Molecular Detection and Characterization of Bovine Diarrhea Virus (BVDV) in Aborted Fetuses and Semen Samples from Paraguay
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Study Population
2.2. RNA Extraction and RT-PCR
2.3. Sequencing
2.4. Bioinformatic Analysis
3. Results
3.1. Bovine Semen and Serum Sample
3.2. Bovine Aborted Fetuses
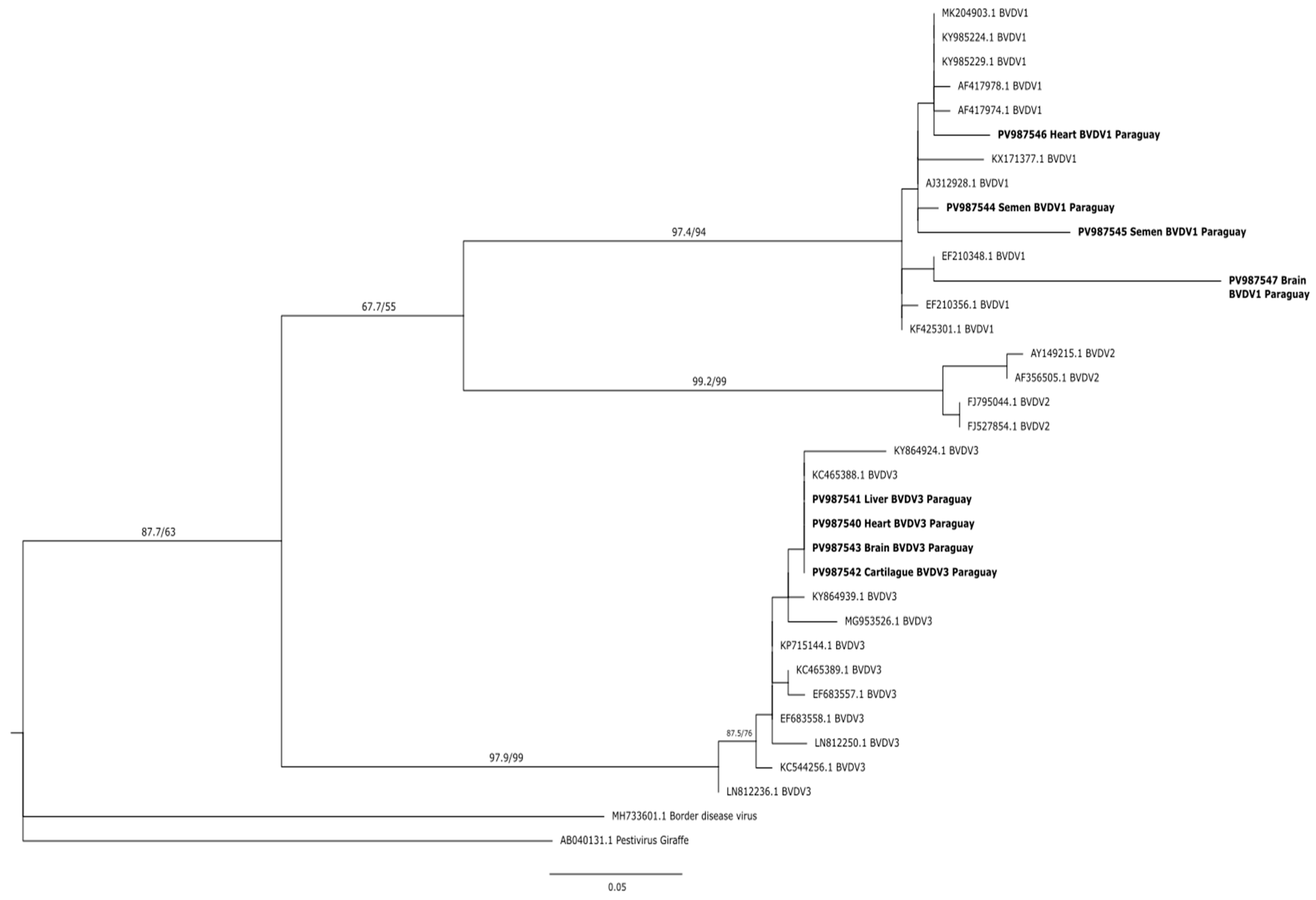
3.3. Sequencing and Bioinformatic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lanyon, S.R.; Hill, F.I.; Reichel, M.P.; Brownlie, J. Bovine viral diarrhoea: Pathogenesis and diagnosis. Vet. J. 2014, 199, 201–209. [Google Scholar] [CrossRef]
- Walz, P.H.; Grooms, D.L.; Passler, T.; Ridpath, J.F.; Tremblay, R.; Step, D.L.; Callan, R.; Givens, M.D.; American College of Veterinary Internal Medicine. Control of Bovine Viral Diarrhea Virus in Ruminants. J. Vet. Intern. Med. 2010, 24, 476–486. [Google Scholar] [CrossRef]
- Smith, D.B.; Meyers, G.; Bukh, J.; Gould, E.A.; Monath, T.; Scott Muerhoff, A.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; Simmonds, P.; et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J. Gen. Virol. 2017, 98, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, M.; Peterhans, E. Pestiviruses. Annu. Rev. Anim. Biosci. 2014, 2, 141–163. [Google Scholar] [CrossRef]
- Yeşilbağ, K.; Alpay, G.; Becher, P. Variability and Global Distribution of Subgenotypes of Bovine Viral Diarrhea Virus. Viruses 2017, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Avalos Ramirez, R.; Orlich, M.; Cedillo Rosales, S.; König, M.; Schweizer, M.; Stalder, H.; Schirrmeier, H.; Thiel, H.-J. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003, 311, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Bauermann, F.V.; Ridpath, J.F. Epidemiology of Pestivirus H in Brazil and Its Control Implications. Front. Vet. Sci. 2021, 8, 693041. [Google Scholar] [CrossRef]
- Pinior, B.; Firth, C.L.; Richter, V.; Lebl, K.; Trauffler, M.; Dzieciol, M.; Hutter, S.E.; Burgstaller, J.; Obritzhauser, W.; Winter, P.; et al. A systematic review of financial and economic assessments of bovine viral diarrhea virus (BVDV) prevention and mitigation activities worldwide. J. Prev. Vet. Med. 2017, 137, 77–92. [Google Scholar] [CrossRef]
- Richter, V.; Lebl, K.; Baumgartner, W.; Obritzhauser, W.; Käsbohrer, A.; Pinior, B. A systematic worldwide review of the direct monetary losses in cattle due to bovine viral diarrhoea virus infection. Vet. J. 2017, 220, 80–87. [Google Scholar] [CrossRef]
- Neill, J.D.; Workman, A.M.; Hesse, R.; Bai, J.; Porter, E.P.; Meadors, B.; Anderson, J.; Bayles, D.O.; Falkenberg, S.M. Identification of BVDV2b and 2c subgenotypes in the United States: Genetic and antigenic characterization. Virology 2019, 528, 19–29. [Google Scholar] [CrossRef]
- Quintero Barbosa, J.; Corredor Figueroa, A.P.; Salas, S.S.; Camargo, H.; Sanchéz, A.; Tobón, J.; Ortiz, D.; Schachtebeck, E.; Gutierrez, M.F. High prevalence of persistently infected animals from bovine viral diarrhea in Colombian cattle. BMC Vet. Res. 2019, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Maya, L.; Macías-Rioseco, M.; Silveira, C.; Giannitti, F.; Castells, M.; Salvo, M.; Rivero, R.; Cristina, J.; Gianneechini, E.; Puentes, R.; et al. An extensive field study reveals the circulation of new genetic variants of subtype 1a of bovine viral diarrhea virus in Uruguay. Arch. Virol. 2020, 165, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.F.; Cargnelutti, J.F.; Monteiro, F.L.; Bauermann, F.V.; Ridpath, J.F.; Weiblen, R. A genetic profile of bovine pestiviruses circulating in Brazil (1998–2018). Anim. Health Res. Rev. 2018, 19, 134–141. [Google Scholar] [CrossRef]
- Schirrmeier, H.; Strebelow, G.; Depner, K.; Hoffmann, B.; Beer, M. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J. Gen. Virol. 2004, 85, 3647–3652. [Google Scholar] [CrossRef]
- Pecora, A.; Perez Aguirreburualde, M.S.; Ridpath, J.F.; Dus Santos, M.J. Molecular Characterization of Pestiviruses in Fetal Bovine Sera Originating From Argentina: Evidence of Circulation of HoBi-Like Viruses. Front. Vet. Sci. 2019, 6, 359. [Google Scholar] [CrossRef]
- Vista de Comercio Internacional y Competitividad de la Producción Ganadera en Paraguay. Available online: https://revistascientificas.una.py/index.php/RE/article/view/322/331 (accessed on 3 July 2025).
- Cáceres Fernández, L.D. Diagnóstico de las Principales Enfermedades Reproductivas en Toros de Diferentes Establecimientos Ganaderos del Paraguay. Tesis para Especialista en Diagnóstico Veterinario de Laboratorio, Universidad Nacional de la Plata, La Plata, Argentina, 2018. [Google Scholar]
- Monteiro, F.L.; Cargnelutti, J.F.; Martins, B.; Noll, J.G.; Weiblen, R.; Flores, E.F. Detection of bovine pestiviruses in sera of beef calves by a RT-PCR based on a newly designed set of pan–bovine pestivirus primers. J. Vet. Diagn. Invest. 2019, 31, 255–258. [Google Scholar] [CrossRef]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Wertheim-van Dillen, P.M.; Van der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef]
- Alarcón-Zúñiga, B.; Zepeda-Batista, J.L.; Ruíz-Flores, A.; Gómez-Meza, L.J.; García-Muñiz, J.G.; Núñez-Domínguez, R.; Ramírez-Valverde, R.; Villegas-Velázquez, I. Modificación del método de tiocianato de guanidina para extraer ADN de semen para análisis genómico en mamíferos. Rev. Mex. Cienc. Pecu. 2016, 7, 405. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Givens, M.D.; Marley, M.S. Immunology of chronic BVDV infections. Biologicals 2013, 41, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Mascitti, A.K.; Fraga, A.P.; Abreu, J.P.R.; Weber, M.N.; Salla, P.F.; Corrêa, M.V.S.; Ikuta, N.; Canal, C.; Lunge, V. Pesquisa do vírus da diarreia viral bovina em touros mantidos a campo no estado do Rio Grande do Sul. Arq. Bras. Med. Veterinária Zootec. 2017, 69, 766–770. [Google Scholar] [CrossRef]
- Mishra, N.; Semmannan, K.; Mallinath, K.C.; Rajukumar, K.; Khetan, R.; Gautam, S.; Venkatesha, M.D.; Byregowda, S.M. Identification of Bovine Viral Diarrhoea Virus Type 2 in Cattle Bull Semen from Southern India and its Genetic Characterization. Curr. Sci. 2018, 114, 666–670. [Google Scholar] [CrossRef]
- Sharifzadeh, A.; Doosti, A.; Ghasemi-Dehkordi, P. Reverse Transcriptase PCR Assay for Detection of Bovine viral diarrhea virus (BVDV) Infection in Iranian Bull’s Semen Samples. Middle East J. Sci. Res. 2011, 9, 132–139. [Google Scholar]
- Givens, M.D.; Waldrop, J.G. Bovine viral diarrhea virus in embryo and semen production systems. Vet. Clin. N. Am. Food Anim. Pract. 2004, 20, 21–38. [Google Scholar] [CrossRef]
- Kirkland, P.D.; Richards, S.G.; Rothwell, J.T.; Stanley, D.F. Replication of bovine viral diarrhoea virus in the bovine reproductive tract and excretion of virus in semen during acute and chronic infections. Vet. Rec. 1991, 128, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Read, A.J.; Gestier, S.; Parrish, K.; Finlaison, D.S.; Gu, X.; O’Connor, T.W.; Kirkland, P.D. Prolonged Detection of Bovine Viral Diarrhoea Virus Infection in the Semen of Bulls. Viruses 2020, 12, 674. [Google Scholar] [CrossRef]
- Givens, M.D.; Meath, A.M.; Carson, R.L.; Brock, K.V.; Edens, M.S.D.; Wenzel, J.G.W.; Stringfellow, D. Analytical sensitivity of assays used for detection of bovine viral diarrhea virus in semen samples from the Southeastern United States. Vet. Microbiol. 2003, 96, 145–155. [Google Scholar]
- Kirkbride, C.A. Viral Agents and Associated Lesions Detected in a 10-Year Study of Bovine Abortions and Stillbirths. J. Vet. Diagn. Investig. 1992, 4, 374–379. [Google Scholar] [CrossRef]
- Da Silva Silveira, C.; Maya, L.; Casaux, M.L.; Schild, C.; Caffarena, D.; Aráoz, V.; da Costa, R.A.; Macías-Rioseco, M.; Perdomo, Y.; Castells, M.; et al. Diseases associated with bovine viral diarrhea virus subtypes 1a and 2b in beef and dairy cattle in Uruguay. Braz. J. Microbiol. 2020, 51, 357–368. [Google Scholar] [CrossRef]
- Campero, C.M.; Moore, D.P.; Odeón, A.C.; Cipolla, A.L.; Odriozola, E. Aetiology of bovine abortion in Argentina. Vet. Res. Commun. 2003, 27, 359–369. [Google Scholar] [CrossRef]
- Spetter, M.J.; Louge Uriarte, E.L.; Armendano, J.I.; Morrell, E.L.; Cantón, G.J.; Verna, A.E.; Dorsch, M.A.; Pereyra, S.B.; Odeón, A.C.; Saliki, J.T.; et al. Detection methods and characterization of bovine viral diarrhea virus in aborted fetuses and neonatal calves over a 22-year period. Braz. J. Microbiol. 2020, 57, 2077–2086. [Google Scholar] [CrossRef] [PubMed]
- Hyndman, L.; Vilcek, S.; Conner, J.; Nettleton, P.F. A novel nested reverse transcription PCR detects bovine viral diarrhoea virus in fluids from aborted bovine fetuses. J. Virol. Methods 1998, 71, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Oem, J.K.; Chung, J.Y.; Roh, I.S.; Kim, H.R.; Bae, Y.C.; Lee, K.H.; Jin, Y.H.; Lee, O.S. Characterization and Phylogenetic Analysis of Bovine Viral Diarrhea Virus in Brain Tissues from Nonambulatory (Downer) Cattle in Korea. J. Vet. Diagn. Invest. 2010, 22, 518–523. [Google Scholar] [CrossRef]
- Decaro, N.; Lucente, M.S.; Mari, V.; Sciarretta, R.; Pinto, P.; Buonavoglia, D.; Martella, V.; Buonavoglia, C. Hobi-like pestivirus in aborted bovine fetuses. J. Clin. Microbiol. 2012, 50, 509–512. [Google Scholar] [CrossRef]
- Spetter, M.J.; Uriarte, E.L.L.; Altamiranda, E.A.G.; Leunda, M.R.; Pereyra, S.B.; Verna, A.E.; Odeón, A.C. Dual natural infection with bovine viral diarrhea virus -1 and -2 in a stillborn calf: Tissue distribution and molecular characterization. Open Vet. J. 2018, 8, 493–497. [Google Scholar] [CrossRef]
- Weber, M.N.; Streck, A.F.; Silveira, S.; Mósena, A.C.S.; Da Silva, M.S.; Canal, C.W. Homologous recombination in pestiviruses: Identification of three putative novel events between different subtypes/genogroups. Infect. Genet. Evol. 2015, 30, 219–224. [Google Scholar] [CrossRef]
- Yilmaz, H.; Altan, E.; Ridpath, J.; Turan, N. Genetic diversity and frequency of bovine viral diarrhea virus (BVDV) detected in cattle in Turkey. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 411–416. [Google Scholar] [CrossRef] [PubMed]
| Seminal Samples | Serum Samples | ||
|---|---|---|---|
| N | Pestivirus A + RT-PCR | ELISA+ | |
| Breed | |||
| Braford | 5 | 5/5 (100%) | 2/5 (40%) |
| Brahman | 12 | 4/12 (33.3%) | 0 |
| Brangus | 41 | 25/41 (61%) | 21/41 (51%) |
| Pampa | 2 | Negative | 2/2 (100%) |
| Senepol | 4 | 3/4 (75%) | 1/4 (25%) |
| Hibrid | 9 | 3/9(33.3%) | 3/9 (33%) |
| Age | |||
| 0–5 years | 16 | 11/16 (69%) | 5/16 (31%) |
| 6–9 years | 23 | 8/23 (35%) | 11/23 (48%) |
| Adults | 33 | 21/33 (64%) | 13/33 (39%) |
| NDA * | 1 | Negative | 0 |
| Department | |||
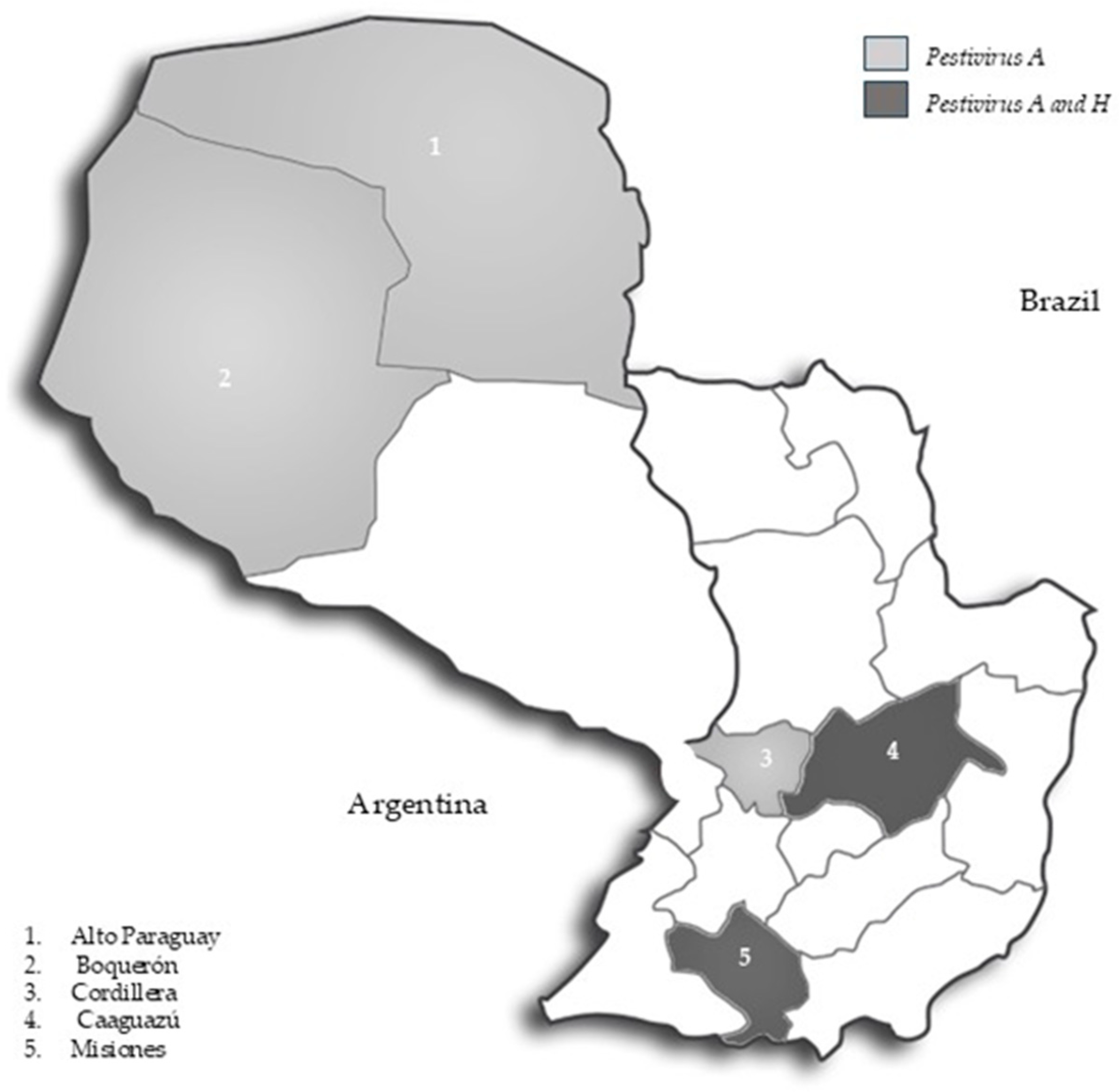
| Alto Paraguay | 10 | 10/10 (100%) | 4/10 (40%) |
| Boquerón | 33 | 8/33 (24.2%) | 15/33 (45%) |
| Caaguazú | 10 | 9/10 (90%) | 2/10 (20%) |
| Cordillera | 20 | 13/20 (65%) | 8/20 (40%) |
| Organs | Fetus 1 | Fetus 7 | ||||
|---|---|---|---|---|---|---|
| Pestivirus | Pestivirus | |||||
| A | B | H | A | B | H | |
| Heart | + | − | + | − | − | − |
| Lung | + | − | + | + | − | − |
| Liver | + | − | + | − | − | + |
| Brain | * | * | * | + | − | + |
| Auricular cartilage | + | − | + | − | − | + |
| Abdominal cavity liquid | − | − | − | * | * | * |
| Pericardiac Liquid | − | − | − | * | * | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez Valinotti, M.F.; Nara Pereira, E.M.; Martinez Pereira, M.; Rodriguez Valinotti, R.; Rodriguez Sanchez, A. Molecular Detection and Characterization of Bovine Diarrhea Virus (BVDV) in Aborted Fetuses and Semen Samples from Paraguay. Viruses 2025, 17, 1295. https://doi.org/10.3390/v17101295
Rodriguez Valinotti MF, Nara Pereira EM, Martinez Pereira M, Rodriguez Valinotti R, Rodriguez Sanchez A. Molecular Detection and Characterization of Bovine Diarrhea Virus (BVDV) in Aborted Fetuses and Semen Samples from Paraguay. Viruses. 2025; 17(10):1295. https://doi.org/10.3390/v17101295
Chicago/Turabian StyleRodriguez Valinotti, María Fátima, Eva Megumi Nara Pereira, Magaly Martinez Pereira, Rosmary Rodriguez Valinotti, and Antonio Rodriguez Sanchez. 2025. "Molecular Detection and Characterization of Bovine Diarrhea Virus (BVDV) in Aborted Fetuses and Semen Samples from Paraguay" Viruses 17, no. 10: 1295. https://doi.org/10.3390/v17101295
APA StyleRodriguez Valinotti, M. F., Nara Pereira, E. M., Martinez Pereira, M., Rodriguez Valinotti, R., & Rodriguez Sanchez, A. (2025). Molecular Detection and Characterization of Bovine Diarrhea Virus (BVDV) in Aborted Fetuses and Semen Samples from Paraguay. Viruses, 17(10), 1295. https://doi.org/10.3390/v17101295






