Isolation and Phylogenetic Analysis of Atypical Porcine Pestivirus Isolates Identified in Russian Swine Herds
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing and RNA Extraction
2.3. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.4. Reverse Transcriptase PCR and Sequencing
2.5. Phylogenetic Analysis
2.6. Cell Lines
2.7. Virus Isolation
2.8. Statistical Analysis
3. Results
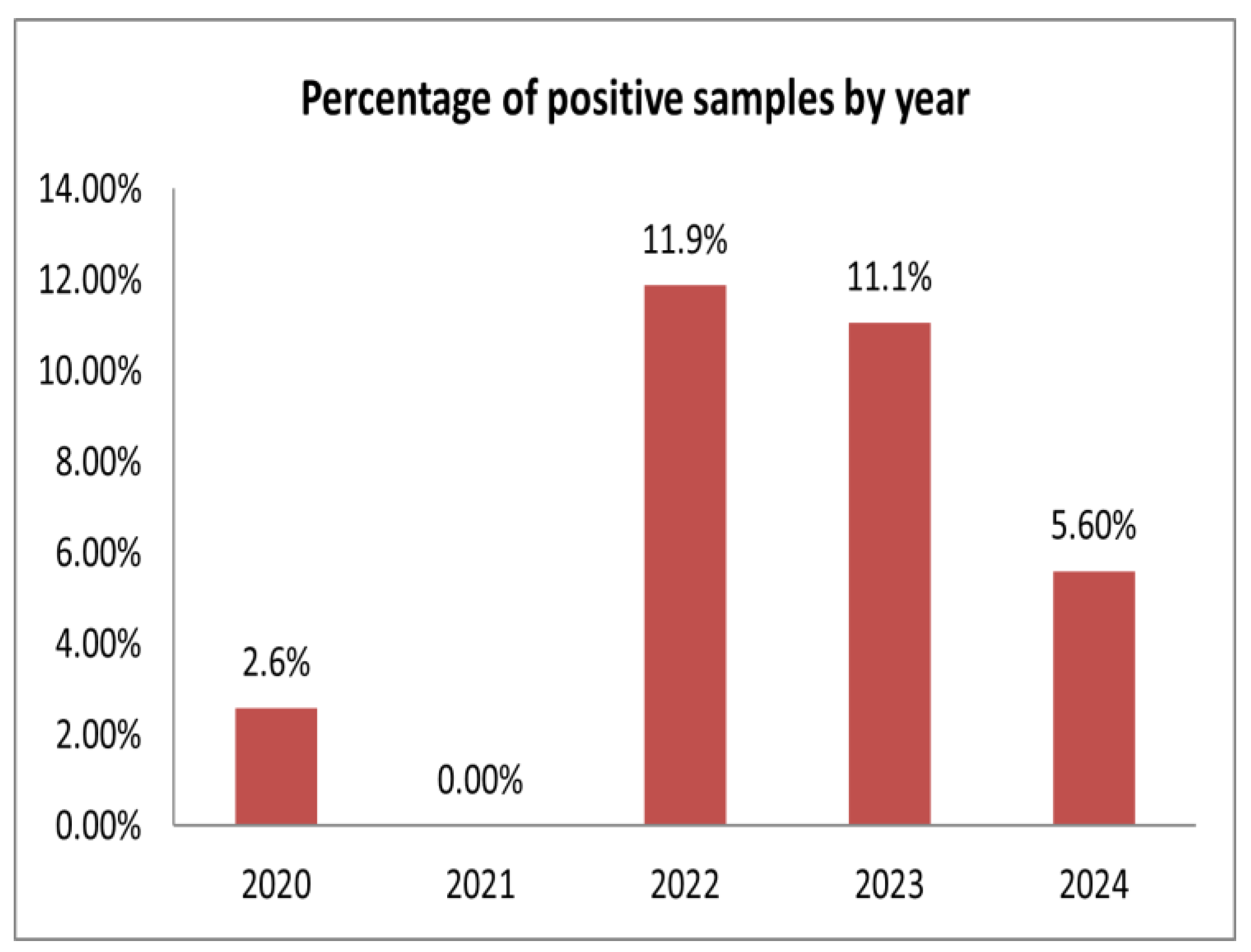
3.1. APPV Detection Rates in Swine Herds in Russia
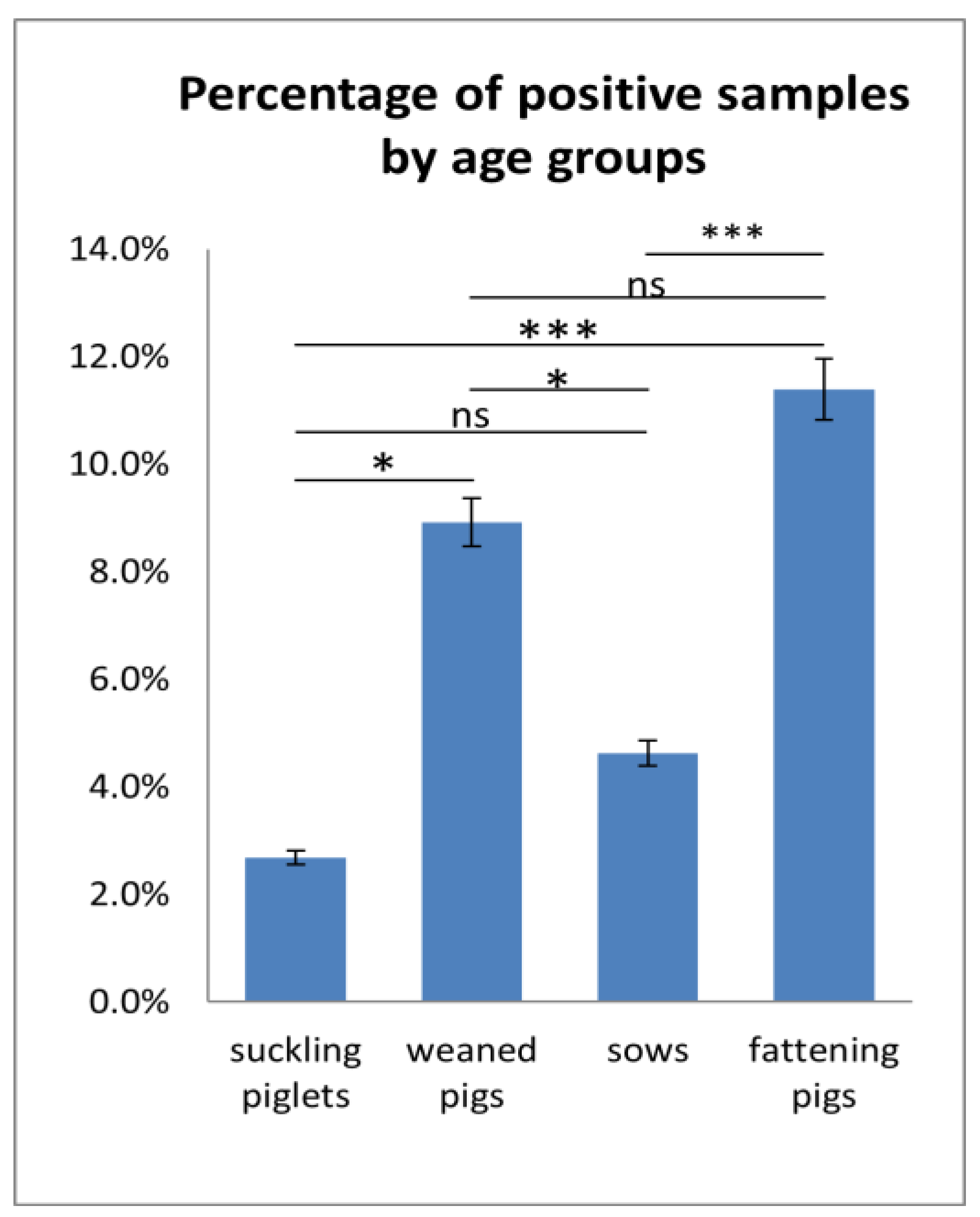
APPV Detection Rates in Specimen Types and Different Age Groups
3.2. Phylogenetic Analysis and Nucleotide Identity of APPV
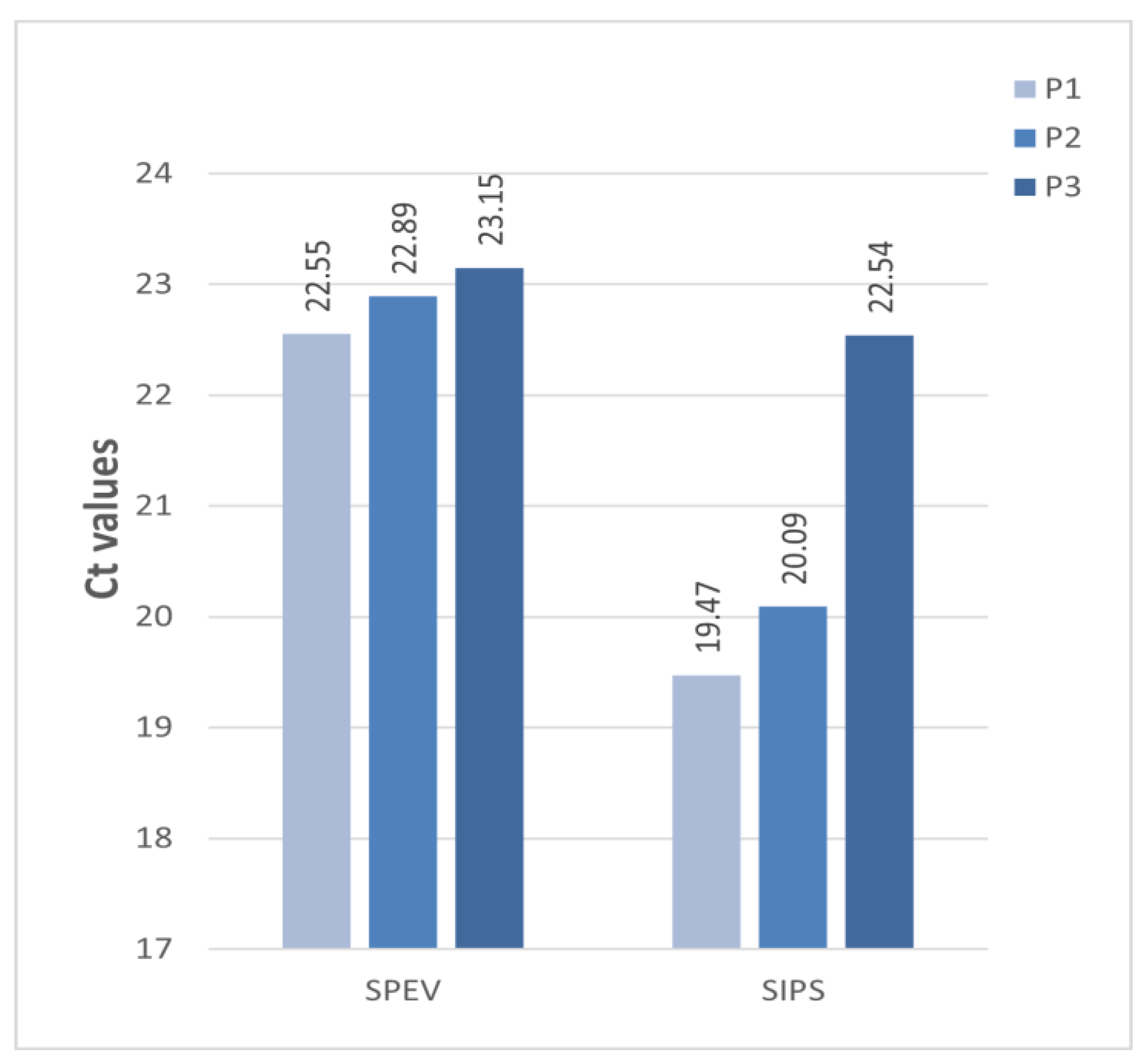
3.3. APPV Isolation in Cell Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, D.B.; Meyers, G.; Bukh, J.; Gould, E.A.; Monath, T.; Scott Muerhoff, A.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; Simmonds, P.; et al. Proposed Revision to the Taxonomy of the Genus Pestivirus, Family Flaviviridae. J. Gen. Virol. 2017, 98, 2106–2112. [Google Scholar] [CrossRef]
- Genus: Pestivirus|ICTV. Available online: https://ictv.global/report/chapter/flaviviridaeport/flaviviridaeport/flaviviridae/pestivirus (accessed on 17 October 2024).
- Tautz, N.; Tews, B.A.; Meyers, G. The Molecular Biology of Pestiviruses. Adv. Virus Res. 2015, 93, 47–160. [Google Scholar] [CrossRef] [PubMed]
- Jo, W.K.; van Elk, C.; van de Bildt, M.; van Run, P.; Petry, M.; Jesse, S.T.; Jung, K.; Ludlow, M.; Kuiken, T.; Osterhaus, A. An Evolutionary Divergent Pestivirus Lacking the Npro Gene Systemically Infects a Whale Species. Emerg. Microbes Infect. 2019, 8, 1383–1392. [Google Scholar] [CrossRef]
- Kirkland, P.D.; Frost, M.J.; Finlaison, D.S.; King, K.R.; Ridpath, J.F.; Gu, X. Identification of a Novel Virus in Pigs--Bungowannah Virus: A Possible New Species of Pestivirus. Virus Res. 2007, 129, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Lamp, B.; Schwarz, L.; Högler, S.; Riedel, C.; Sinn, L.; Rebel-Bauder, B.; Weissenböck, H.; Ladinig, A.; Rümenapf, T. Novel Pestivirus Species in Pigs, Austria, 2015. Emerg. Infect. Dis. 2017, 23, 1176–1179. [Google Scholar] [CrossRef] [PubMed]
- Kiesler, A.; Seitz, K.; Schwarz, L.; Buczolich, K.; Petznek, H.; Sassu, E.; Dürlinger, S.; Högler, S.; Klang, A.; Riedel, C.; et al. Clinical and Serological Evaluation of LINDA Virus Infections in Post-Weaning Piglets. Viruses 2019, 11, 975. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Collin, E.A.; Peddireddi, L.; Yuan, F.; Chen, Z.; Hesse, R.A.; Gauger, P.C.; Clement, T.; Fang, Y.; Anderson, G. Discovery of a Novel Putative Atypical Porcine Pestivirus in Pigs in the USA. J. Gen. Virol. 2015, 96, 2994–2998. [Google Scholar] [CrossRef] [PubMed]
- Arruda, B.L.; Arruda, P.H.; Magstadt, D.R.; Schwartz, K.J.; Dohlman, T.; Schleining, J.A.; Patterson, A.R.; Visek, C.A.; Victoria, J.G. Identification of a Divergent Lineage Porcine Pestivirus in Nursing Piglets with Congenital Tremors and Reproduction of Disease Following Experimental Inoculation. PLoS ONE 2016, 11, e0150104. [Google Scholar] [CrossRef]
- Postel, A.; Hansmann, F.; Baechlein, C.; Fischer, N.; Alawi, M.; Grundhoff, A.; Derking, S.; Tenhündfeld, J.; Pfankuche, V.M.; Herder, V.; et al. Presence of Atypical Porcine Pestivirus (APPV) Genomes in Newborn Piglets Correlates with Congenital Tremor. Sci. Rep. 2016, 6, 27735. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.; Röntgen, M.; Maak, S. Congenital Splay Leg Syndrome in Piglets—Current Knowledge and a New Approach to Etiology. Front. Vet. Sci. 2021, 8, 609883. [Google Scholar] [CrossRef] [PubMed]
- Dall Agnol, A.M.; Alfieri, A.F.; Alfieri, A.A. Pestivirus K (Atypical Porcine Pestivirus): Update on the Virus, Viral Infection, and the Association with Congenital Tremor in Newborn Piglets. Viruses 2020, 12, 903. [Google Scholar] [CrossRef] [PubMed]
- de Groof, A.; Deijs, M.; Guelen, L.; van Grinsven, L.; van Os-Galdos, L.; Vogels, W.; Derks, C.; Cruijsen, T.; Geurts, V.; Vrijenhoek, M.; et al. Atypical Porcine Pestivirus: A Possible Cause of Congenital Tremor Type A-II in Newborn Piglets. Viruses 2016, 8, 271. [Google Scholar] [CrossRef]
- Dessureault, F.G.; Choinière, M.; Provost, C.; Gagnon, C.A. First Report of Atypical Porcine Pestivirus in Piglets with Congenital Tremor in Canada. Can. Vet. J. Rev. Vet. Can. 2018, 59, 429–432. [Google Scholar]
- Sutton, K.M.; Lahmers, K.K.; Harris, S.P.; Wijesena, H.R.; Mote, B.E.; Kachman, S.D.; Borza, T.; Ciobanu, D.C. Detection of Atypical Porcine Pestivirus Genome in Newborn Piglets Affected by Congenital Tremor and High Preweaning Mortality1. J. Anim. Sci. 2019, 97, 4093–4100. [Google Scholar] [CrossRef]
- Gatto, I.R.H.; Arruda, P.H.; Visek, C.A.; Victoria, J.G.; Patterson, A.R.; Krull, A.C.; Schwartz, K.J.; de Oliveira, L.G.; Arruda, B.L. Detection of Atypical Porcine Pestivirus in Semen from Commercial Boar Studs in the United States. Transbound. Emerg. Dis. 2018, 65, e339–e343. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, L.; Riedel, C.; Högler, S.; Sinn, L.J.; Voglmayr, T.; Wöchtl, B.; Dinhopl, N.; Rebel-Bauder, B.; Weissenböck, H.; Ladinig, A.; et al. Congenital Infection with Atypical Porcine Pestivirus (APPV) Is Associated with Disease and Viral Persistence. Vet. Res. 2017, 48, 1. [Google Scholar] [CrossRef]
- Yuan, J.; Han, Z.; Li, J.; Huang, Y.; Yang, J.; Ding, H.; Zhang, J.; Zhu, M.; Zhang, Y.; Liao, J.; et al. Atypical Porcine Pestivirus as a Novel Type of Pestivirus in Pigs in China. Front. Microbiol. 2017, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, X.; Su, D.; Feng, J.; Wei, L.; Cai, W.; Li, J.; Lin, S.; Yan, H.; He, D. Detection and Genetic Characterization of Atypical Porcine Pestivirus in Piglets With Congenital Tremors in Southern China. Front. Microbiol. 2019, 10, 1406. [Google Scholar] [CrossRef]
- Muñoz-González, S.; Canturri, A.; Pérez-Simó, M.; Bohórquez, J.A.; Rosell, R.; Cabezón, O.; Segalés, J.; Domingo, M.; Ganges, L. First Report of the Novel Atypical Porcine Pestivirus in Spain and a Retrospective Study. Transbound. Emerg. Dis. 2017, 64, 1645–1649. [Google Scholar] [CrossRef]
- (PDF) Detection of Atypical Porcine Pestivirus (APPV) from a Case of Congenital Tremor in Korea. Available online: https://www.researchgate.net/publication/323336067_Detection_of_atypical_porcine_pestivirus_APPV_from_a_case_of_congenital_tremor_in_Korea (accessed on 17 October 2024).
- Mósena, A.C.S.; Weber, M.N.; da Cruz, R.a.S.; Cibulski, S.P.; da Silva, M.S.; Puhl, D.E.; Hammerschmitt, M.E.; Takeuti, K.L.; Driemeier, D.; de Barcellos, D.E.S.N.; et al. Presence of Atypical Porcine Pestivirus (APPV) in Brazilian Pigs. Transbound. Emerg. Dis. 2018, 65, 22–26. [Google Scholar] [CrossRef]
- Hill, H. Impact of Atypical Porcine Pestivirus (APPV) in Great Britain. In The impact of Atypical Porcine Pestivirus (APPV) in Great Britain; The University of Edinburgh: Edinburgh, UK, 2021. [Google Scholar] [CrossRef]
- Postel, A.; Meyer, D.; Cagatay, G.N.; Feliziani, F.; De Mia, G.M.; Fischer, N.; Grundhoff, A.; Milićević, V.; Deng, M.-C.; Chang, C.-Y.; et al. High Abundance and Genetic Variability of Atypical Porcine Pestivirus in Pigs from Europe and Asia. Emerg. Infect. Dis. 2017, 23, 2104–2107. [Google Scholar] [CrossRef] [PubMed]
- Dénes, L.; Biksi, I.; Albert, M.; Szeredi, L.; Knapp, D.G.; Szilasi, A.; Bálint, Á.; Balka, G. Detection and Phylogenetic Characterization of Atypical Porcine Pestivirus Strains in Hungary. Transbound. Emerg. Dis. 2018, 65, 2039–2042. [Google Scholar] [CrossRef]
- Kasahara-Kamiie, M.; Kagawa, M.; Shiokawa, M.; Sunaga, F.; Fukase, Y.; Aihara, N.; Shiga, T.; Kamiie, J.; Aoki, H.; Nagai, M. Detection and Genetic Analysis of a Novel Atypical Porcine Pestivirus from Piglets with Congenital Tremor in Japan. Transbound. Emerg. Dis. 2022, 69, 1761–1769. [Google Scholar] [CrossRef]
- Sozzi, E.; Salogni, C.; Lelli, D.; Barbieri, I.; Moreno, A.; Alborali, G.L.; Lavazza, A. Molecular Survey and Phylogenetic Analysis of Atypical Porcine Pestivirus (APPV) Identified in Swine and Wild Boar from Northern Italy. Viruses 2019, 11, 1142. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, H.; Leveringhaus, E.; Malmsten, A.; Dalin, A.-M.; Postel, A.; Malmberg, M. Atypical Porcine Pestivirus-A Widespread Virus in the Swedish Wild Boar Population. Transbound. Emerg. Dis. 2022, 69, 2349–2360. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, C.; Stalder, H.; Sidler, X.; Renzullo, S.; Gurtner, C.; Grahofer, A.; Schweizer, M. Long-Term Circulation of Atypical Porcine Pestivirus (APPV) within Switzerland. Viruses 2019, 11, 653. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.; Kristensen, C.S.; Strandbygaard, B.; Bøtner, A.; Rasmussen, T.B. Detection of Atypical Porcine Pestivirus in Piglets from Danish Sow Herds. Viruses 2021, 13, 717. [Google Scholar] [CrossRef]
- Michelitsch, A.; Dalmann, A.; Wernike, K.; Reimann, I.; Beer, M. Seroprevalences of Newly Discovered Porcine Pestiviruses in German Pig Farms. Vet. Sci. 2019, 6, 86. [Google Scholar] [CrossRef]
- Colom-Cadena, A.; Ganges, L.; Muñoz-González, S.; Castillo-Contreras, R.; Bohórquez, J.A.; Rosell, R.; Segalés, J.; Marco, I.; Cabezon, O. Atypical Porcine Pestivirus in Wild Boar (Sus Scrofa), Spain. Vet. Rec. 2018, 183, 569. [Google Scholar] [CrossRef] [PubMed]
- Cagatay, G.N.; Antos, A.; Meyer, D.; Maistrelli, C.; Keuling, O.; Becher, P.; Postel, A. Frequent Infection of Wild Boar with Atypical Porcine Pestivirus (APPV). Transbound. Emerg. Dis. 2018, 65, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.C.; Mora-Díaz, J.-C.; Magtoto, R.L.; Hulzen, A.V.; Ferreyra, F.M.; Falkenberg, S.M.; Giménez-Lirola, L.G.; Arruda, B.L. Dynamics of Infection of Atypical Porcine Pestivirus in Commercial Pigs from Birth to Market: A Longitudinal Study. Viruses 2023, 15, 1767. [Google Scholar] [CrossRef]
- Song, H.; Gao, X.; Fu, Y.; Li, J.; Fan, G.; Shao, L.; Zhang, J.; Qiu, H.-J.; Luo, Y. Isolation and Molecular Characterization of Atypical Porcine Pestivirus Emerging in China. Viruses 2023, 15, 2149. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Feng, Y.; Bai, J.; Liu, X.; Arruda, B.; Anbalagan, S.; Peddireddi, L. Genetic Diversity and Prevalence of Atypical Porcine Pestivirus in the Midwest of US Swine Herds during 2016–2018. Transbound. Emerg. Dis. 2022, 69, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Gatto, I.R.H.; Harmon, K.; Bradner, L.; Silva, P.; Linhares, D.C.L.; Arruda, P.H.; de Oliveira, L.G.; Arruda, B.L. Detection of Atypical Porcine Pestivirus in Brazil in the Central Nervous System of Suckling Piglets with Congenital Tremor. Transbound. Emerg. Dis. 2018, 65, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Cagatay, G.N.; Meyer, D.; Wendt, M.; Becher, P.; Postel, A. Characterization of the Humoral Immune Response Induced after Infection with Atypical Porcine Pestivirus (APPV). Viruses 2019, 11, 880. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Wang, L. Genotyping Atypical Porcine Pestivirus Using NS5a. Infect. Genet. Evol. 2021, 92, 104866. [Google Scholar] [CrossRef]
- Shiokawa, M.; Morita, Y.; Nagai, M.; Haritani, M.; Aoki, H. Isolation and Artificial Production of Atypical Porcine Pestivirus, Using the Swine-Kidney-Derived Cell Line SK-L. Arch. Virol. 2023, 168, 294. [Google Scholar] [CrossRef] [PubMed]
- Cagatay, G.N.; Antos, A.; Suckstorff, O.; Isken, O.; Tautz, N.; Becher, P.; Postel, A. Porcine Complement Regulatory Protein CD46 Is a Major Receptor for Atypical Porcine Pestivirus but Not for Classical Swine Fever Virus. J. Virol. 2021, 95, e02186-20. [Google Scholar] [CrossRef] [PubMed]
- Beer, M.; Wernike, K.; Dräger, C.; Höper, D.; Pohlmann, A.; Bergermann, C.; Schröder, C.; Klinkhammer, S.; Blome, S.; Hoffmann, B. High Prevalence of Highly Variable Atypical Porcine Pestiviruses Found in Germany. Transbound. Emerg. Dis. 2017, 64, e22–e26. [Google Scholar] [CrossRef] [PubMed]

| Investigated Area | Farm’s ID | 2020 | 2021 | 2022 | 2023 | 2024 | Total by Farm/Region | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive/Total Tested | Positive % | Positive/Total Tested | Positive % | Positive/Total Tested | Positive % | Positive/Total Tested | Positive % | Positive/Total Tested | Positive % | Positive/Total Tested | Positive % | ||
| Krasnoyarsk Krai (Farms/Total) | KKA | 0/27 | 0% | 0/20 | 0% | 13/114 | 11.4% | 0/21 | 0% | 7/62 | 11.3% | 20/244 | 8.2% |
| KKB | 1/34 | 2.9% | 0/20 | 0% | 11/138 | 7.9% | 6/43 | 13.9% | 18/235 | 7.7% | |||
| KKC | 1/19 | 5.3% | 0/9 | 0% | 7/64 | 10.9% | 5/35 | 14.3% | 13/127 | 10.2% | |||
| KKD | 1/36 | 2.8% | 0/20 | 0% | 9/107 | 8.4% | 0/8 | 0% | 1/76 | 1.3% | 11/247 | 4.5% | |
| Total | 3/116 | 2.6% | 0/69 | 0% | 40/423 | 9.5% | 0/29 | 0% | 19/216 | 8.8% | 62/853 | 7.3% | |
| Republic of Buryatia | RB | 0/67 | 0% | 0/143 | 0% | 8/125 | 6.4% | 6/120 | 5% | 14/455 | 3.1% | ||
| Tomsk Region (Farms/Total) | TA | 0/42 | 0% | 0/30 | 0% | 0/72 | 0% | ||||||
| TB | 1/10 | 10% | 1/30 | 3.3% | 2/40 | 5% | |||||||
| TC | 0/39 | 0% | 0/28 | 0% | 0/67 | 0% | |||||||
| TD | 0/40 | 0% | 2/30 | 6.7% | 2/70 | 2.9% | |||||||
| Total | 1/131 | 0.8% | 3/118 | 2.5% | 4/249 | 1.6% | |||||||
| Vologda Region | VR | 0/48 | 0% | 0/48 | 0% | ||||||||
| Kemerovo Region (Farms/Total) | KA | 0/10 | 0% | 0/19 | 0% | 0/29 | 0% | ||||||
| KB | 0/16 | 0% | 0/16 | 0% | |||||||||
| Total | 0/10 | 0% | 0/35 | 0% | 0/45 | 0% | |||||||
| Kirov Region (Farms/Total | KRA | 0/19 | 0% | 0/19 | 0% | ||||||||
| KRB | 0/11 | 0% | 0/11 | 0% | |||||||||
| Total | 0/30 | 0% | 0/30 | 0% | |||||||||
| Tyumen Region | TR | 0/15 | 0% | 0/15 | 0% | ||||||||
| Belgorod Region | BR | 0/50 | 0% | 97/332 | 29.2% | 3/14 | 21.4% | 100/396 | 25.3% | ||||
| Kursk Region | KR | 2/12 | 16.7% | 2/12 | 16.7% | ||||||||
| Republic of Mordovia | RM | 14/160 | 8.8% | 14/160 | 8.8% | ||||||||
| Moscow Region (Farms/Total) | MA | 0/44 | 0% | 0/44 | 0% | ||||||||
| MB | 0/5 | 0% | 0/5 | 0% | |||||||||
| MC | 12/56 | 21.4% | 1/14 | 7.1% | 13/70 | 18.6% | |||||||
| Total | 0/44 | 0% | 0/5 | 0% | 12/56 | 21.4% | 1/14 | 7.1% | 13/119 | 10.9% | |||
| Novosibirsk Region | NR | 0/4 | 0% | 0/23 | 0% | 0/27 | 0% | ||||||
| Pskov Region | PR | 23/138 | 16.7% | 23/138 | 16.7% | ||||||||
| Sverdlovsk Region | SR | 0/56 | 0% | 0/27 | 0% | 0/83 | 0% | ||||||
| Total by year | 12/24 (50%) | 3/116 | 2.6% | 0/282 | 0% | 152/1280 | 11.9% | 48/434 | 11.1% | 29/518 | 5.6% | 232/2630 | 8.8% |
| Specimen Type | Total Investigated | Number of Positive | % |
|---|---|---|---|
| Serum | 2420 | 217 | 9% |
| Semen | 10 | 0 | 0% |
| Feces | 53 | 0 | 0% |
| BAL | 4 | 0 | 0% |
| Tissue specimens | 143 | 15 | 10.5% |
| Total | 2630 | 232 | 8.8% |
| Russian Isolates | APPV Strains from GenBank | ||||
|---|---|---|---|---|---|
| Hungary OQ190181 | Hungary OQ190177 | USA MW183242 | USA MF590069 | South Korea MF979135 | |
| Belgorod_2022_ | 98.8 | 98.6 | 93.5 | 85.9 | 89.6 |
| Belgorod 151 2023 | 98.8 | 98.6 | 93.5 | 85.9 | 89.6 |
| Tomskii_3.6/VIEV_2022 | 95.0 | 94.3 | 88.4 | 84.6 | 87.9 |
| Kranoyarski_2.9/VIEV_2024 | 96.5 | 95.9 | 94.5 | 85.9 | 88.8 |
| F1_APPV_Krasnoyarsk_2022 | 96.3 | 95.7 | 89.3 | 86.3 | 88.5 |
| Ramenskoe_2023 | 90.3 | 89.5 | 92.4 | 94.3 | 89.8 |
| Pskov_2023 | 88.6 | 88.7 | 89.2 | 93.6 | 87.5 |
| Mordovia_2022 | 91.0 | 91.2 | 90.5 | 89.9 | 97.1 |
| Kursk_2023 | 99.1 | 98.9 | 93.9 | 86.2 | 90.1 |
| Buryatiya_18/VIEV_2024 | 96.1 | 95.5 | 94.3 | 85.7 | 88.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anoyatbekova, A.; Yuzhakov, A. Isolation and Phylogenetic Analysis of Atypical Porcine Pestivirus Isolates Identified in Russian Swine Herds. Viruses 2025, 17, 2. https://doi.org/10.3390/v17010002
Anoyatbekova A, Yuzhakov A. Isolation and Phylogenetic Analysis of Atypical Porcine Pestivirus Isolates Identified in Russian Swine Herds. Viruses. 2025; 17(1):2. https://doi.org/10.3390/v17010002
Chicago/Turabian StyleAnoyatbekova, Afshona, and Anton Yuzhakov. 2025. "Isolation and Phylogenetic Analysis of Atypical Porcine Pestivirus Isolates Identified in Russian Swine Herds" Viruses 17, no. 1: 2. https://doi.org/10.3390/v17010002
APA StyleAnoyatbekova, A., & Yuzhakov, A. (2025). Isolation and Phylogenetic Analysis of Atypical Porcine Pestivirus Isolates Identified in Russian Swine Herds. Viruses, 17(1), 2. https://doi.org/10.3390/v17010002







