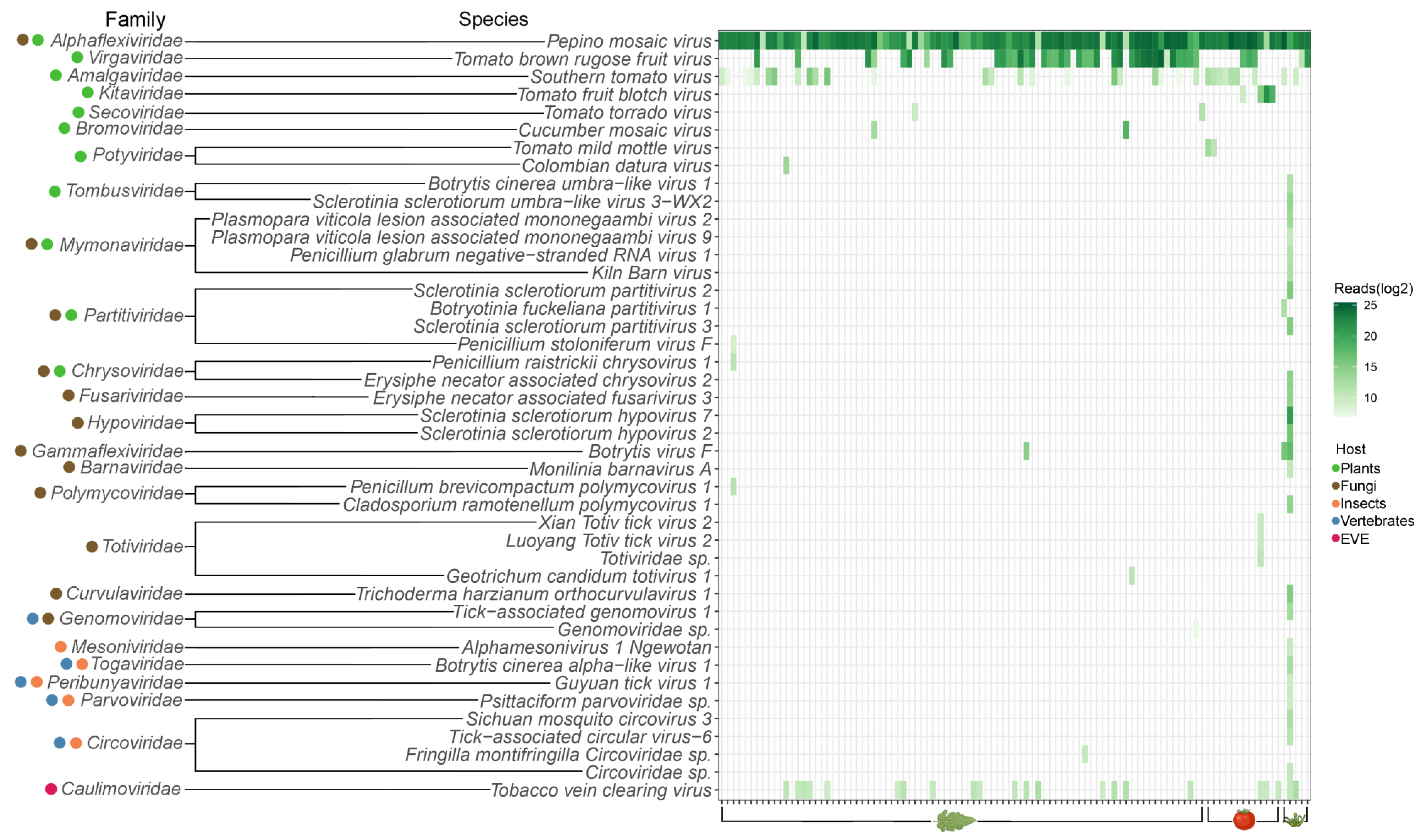
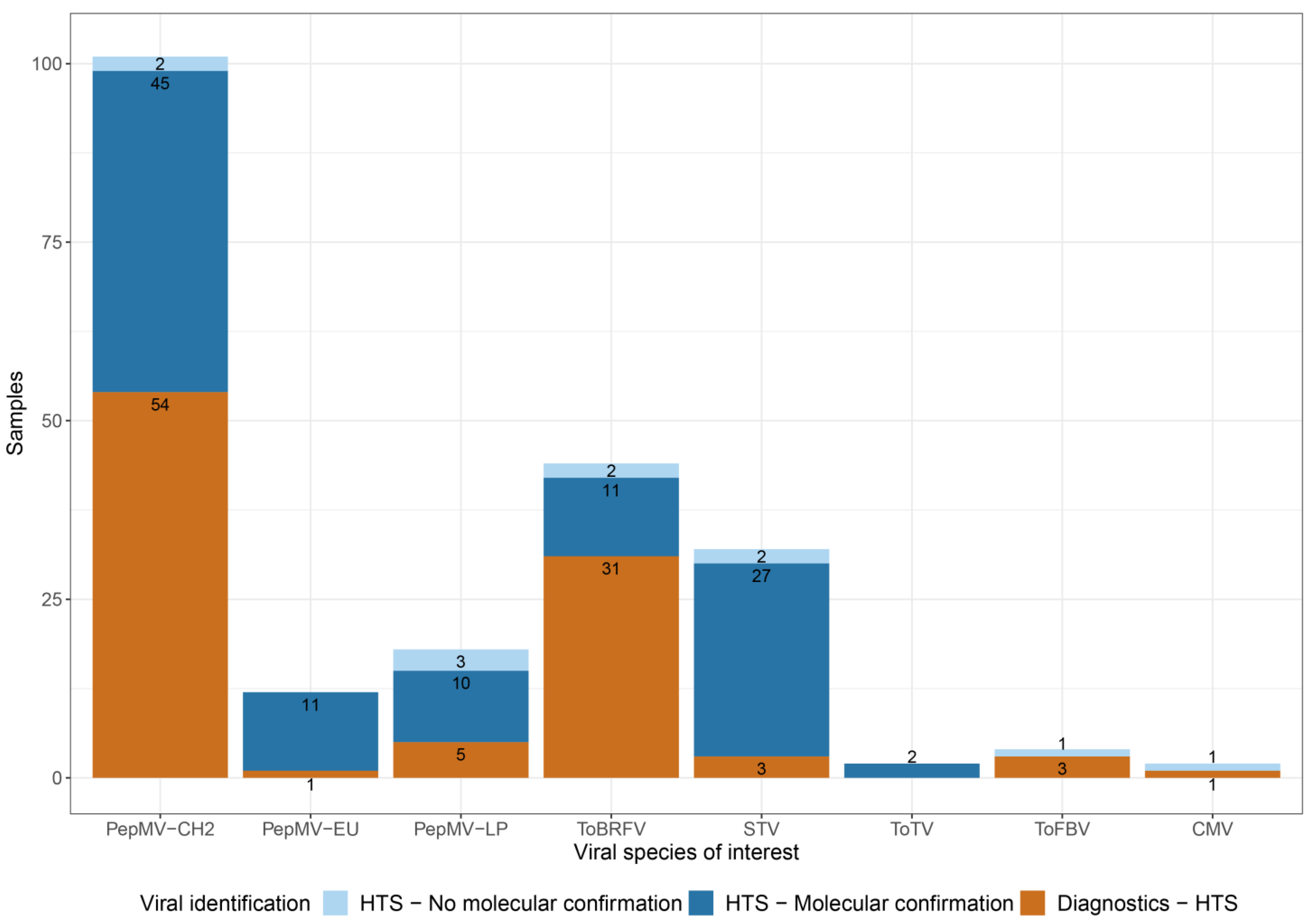
3.1. Commercial Tomato Virome Overview
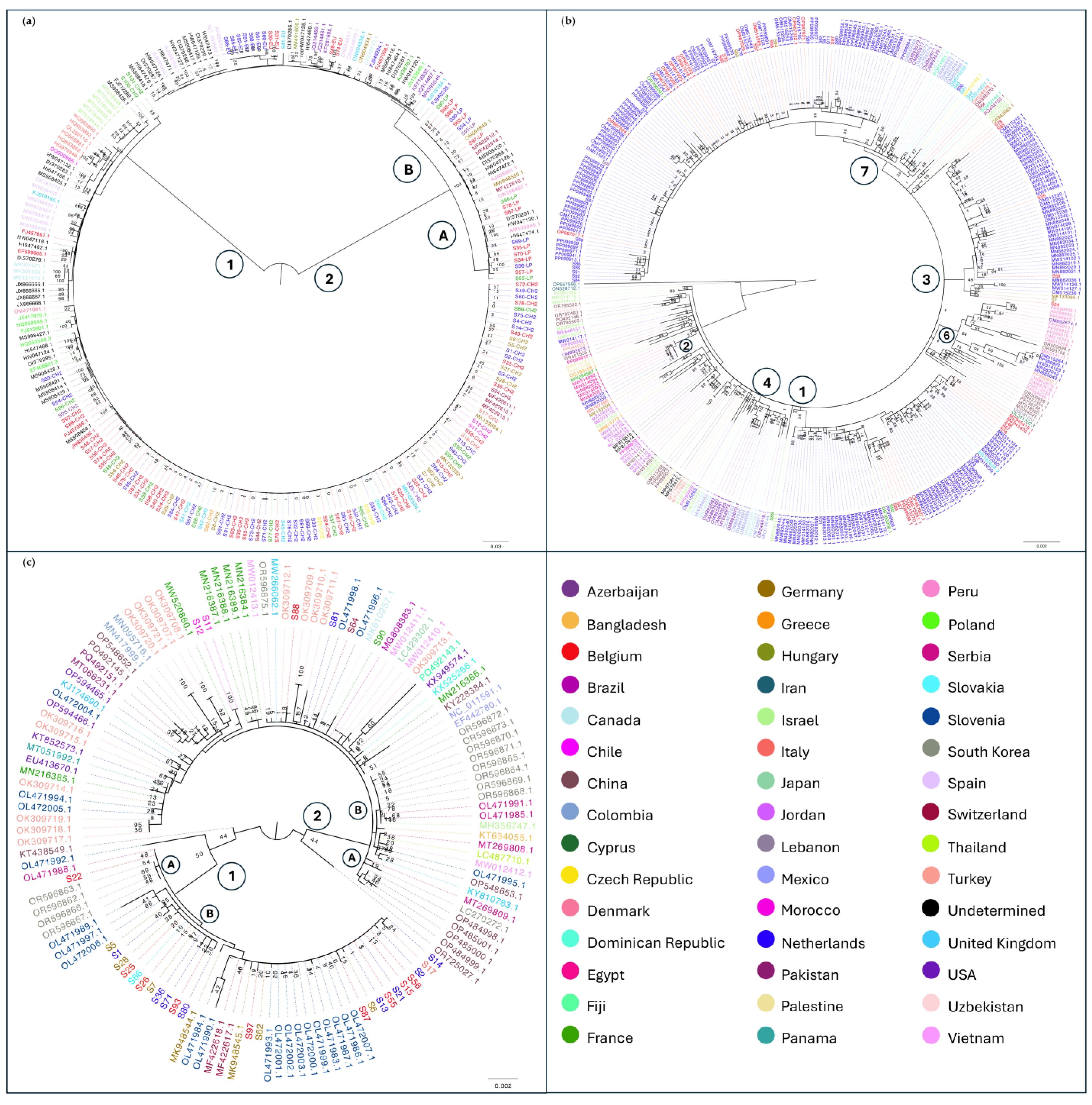
3.2. Phylogenetic Analysis of Viral Species of Interest for Tomato Cultivation
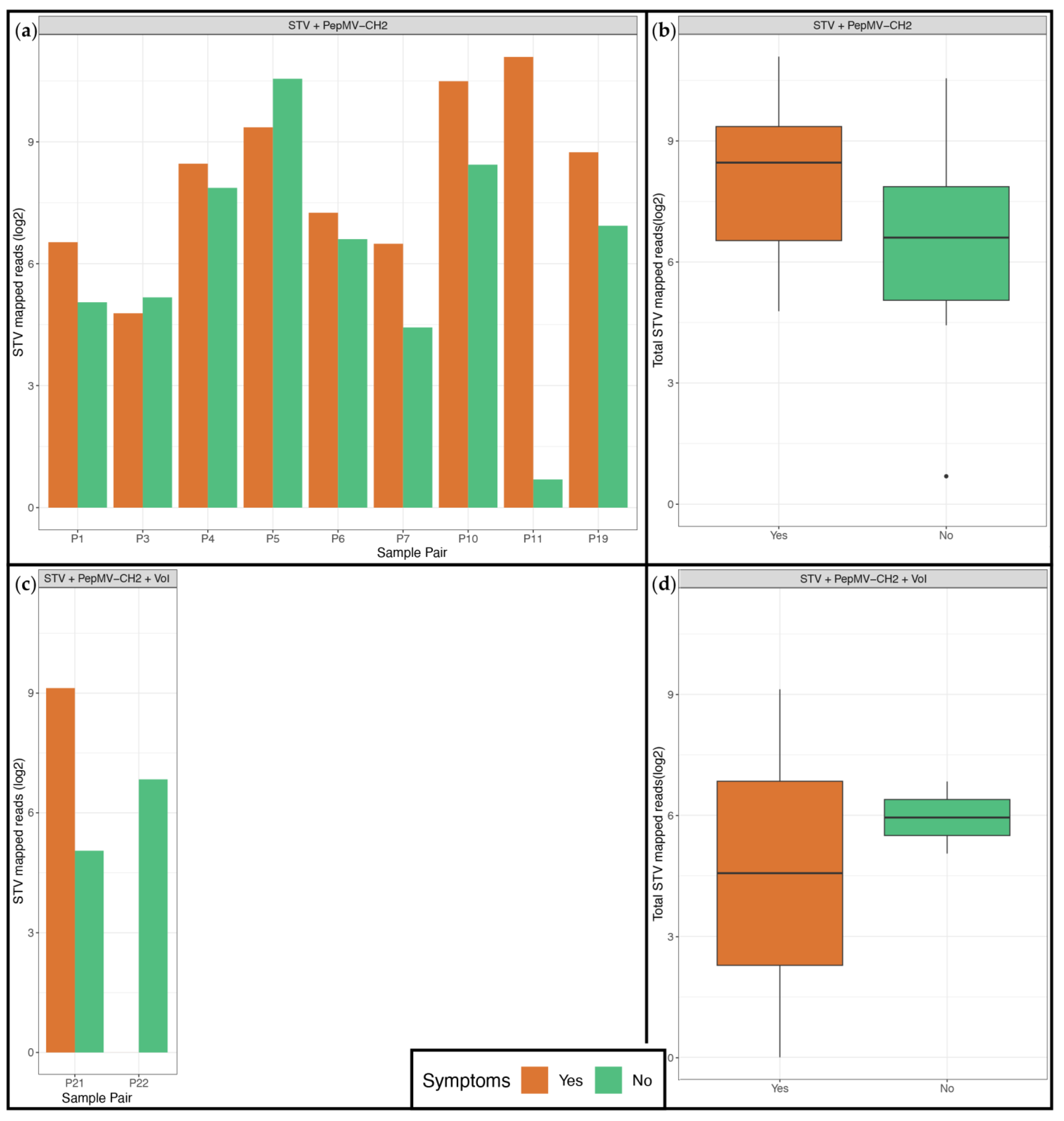
3.3. Viral Identification and Disease Etiology
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rivarez, M.P.S.; Vučurović, A.; Mehle, N.; Ravnikar, M.; Kutnjak, D. Global Advances in Tomato Virome Research: Current Status and the Impact of High-Throughput Sequencing. Front. Microbiol. 2021, 12, 671925. [Google Scholar] [CrossRef]
- Jones, R.A.C. Global Plant Virus Disease Pandemics and Epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef]
- Roossinck, M.J.; García-Arenal, F. Ecosystem Simplification, Biodiversity Loss and Plant Virus Emergence. Curr. Opin. Virol. 2015, 10, 56–62. [Google Scholar] [CrossRef]
- Dombrovsky, A.; Smith, E. Seed Transmission of Tobamoviruses: Aspects of Global Disease Distribution. In Advances in Seed Biology; IntechOpen Limited: London, UK, 2017. [Google Scholar]
- Jones, R.A.C. Future Scenarios for Plant Virus Pathogens as Climate Change Progresses. Adv. Virus Res. 2016, 95, 87–147. [Google Scholar] [PubMed]
- FAOSTAT (Food and Agriculture Organization of the United Nations). Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 4 December 2024).
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P.H.J. Emerging Viral Diseases of Tomato Crops. Mol. Plant-Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Wai, A.H.; Naing, A.H.; Lee, D.-J.; Kim, C.K.; Chung, M.-Y. Molecular Genetic Approaches for Enhancing Stress Tolerance and Fruit Quality of Tomato. Plant Biotechnol. Rep. 2020, 14, 515–537. [Google Scholar] [CrossRef]
- Shahriari, Z.; Su, X.; Zheng, K.; Zhang, Z. Advances and Prospects of Virus-Resistant Breeding in Tomatoes. Int. J. Mol. Sci. 2023, 24, 15448. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Thomma, B.P.H.J. Pepino Mosaic Virus: A Successful Pathogen That Rapidly Evolved from Emerging to Endemic in Tomato Crops. Mol. Plant Pathol. 2010, 11, 179–189. [Google Scholar] [CrossRef]
- van der Vlugt, R.A.A.; Stijger, C.C.M.M.; Verhoeven, J.T.J.; Lesemann, D.-E. First Report of Pepino Mosaic Virus on Tomato. Plant Dis. 2000, 84, 103. [Google Scholar] [CrossRef]
- Moreno-Pérez, M.G.; Pagán, I.; Aragón-Caballero, L.; Cáceres, F.; Fraile, A.; García-Arenal, F. Ecological and Genetic Determinants of Pepino Mosaic Virus Emergence. J. Virol. 2014, 88, 3359–3368. [Google Scholar] [CrossRef]
- EPPO Global Database. Available online: https://gd.eppo.int/taxon/PEPMV0/distribution (accessed on 4 December 2024).
- Spence, N.J.; Basham, J.; Mumford, R.A.; Hayman, G.; Edmondson, R.; Jones, D.R. Effect of Pepino Mosaic Virus on the Yield and Quality of Glasshouse-Grown Tomatoes in the UK. Plant Pathol. 2006, 55, 595–606. [Google Scholar] [CrossRef]
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I.; Elad, N.; Tam, Y.; Sela, N.; Abu-Ras, A.; et al. A New Israeli Tobamovirus Isolate Infects Tomato Plants Harboring Tm-22 Resistance Genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef]
- Avni, B.; Gelbart, D.; Sufrin-Ringwald, T.; Zinger, A.; Chen, L.; MacHbash, Z.; Bekelman, I.; Segoli, M.; Dombrovsky, A.; Kamenetsky, R.; et al. Tomato Genetic Resistance to Tobamoviruses Is Compromised. Acta Hortic. 2021, 1316, 89–98. [Google Scholar] [CrossRef]
- Prasad, A.; Sharma, N.; Hari-Gowthem, G.; Muthamilarasan, M.; Prasad, M. Tomato Yellow Leaf Curl Virus: Impact, Challenges, and Management. Trends Plant Sci. 2020, 25, 897–911. [Google Scholar] [CrossRef]
- Zaidi, S.S.-E.-A.; Martin, D.P.; Amin, I.; Farooq, M.; Mansoor, S. Tomato Leaf Curl New Delhi Virus: A Widespread Bipartite Begomovirus in the Territory of Monopartite Begomoviruses. Mol. Plant Pathol. 2017, 18, 901–911. [Google Scholar] [CrossRef]
- Qi, S.; Zhang, S.; Islam, M.M.; El-Sappah, A.H.; Zhang, F.; Liang, Y. Natural Resources Resistance to Tomato Spotted Wilt Virus (TSWV) in Tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2021, 22, 10978. [Google Scholar] [CrossRef] [PubMed]
- Kanapiya, A.; Amanbayeva, U.; Tulegenova, Z.; Abash, A.; Zhangazin, S.; Dyussembayev, K.; Mukiyanova, G. Recent Advances and Challenges in Plant Viral Diagnostics. Front. Plant Sci. 2024, 15, 1451790. [Google Scholar] [CrossRef] [PubMed]
- González-Pérez, E.; Chiquito-Almanza, E.; Villalobos-Reyes, S.; Canul-Ku, J.; Anaya-López, J.L. Diagnosis and Characterization of Plant Viruses Using HTS to Support Virus Management and Tomato Breeding. Viruses 2024, 16, 888. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant Virus Metagenomics: Advances in Virus Discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef]
- Bassi, C.; Guerriero, P.; Pierantoni, M.; Callegari, E.; Sabbioni, S. Novel Virus Identification through Metagenomics: A Systematic Review. Life 2022, 12, 2048. [Google Scholar] [CrossRef]
- Adams, I.; Fox, A. Diagnosis of Plant Viruses Using Next-Generation Sequencing and Metagenomic Analysis. In Current Research Topics in Plant Virology; Wang, A., Zhou, X., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 323–335. ISBN 978-3-319-32919-2. [Google Scholar]
- Conceição-Neto, N.; Zeller, M.; Lefrère, H.; De Bruyn, P.; Beller, L.; Deboutte, W.; Yinda, C.K.; Lavigne, R.; Maes, P.; Ranst, M.V.; et al. Modular Approach to Customise Sample Preparation Procedures for Viral Metagenomics: A Reproducible Protocol for Virome Analysis. Sci. Rep. 2015, 5, 16532. [Google Scholar] [CrossRef]
- De Coninck, L. ViPER. Zenodo. 2021.
- Gutiérrez-Aguirre, I.; Mehle, N.; Delić, D.; Gruden, K.; Mumford, R.; Ravnikar, M. Real-Time Quantitative PCR Based Sensitive Detection and Genotype Discrimination of Pepino Mosaic Virus. J. Virol. Methods 2009, 162, 46–55. [Google Scholar] [CrossRef] [PubMed]
- ISF-ISHI-Veg. Detection of Infectious Tomato Brown Rugose Fruit Virus (ToBRFV) in Tomato and Pepper Seeds; International Seed Federation: Nyon, Switzerland, 2020. [Google Scholar]
- Elvira González, L.; Peiró, R.; Rubio, L.; Galipienso, L. Persistent Southern Tomato Virus (STV) Interacts with Cucumber Mosaic and/or Pepino Mosaic Virus in Mixed- Infections Modifying Plant Symptoms, Viral Titer and Small RNA Accumulation. Microorganisms 2021, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Vásquez, J.A.; Rubio, L.; Alfaro-Fernández, A.; Debreczeni, D.E.; Font-San-Ambrosio, I.; Falk, B.W.; Ferriol, I. Detection and Absolute Quantitation of Tomato Torrado Virus (ToTV) by Real Time RT-PCR. J. Virol. Methods 2015, 221, 90–94. [Google Scholar] [CrossRef]
- Maachi, A.; Torre, C.; Sempere, R.N.; Hernando, Y.; Aranda, M.A.; Donaire, L. Use of High-Throughput Sequencing and Two RNA Input Methods to Identify Viruses Infecting Tomato Crops. Microorganisms 2021, 9, 1043. [Google Scholar] [CrossRef]
- Bertolini, E.; Olmos, A.; López, M.M.; Cambra, M. Multiplex Nested Reverse Transcription-Polymerase Chain Reaction in a Single Tube for Sensitive and Simultaneous Detection of Four RNA Viruses and Pseudomonas savastanoi pv. savastanoi in Olive Trees. Phytopathology 2003, 93, 286–292. [Google Scholar] [CrossRef]
- Feng, J.-L.; Chen, S.-N.; Tang, X.-S.; Ding, X.-F.; Du, Z.-Y.; Chen, J.-S.; Xu, M.-H. Quantitative Determination of Cucumber Mosaic Virus Genome RNAs in Virions by Real-Time Reverse Transcription-Polymerase Chain Reaction. Acta Biochim. Biophys. Sin. 2006, 38, 669–676. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Paeleman, A.; Wittemans, L.; Goen, K.; Lievens, B.; Bragard, C.; Vanachter, A.C.R.C.; Thomma, B.P.H.J. Genetic Characterization of Pepino Mosaic Virus Isolates from Belgian Greenhouse Tomatoes Reveals Genetic Recombination. Eur. J. Plant Pathol. 2008, 121, 131–146. [Google Scholar] [CrossRef]
- Quek, S.; Hadermann, A.; Wu, Y.; De Coninck, L.; Hegde, S.; Boucher, J.R.; Cresswell, J.; Foreman, E.; Steven, A.; LaCourse, E.J.; et al. Diverse RNA Viruses of Parasitic Nematodes Can Elicit Antibody Responses in Vertebrate Hosts. Nat. Microbiol. 2024, 9, 2488–2505. [Google Scholar] [CrossRef]
- Mbigha Donfack, K.C.; De Coninck, L.; Ghogomu, S.M.; Matthijnssens, J. Aedes Mosquito Virome in Southwestern Cameroon: Lack of Core Virome, But a Very Rich and Diverse Virome in Ae. Africanus Compared to Other Aedes Species. Viruses 2024, 16, 1172. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-Metagenomes from Highly Chimeric Reads. In Research in Computational Molecular Biology, Proceedings of the 17th Annual International Conference, RECOMB 2013, Beijing, China, 7–10 April 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 158–170. [Google Scholar]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Camargo, A.P.; Schulz, F.; Eloe-Fadrosh, E.; Roux, S.; Kyrpides, N.C. CheckV Assesses the Quality and Completeness of Metagenome-Assembled Viral Genomes. Nat. Biotechnol. 2021, 39, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive Metagenomic Visualization in a Web Browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- De Castro, E.; Hulo, C.; Masson, P.; Auchincloss, A.; Bridge, A.; Le Mercier, P. ViralZone 2024 Provides Higher-Resolution Images and Advanced Virus-Specific Resources. Nucleic Acids Res. 2024, 52, D817–D821. [Google Scholar] [CrossRef]
- Diop, S.I.; Geering, A.D.W.; Alfama-Depauw, F.; Loaec, M.; Teycheney, P.-Y.; Maumus, F. Tracheophyte Genomes Keep Track of the Deep Evolution of the Caulimoviridae. Sci. Rep. 2018, 8, 572. [Google Scholar] [CrossRef]
- van de Vossenberg, B.T.L.H.; Visser, M.; Bruinsma, M.; Koenraadt, H.M.S.; Westenberg, M.; Botermans, M. Real-Time Tracking of Tomato Brown Rugose Fruit Virus (ToBRFV) Outbreaks in the Netherlands Using Nextstrain. PLoS ONE 2020, 15, e0234671. [Google Scholar] [CrossRef] [PubMed]
- de Koning, P.P.M.; Fowkes, A.R.; Zisi, Z.; Beris, D.; Oplaat, C.; McGreig, S.; Skelton, A.; Adams, I.P.; van Gemert, J.; de Krom, C.; et al. Tomato Brown Rugose Fruit Virus Nextstrain Build Version 4: Pathways of Introduction and Local Spread. PhytoFrontiersTM. [CrossRef]
- Zisi, Z.; Ghijselings, L.; Vogel, E.; Vos, C.; Matthijnssens, J. Single Amino Acid Change in Tomato Brown Rugose Fruit Virus Breaks Virus-Specific Resistance in New Resistant Tomato Cultivar. Front. Plant Sci. 2024, 15, 1382862. [Google Scholar] [CrossRef] [PubMed]
- van der Vlugt, R.A.A.; Verbeek, M.; Dullemans, A.M.; Wintermantel, W.M.; Cuellar, W.J.; Fox, A.; Thompson, J.R. Torradoviruses. Annu. Rev. Phytopathol. 2015, 53, 485–512. [Google Scholar] [CrossRef]
- Anikina, I.; Kamarova, A.; Issayeva, K.; Issakhanova, S.; Mustafayeva, N.; Insebayeva, M.; Mukhamedzhanova, A.; Khan, S.M.; Ahmad, Z.; Lho, L.H.; et al. Plant Protection from Virus: A Review of Different Approaches. Front. Plant Sci. 2023, 14, 1163270. [Google Scholar] [CrossRef]
- Giesbers, A.K.J.; Vogel, E.; Skelton, A.; Zisi, Z.; Wildhagen, M.; Loh, Y.L.; Ghijselings, L.; Groothuismink, J.; Westenberg, M.; Matthijnssens, J.; et al. Detection of Tomato Brown Rugose Fruit Virus in Environmental Residues: The Importance of Contextualizing Test Results. Plant Pathol. 2024, 73, 2071–2083. [Google Scholar] [CrossRef]
- Skelton, A.; van Gemert, J.; Fowkes, A.; Frew, L.; Alraiss, K.; Hodgson, R.; Cressey, J.; Barnhoorn, R.; Macarthur, R.; Stijger, I.; et al. Detection of Tomato Brown Rugose Fruit Virus Is Influenced by Infection at Different Growth Stages and Sampling from Different Plant Parts. Plant Pathol. 2023, 72, 1491–1504. [Google Scholar] [CrossRef]
- EPPO Global Database. Available online: https://gd.eppo.int/taxon/TOBRFV/distribution (accessed on 4 December 2024).
- Hanssen, I.M.; Gutiérrez-Aguirre, I.; Paeleman, A.; Goen, K.; Wittemans, L.; Lievens, B.; Vanachter, A.C.R.C.; Ravnikar, M.; Thomma, B.P.H.J. Cross-Protection or Enhanced Symptom Display in Greenhouse Tomato Co-Infected with Different Pepino Mosaic Virus Isolates. Plant Pathol. 2010, 59, 13–21. [Google Scholar] [CrossRef]
- Pagán, I.; del Carmen Córdoba-Sellés, M.; Martínez-Priego, L.; Fraile, A.; Malpica, J.M.; Jordá, C.; García-Arenal, F. Genetic Structure of the Population of Pepino Mosaic Virus Infecting Tomato Crops in Spain. Phytopathology 2006, 96, 274–279. [Google Scholar] [CrossRef]
- Botermans, M.; de Koning, P.P.M.; Oplaat, C.; Fowkes, A.R.; McGreig, S.; Skelton, A.; Adams, I.P.; Fox, A.; De Jonghe, K.; Demers, J.E.; et al. Tomato Brown Rugose Fruit Virus Nextstrain Build Version 3: Rise of a Novel Clade. PhytoFrontiers 2022, 3, 442–446. [Google Scholar] [CrossRef]
- Randa-Zelyüt, F.; Fox, A.; Karanfil, A. Population Genetic Dynamics of Southern Tomato Virus from Turkey. Journal of Plant Pathology 2023, 105, 211–224. [Google Scholar] [CrossRef]
- Sabanadzovic, S.; Valverde, R.A.; Brown, J.K.; Martin, R.R.; Tzanetakis, I.E. Southern Tomato Virus: The Link between the Families Totiviridae and Partitiviridae. Virus Res. 2009, 140, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Gómez, P.; Sempere, R.N.; Aranda, M.A. Chapter 14—Pepino Mosaic Virus and Tomato Torrado Virus: Two Emerging Viruses Affecting Tomato Crops in the Mediterranean Basin. In Advances in Virus Research; Loebenstein, G., Lecoq, H., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 84, pp. 505–532. ISBN 0065-3527. [Google Scholar]
- Pospieszny, H.; Borodynko, N.; Obrępalska-Stęplowska, A.; Hasiów, B. The First Report of Tomato Torrado Virus in Poland. Plant Dis. 2007, 91, 1364. [Google Scholar] [CrossRef] [PubMed]
- Verdin, E.; Gognalons, P.; Wipf-Scheibel, C.; Bornard, I.; Ridray, G.; Schoen, L.; Lecoq, H. First Report of Tomato Torrado Virus in Tomato Crops in France. Plant Dis. 2009, 93, 1352. [Google Scholar] [CrossRef]
- Alfaro-Fernández, A.; Bese, G.; Córdoba-Sellés, C.; Cebrián, M.C.; Herrera-Vásquez, J.A.; Forray, A.; Jordá, C. First Report of Tomato Torrado Virus Infecting Tomato in Hungary. Plant Dis. 2009, 93, 554. [Google Scholar] [CrossRef]
- Davino, S.; Bivona, L.; Iacono, G.; Davino, M. First Report of Tomato Torrado Virus Infecting Tomato in Italy. Plant Dis. 2010, 94, 1172. [Google Scholar] [CrossRef]
- Alfaro-Fernández, A.; Córdoba-Sellés, M.D.C.; Juárez, M.; Herrera-Vásquez, J.; Sánchez-Navarro, J.; Cebrián, M.D.C.; Font, M.I.; Jordá, C. Occurrence and Geographical Distribution of the ‘Torrado’ Disease in Spain. J. Phytopathol. 2010, 158, 457–469. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Lapidot, M. Chapter 2—Major Tomato Viruses in the Mediterranean Basin. In Advances in Virus Research; Loebenstein, G., Lecoq, H., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 84, pp. 31–66. ISBN 0065-3527. [Google Scholar]
- EPPO Global Database. Available online: https://gd.eppo.int/taxon/TOTV00/distribution (accessed on 4 December 2024).
- Salem, N.M.; Jewehan, A.; Aranda, M.A.; Fox, A. Tomato Brown Rugose Fruit Virus Pandemic. Annu. Rev. Phytopathol. 2023, 61, 137–164. [Google Scholar] [CrossRef]
- Moodley, V.; Gubba, A.; Mafongoya, P.L. Emergence and Full Genome Analysis of Tomato Torrado Virus in South Africa. Viruses 2020, 12, 1167. [Google Scholar] [CrossRef] [PubMed]
- Inoue-Nagata, A.K.; Jordan, R.; Kreuze, J.; Li, F.; López-Moya, J.J.; Mäkinen, K.; Ohshima, K.; Wylie, S.J.; Consortium, I.R. ICTV Virus Taxonomy Profile: Potyviridae 2022. J. Gen. Virol. 2022, 103, 1738. [Google Scholar] [CrossRef] [PubMed]
- Walkey, D.G.A.; Spence, N.J.; Clay, C.M.; Miller, A. A Potyvirus Isolated from Solanaceous Hosts. Plant Pathol. 1994, 43, 931–937. [Google Scholar] [CrossRef]
- Luigi, M.; Tiberini, A.; Taglienti, A.; Bertin, S.; Dragone, I.; Sybilska, A.; Tarchi, F.; Goggioli, D.; Lewandowski, M.; Simoni, S.; et al. Molecular Methods for the Simultaneous Detection of Tomato Fruit Blotch Virus and Identification of Tomato Russet Mite, a New Potential Virus–Vector System Threatening Solanaceous Crops Worldwide. Viruses 2024, 16, 806. [Google Scholar] [CrossRef]
- Nakasu, E.Y.T.; Nagata, T.; Inoue-Nagata, A.K. First Report of Tomato Fruit Blotch Virus Infecting Tomatoes in Brazil. Plant Dis. 2022, 106, 2271. [Google Scholar] [CrossRef]
- Beris, D.; Galeou, A.; Kektsidou, O.; Varveri, C. First Report of Tomato Fruit Blotch Fruit Virus Infecting Tomato in Greece. New Dis. Rep. 2023, 48, e12219. [Google Scholar] [CrossRef]
- Duarte, M.E.; Lewandowski, M.; de Mendonça, R.S.; Simoni, S.; Navia, D. Genetic Analysis of the Tomato Russet Mite Provides Evidence of Oligophagy and a Widespread Pestiferous Haplotype. Exp. Appl. Acarol. 2023, 89, 171–199. [Google Scholar] [CrossRef]
- Perring, T.; Farrar, C. Historical Perspective and Current World Status of the Tomato Russet Mite (Acari: Eriophyidae); Entomological Society of America: College Park, MD, USA, 1986; Volume 63, pp. 1–19. [Google Scholar]
- Mochizuki, T.; Ohki, S.T. Cucumber Mosaic Virus: Viral Genes as Virulence Determinants. Mol. Plant Pathol. 2012, 13, 217–225. [Google Scholar] [CrossRef]
- Palukaitis, P.; Roossinck, M.J.; Dietzgen, R.G.; Francki, R.I.B. Cucumber Mosaic Virus. In Advances in Virus Research; Maramorosch, K., Murphy, F.A., Shatkin, A.J., Eds.; Academic Press: Cambridge, MA, USA, 1992; Volume 41, pp. 281–348. ISBN 0065-3527. [Google Scholar]
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathology 2022, 113, 117–141. [Google Scholar] [CrossRef]
- Pringle, C.R. The Universal System of Virus Taxonomy of the International Committee on Virus Taxonomy (ICTV), Including New Proposals Ratified since Publication of the Sixth ICTV Report in 1995. Arch. Virol. 1998, 143, 203–210. [Google Scholar] [CrossRef]
- Lockhart, B.E.; Menke, J.; Dahal, G.; Olszewski, N.E. Characterization and Genomic Analysis of Tobacco Vein Clearing Virus, a Plant Pararetrovirus That Is Transmitted Vertically and Related to Sequences Integrated in the Host Genome. J. Gen. Virol. 2000, 81, 1579–1585. [Google Scholar] [CrossRef]
- Abass, M.; Lahuf, A. High-Throughput Sequencing and Bioinformatic Analysis Reveal Presence of the Endogenous Pararetrovirus Tobacco Vein Clearing Virus Genome in the Tomato (Solanum lycopersicum) Host Genome. Arab. J. Plant Prot. 2023, 41, 77–84. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zisi, Z.; Ruiz Movilla, I.; Basler, N.; Close, L.; Ghijselings, L.; Van der Hoeven, R.; Papadaki, M.I.; Rabbinowitsch, E.; Van Reeth, F.; Swinnen, J.; et al. Metagenomics Study of the Commercial Tomato Virome Focused on Virus Species of Epidemiological Interest. Viruses 2025, 17, 1334. https://doi.org/10.3390/v17101334
Zisi Z, Ruiz Movilla I, Basler N, Close L, Ghijselings L, Van der Hoeven R, Papadaki MI, Rabbinowitsch E, Van Reeth F, Swinnen J, et al. Metagenomics Study of the Commercial Tomato Virome Focused on Virus Species of Epidemiological Interest. Viruses. 2025; 17(10):1334. https://doi.org/10.3390/v17101334
Chicago/Turabian StyleZisi, Zafeiro, Isabel Ruiz Movilla, Nikolas Basler, Lila Close, Lucas Ghijselings, Robby Van der Hoeven, Maria Ioanna Papadaki, Ester Rabbinowitsch, Fiona Van Reeth, Jill Swinnen, and et al. 2025. "Metagenomics Study of the Commercial Tomato Virome Focused on Virus Species of Epidemiological Interest" Viruses 17, no. 10: 1334. https://doi.org/10.3390/v17101334
APA StyleZisi, Z., Ruiz Movilla, I., Basler, N., Close, L., Ghijselings, L., Van der Hoeven, R., Papadaki, M. I., Rabbinowitsch, E., Van Reeth, F., Swinnen, J., Vogel, E., Vos, C., Hanssen, I., & Matthijnssens, J. (2025). Metagenomics Study of the Commercial Tomato Virome Focused on Virus Species of Epidemiological Interest. Viruses, 17(10), 1334. https://doi.org/10.3390/v17101334






