New Coronavirus in Colombian Caribbean Bats: In Silico Analysis Reveals Possible Risk of Interspecific Jumping
Abstract
1. Introduction
2. Materials and Methods
2.1. Capture and Ethical Aspects of the Study
2.2. Sequencing
2.3. Bioinformatics Analysis
2.4. Phylogenetic Analysis
2.5. Evolutionary Analysis
2.6. Analysis and Molecular Docking of S Protein
3. Results
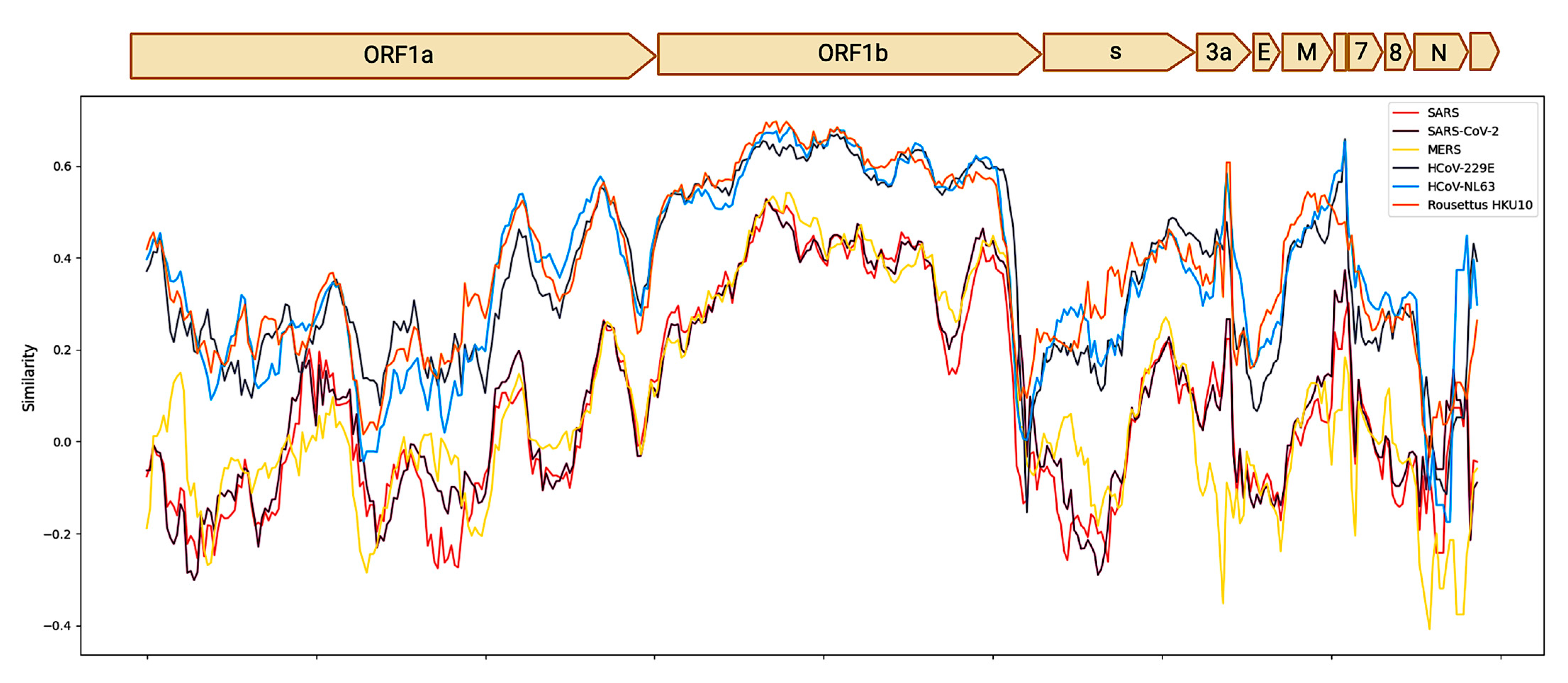
3.1. Genomic, Phylogenetic, and Evolutionary Analyses
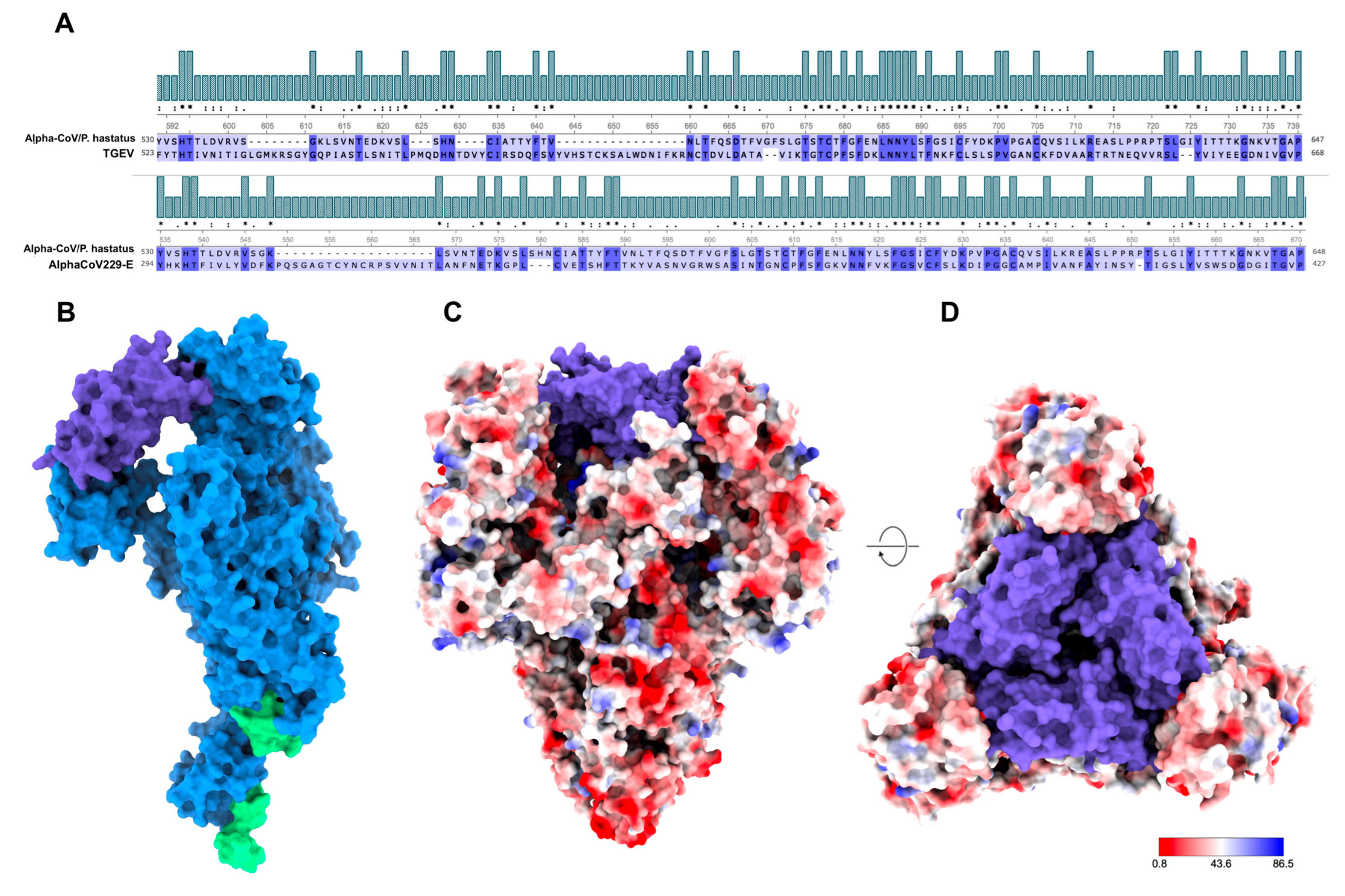
3.2. Antigenic Modeling of the Spike (S) Protein of AlphaCoV/P. hastatus
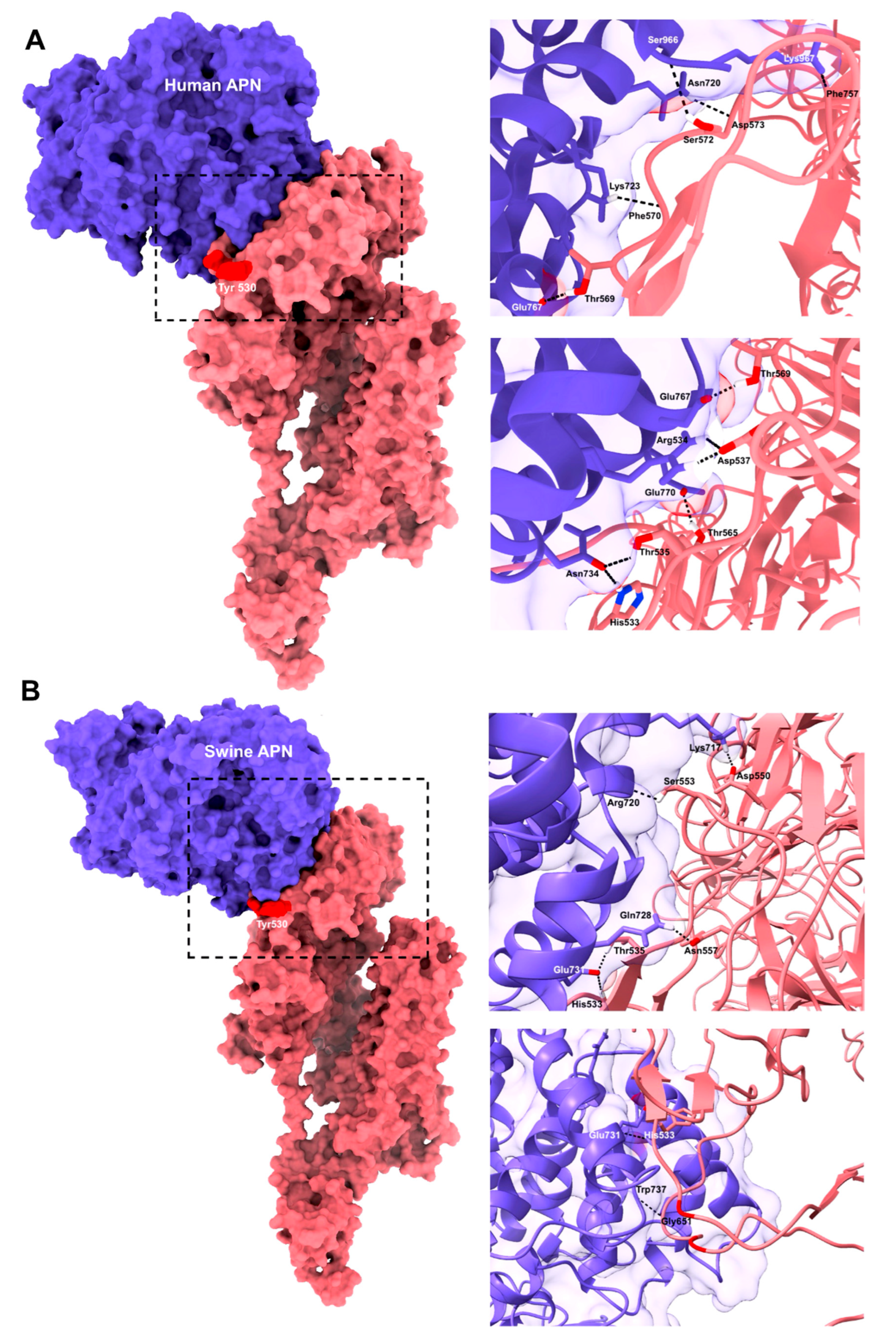
3.3. Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CoV | Coronavirus |
| TGEV | Transmissible gastroenteritis virus |
| NGS | Next generation sequencing |
| RBD | Receptor-binding domain |
| HCoV-229E | Human coronavirus 229E |
| S | Spike protein |
| ANLA | National Environmental Licensing Authority |
References
- Banerjee, A.; Kulcsar, K.; Misra, V.; Frieman, M.; Mossman, K. Bats and Coronaviruses. Viruses 2019, 11, 41. [Google Scholar] [CrossRef]
- Wong, A.C.P.; Lau, S.K.P.; Woo, P.C.Y. Interspecies Jumping of Bat Coronaviruses. Viruses 2021, 13, 2188. [Google Scholar] [CrossRef]
- Letko, M.; Seifert, S.N.; Olival, K.J.; Plowright, R.K.; Munster, V.J. Bat-Borne Virus Diversity, Spillover and Emergence. Nat. Rev. Microbiol. 2020, 18, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Anderson, D.E. Viruses in Bats and Potential Spillover to Animals and Humans. Curr. Opin. Virol. 2019, 34, 79–89. [Google Scholar] [CrossRef]
- Ruiz-Aravena, M.; McKee, C.; Gamble, A.; Lunn, T.; Morris, A.; Snedden, C.E.; Yinda, C.K.; Port, J.R.; Buchholz, D.W.; Yeo, Y.Y.; et al. Ecology, Evolution and Spillover of Coronaviruses from Bats. Nat. Rev. Microbiol. 2021, 20, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Bonilla-Aldana, D.K.; Jimenez-Diaz, S.D.; Arango-Duque, J.S.; Aguirre-Florez, M.; Balbin-Ramon, G.J.; Paniz-Mondolfi, A.; Suárez, J.A.; Pachar, M.R.; Perez-Garcia, L.A.; Delgado-Noguera, L.A.; et al. Bats in Ecosystems and Their Wide Spectrum of Viral Infectious Potential Threats: SARS-CoV-2 and Other Emerging Viruses. Int. J. Infect. Dis. 2021, 102, 87–96. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, B.; Han, Y.; Wang, Y.; Chen, L.; Wu, Z.; Yang, J. ZOVER: The Database of Zoonotic and Vector-Borne Viruses. Nucleic Acids Res. 2022, 50, D943–D949. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Huang, Y.; Yuen, K.Y. Coronavirus Diversity, Phylogeny and Interspecies Jumping. Exp. Biol. Med. 2009, 234, 1117–1127. [Google Scholar] [CrossRef]
- Conceicao, C.; Thakur, N.; Human, S.; Kelly, J.T.; Logan, L.; Bialy, D.; Bhat, S.; Stevenson-Leggett, P.; Zagrajek, A.K.; Hollinghurst, P.; et al. The SARS-CoV-2 Spike Protein Has a Broad Tropism for Mammalian ACE2 Proteins. PLoS Biol. 2020, 18, e3001016. [Google Scholar] [CrossRef]
- Colina, S.E.; Serena, M.S.; Echeverría, M.G.; Metz, G.E. Clinical and Molecular Aspects of Veterinary Coronaviruses. Virus Res. 2021, 297, 198382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Gu, H.; Peng, J.; He, B.; Liu, Y.; Yan, X.; Feng, J.; Liu, Y. Phylogenetic Relationships and Species Composition of Host Community Influence the Transmission of Coronaviruses in Sympatric Bats. Mol. Phylogenet. Evol. 2025, 207, 108343. [Google Scholar] [CrossRef] [PubMed]
- Martínez, C.; Echeverri, D.; Calderón, A.; Hurtado, E.; Gastelbondo, B.; López, Y.; Martínez, J.; López, Y.; Botero, Y.; Guzmán, C.; et al. Bats from an Area of the Colombian Caribbean Reveal the Circulation of Alphacoronavirus. Curr. Res. Parasitol. Vector-Borne Dis. 2025, 7, 100261. [Google Scholar] [CrossRef] [PubMed]
- Díaz, M.; Solari, S.; Gregorin, R.; Aguirre, L.; Barquez, R. Clave de Identificación de Los Murciélagos Neotropicales. 2021. Available online: http://hdl.handle.net/11336/156765 (accessed on 23 September 2022).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank: Update. Nucleic Acids Res. 2004, 32, D23–D26. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Varlamov, A.; Vaskin, Y.; Efremov, I.; German Grehov, O.G.; Kandrov, D.; Rasputin, K.; Syabro, M.; et al. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. ALeaves Facilitates On-Demand Exploration of Metazoan Gene Family Trees on MAFFT Sequence Alignment Server with Enhanced Interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-Likelihood Phylodynamic Analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef] [PubMed]
- Samson, S.; Lord, É.; Makarenkov, V. SimPlot++: A Python Application for Representing Sequence Similarity and Detecting Recombination. Bioinformatics 2022, 38, 3118–3120. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting Modern Challenges in Visualization and Analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.A.; De Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and Continuing Developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful New Tools for Exploring 3D Structures of Biological Macromolecules for Basic and Applied Research and Education in Fundamental Biology, Biomedicine, Biotechnology, Bioengineering and Energy Sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 Update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- Solari, S.; Medellín, R.; Rodríguez-Herrera, B.; Tavares, V.C.; Garbino, G.; Camacho, M.A. Family Phyllostomidae: Handbook of the Mammals of the World; Wilson, D.E., Mittermeier, R.A., Eds.; Lynx Ediciones: Barcelona, Spain, 2019; Volume 9, ISBN 978-84-16728-19-0. [Google Scholar] [CrossRef]
- Cortés-Delgado, N.; Ferbans, L.J. Description of a Refuge Use by Phyllostomus Hastaus (Chiroptera: Phyllostomidae) in the Perijá Serranía, La Guajira, Colombia. Ciencia e Ingeniería. 2014. Available online: https://revistas.uniguajira.edu.co/rev/index.php/cei/article/view/e011 (accessed on 14 January 2025).
- Wilkinson, G.S.; Carter, G.; Bohn, K.M.; Caspers, B.; Chaverri, G.; Farine, D.; Günther, L.; Kerth, G.; Knörnschild, M.; Mayer, F.; et al. Kinship, Association, and Social Complexity in Bats. Behav. Ecol. Sociobiol. 2019, 73, 7. [Google Scholar] [CrossRef]
- Sampedro-Marin, A.C.; Martínez, C.M.; Mercado, A.M.; Osorio, S.C.; Otero, Y.L.; Santos, L.M. Refugios, Período Reproductivo y Composición Social de Las Poblaciones de Desmodus rotundus (Geoffroy, 1810) (Chiroptera: Phyllostomidae), En Zonas Rurales Del Departamento de Sucre, Colombia. Caldasia 2008, 30, 127–134. [Google Scholar]
- Plowright, R.K.; Peel, A.J.; Streicker, D.G.; Gilbert, A.T.; McCallum, H.; Wood, J.; Baker, M.L.; Restif, O. Transmission or Within-Host Dynamics Driving Pulses of Zoonotic Viruses in Reservoir–Host Populations. PLoS Negl. Trop. Dis. 2016, 10, e0004796. [Google Scholar] [CrossRef]
- Caraballo, D.A. Cross-Species Transmission of Bat Coronaviruses in the Americas: Contrasting Patterns between Alphacoronavirus and Betacoronavirus. Microbiol. Spectr. 2022, 10, e0141122. [Google Scholar] [CrossRef]
- Ramírez-Chaves, H.; Morales-Martínez, D.M.; Rodríguez-Posada, M.E.; Suárez-Castro, A.F. Checklist of the Mammals (Mammalia) of Colombia. Mammal. Notes 2022, 7, 253. [Google Scholar] [CrossRef]
- Bittar, C.; Machado, R.R.G.; Comelis, M.T.; Bueno, L.M.; Beguelini, M.R.; Morielle-Versute, E.; Nogueira, M.L.; Rahal, P. Alphacoronavirus Detection in Lungs, Liver, and Intestines of Bats from Brazil. Microb. Ecol. 2020, 79, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Bueno, L.M.; Rizotto, L.S.; Viana, A.d.O.; Silva, L.M.N.; de Moraes, M.V.d.S.; Benassi, J.C.; Scagion, G.P.; Dorlass, E.G.; Lopes, B.L.T.; Cunha, I.N.; et al. High Genetic Diversity of Alphacoronaviruses in Bat Species (Mammalia: Chiroptera) from the Atlantic Forest in Brazil. Transbound. Emerg. Dis. 2022, 69, E2863–E2875. [Google Scholar] [CrossRef]
- Alves, R.S.; do Canto Olegário, J.; Weber, M.N.; da Silva, M.S.; Canova, R.; Sauthier, J.T.; Baumbach, L.F.; Witt, A.A.; Varela, A.P.M.; Mayer, F.Q.; et al. Detection of Coronavirus in Vampire Bats (Desmodus rotundus) in Southern Brazil. Transbound. Emerg. Dis. 2022, 69, 2384–2389. [Google Scholar] [CrossRef]
- Peixoto, F.P.; Braga, P.H.P.; Mendes, P. A Synthesis of Ecological and Evolutionary Determinants of Bat Diversity across Spatial Scales. BMC Ecol. 2018, 18, 18. [Google Scholar] [CrossRef]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953-16. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and Evolution of Pathogenic Coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zeng, L.P.; Yang, X.L.; Ge, X.Y.; Zhang, W.; Li, B.; Xie, J.Z.; Shen, X.R.; Zhang, Y.Z.; Wang, N.; et al. Discovery of a Rich Gene Pool of Bat SARS-Related Coronaviruses Provides New Insights into the Origin of SARS Coronavirus. PLoS Pathog. 2017, 13, e1006698. [Google Scholar] [CrossRef]
- Jia, Y.; Cao, J.; Wei, Z. Bioinformatics Analysis of Spike Proteins of Porcine Enteric Coronaviruses. Biomed. Res. Int. 2021, 2021, 6689471. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Zambrano-Moreno, C.; Pineda, P.; Calderón, C.; Rincón-Monroy, M.A.; Diaz, A.; Marthaler, D.G. Several Lineages of Porcine Epidemic Diarrhea Virus in Colombia during the 2014 and 2016 Epidemic. Transbound. Emerg. Dis. 2021, 68, 2465–2476. [Google Scholar] [CrossRef]
- Artika, I.M.; Dewantari, A.K.; Wiyatno, A. Molecular Biology of Coronaviruses: Current Knowledge. Heliyon 2020, 6, e04743. [Google Scholar] [CrossRef]
- Wong, A.H.M.; Tomlinson, A.C.A.; Zhou, D.; Satkunarajah, M.; Chen, K.; Sharon, C.; Desforges, M.; Talbot, P.J.; Rini, J.M. Receptor-Binding Loops in Alphacoronavirus Adaptation and Evolution. Nat. Commun. 2017, 8, 1735. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, P.; Liu, T.; Shi, C.; Wang, B.; Xu, Y.; Jin, T. Structural Requirements and Plasticity of Receptor-Binding Domain in Human Coronavirus Spike. Front. Mol. Biosci. 2022, 9, 930931. [Google Scholar] [CrossRef]
- Eby, P.; Peel, A.J.; Hoegh, A.; Madden, W.; Giles, J.R.; Hudson, P.J.; Plowright, R.K. Pathogen spillover driven by rapid changes in bat ecology. Nature 2023, 12, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Leopardi, S.; Priori, P.; Zecchin, B.; Zamperin, G.; Milani, A.; Tonon, F.; Giorgiutti, M.; Beato, M.S.; De Benedictis, P. Interface between Bats and Pigs in Heavy Pig Production. Viruses 2021, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Reguera, J.; Santiago, C.; Mudgal, G.; Ordoño, D.; Enjuanes, L.; Casasnovas, J.M. Structural Bases of Coronavirus Attachment to Host Aminopeptidase N and Its Inhibition by Neutralizing Antibodies. PLoS Pathog. 2012, 8, e1002859. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, M.C.; Lam, H.C.; Rovira, A.; Marthaler, D.G. Complete Genome Sequence of Porcine Epidemic Diarrhea Virus Strain COL/Cundinamarca/2014 from Colombia. Genome Announc. 2016, 4, e00239-16. [Google Scholar] [CrossRef]
- Chen, F.; Knutson, T.P.; Rossow, S.; Saif, L.J.; Marthaler, D.G. Decline of Transmissible Gastroenteritis Virus and Its Complex Evolutionary Relationship with Porcine Respiratory Coronavirus in the United States. Sci. Rep. 2019, 9, 3953. [Google Scholar] [CrossRef]
- Sun, Y.; Xing, J.; Xu, Z.Y.; Gao, H.; Xu, S.J.; Liu, J.; Zhu, D.H.; Guo, Y.F.; Yang, B.S.; Chen, X.N.; et al. Re-Emergence of Severe Acute Diarrhea Syndrome Coronavirus (SADS-CoV) in Guangxi, China, 2021. J. Infect. 2022, 8, e130–e133. [Google Scholar] [CrossRef]
- Mattar, S.; González-Tous, M.; Salgado, L. Virus Nipah, Un Paramixovirus Que Emerge de Los Hospedadores de Vida Silvestre y Representa Una Amenaza Para La Salud Humana. Rev. MVZ Córdoba 2018, 24, 7089–7090. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez, C.; Echeverri-De la Hoz, D.; Calderón, A.; López, Y.; Guzmán, C.; Galeano, K.; Bertel, V.; Gastelbondo-Pastrana, B.; Mattar, S. New Coronavirus in Colombian Caribbean Bats: In Silico Analysis Reveals Possible Risk of Interspecific Jumping. Viruses 2025, 17, 1320. https://doi.org/10.3390/v17101320
Martínez C, Echeverri-De la Hoz D, Calderón A, López Y, Guzmán C, Galeano K, Bertel V, Gastelbondo-Pastrana B, Mattar S. New Coronavirus in Colombian Caribbean Bats: In Silico Analysis Reveals Possible Risk of Interspecific Jumping. Viruses. 2025; 17(10):1320. https://doi.org/10.3390/v17101320
Chicago/Turabian StyleMartínez, Caty, Daniel Echeverri-De la Hoz, Alfonso Calderón, Yésica López, Camilo Guzmán, Ketty Galeano, Valeria Bertel, Bertha Gastelbondo-Pastrana, and Salim Mattar. 2025. "New Coronavirus in Colombian Caribbean Bats: In Silico Analysis Reveals Possible Risk of Interspecific Jumping" Viruses 17, no. 10: 1320. https://doi.org/10.3390/v17101320
APA StyleMartínez, C., Echeverri-De la Hoz, D., Calderón, A., López, Y., Guzmán, C., Galeano, K., Bertel, V., Gastelbondo-Pastrana, B., & Mattar, S. (2025). New Coronavirus in Colombian Caribbean Bats: In Silico Analysis Reveals Possible Risk of Interspecific Jumping. Viruses, 17(10), 1320. https://doi.org/10.3390/v17101320






