Mechanisms of Flavivirus Cross-Protection against Yellow Fever in a Mouse Model
Abstract
1. Introduction
2. Materials and Methods
3. Results
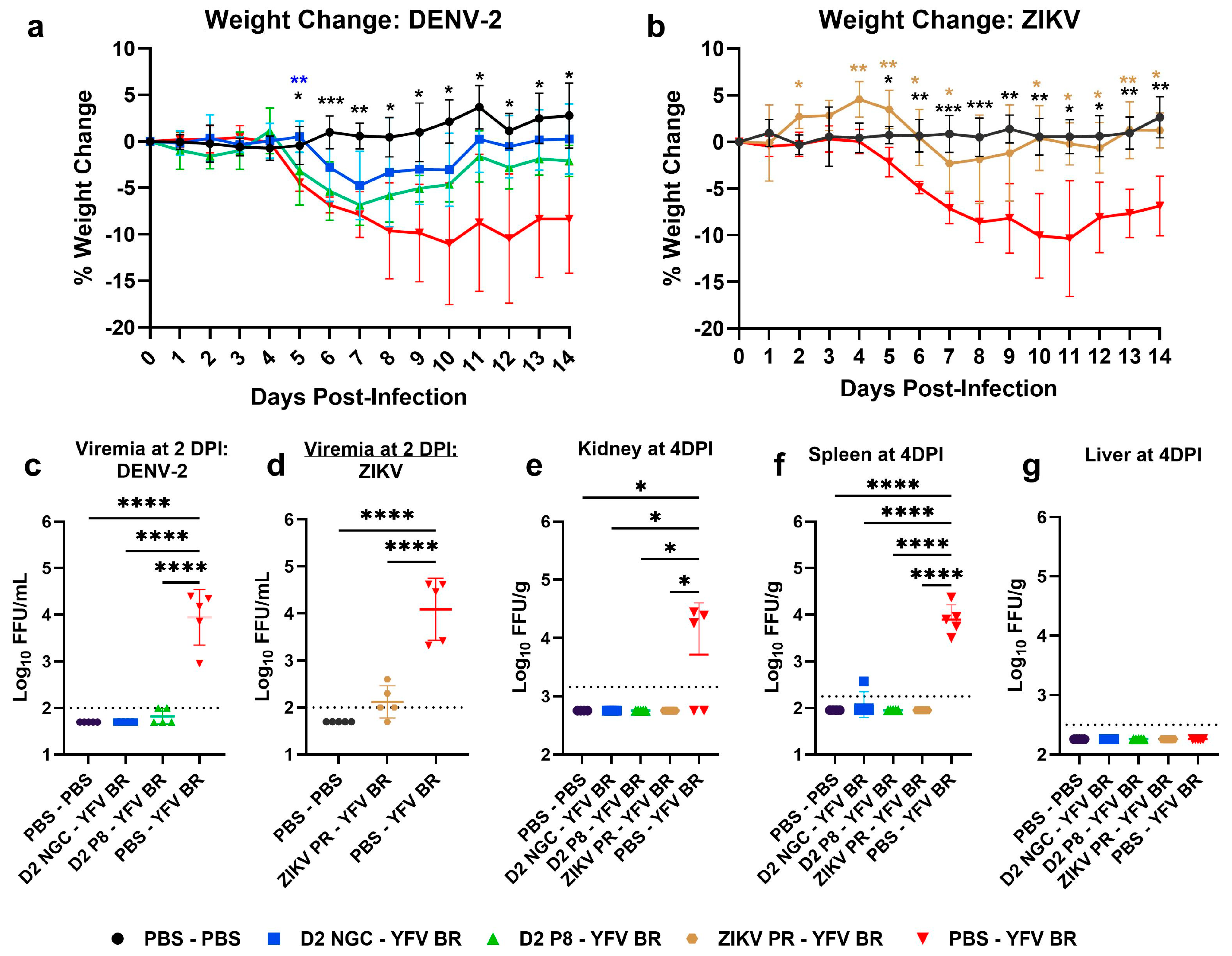
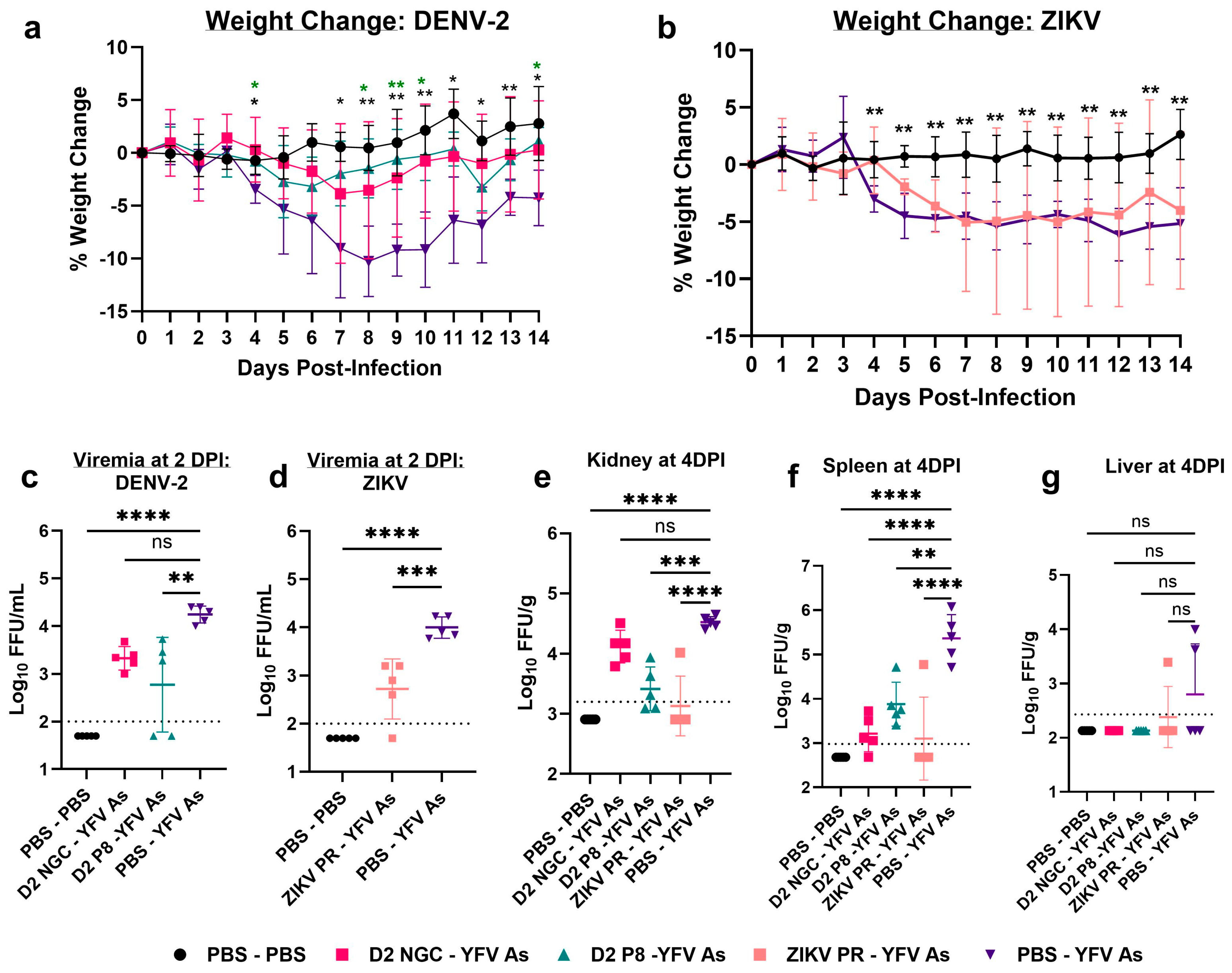
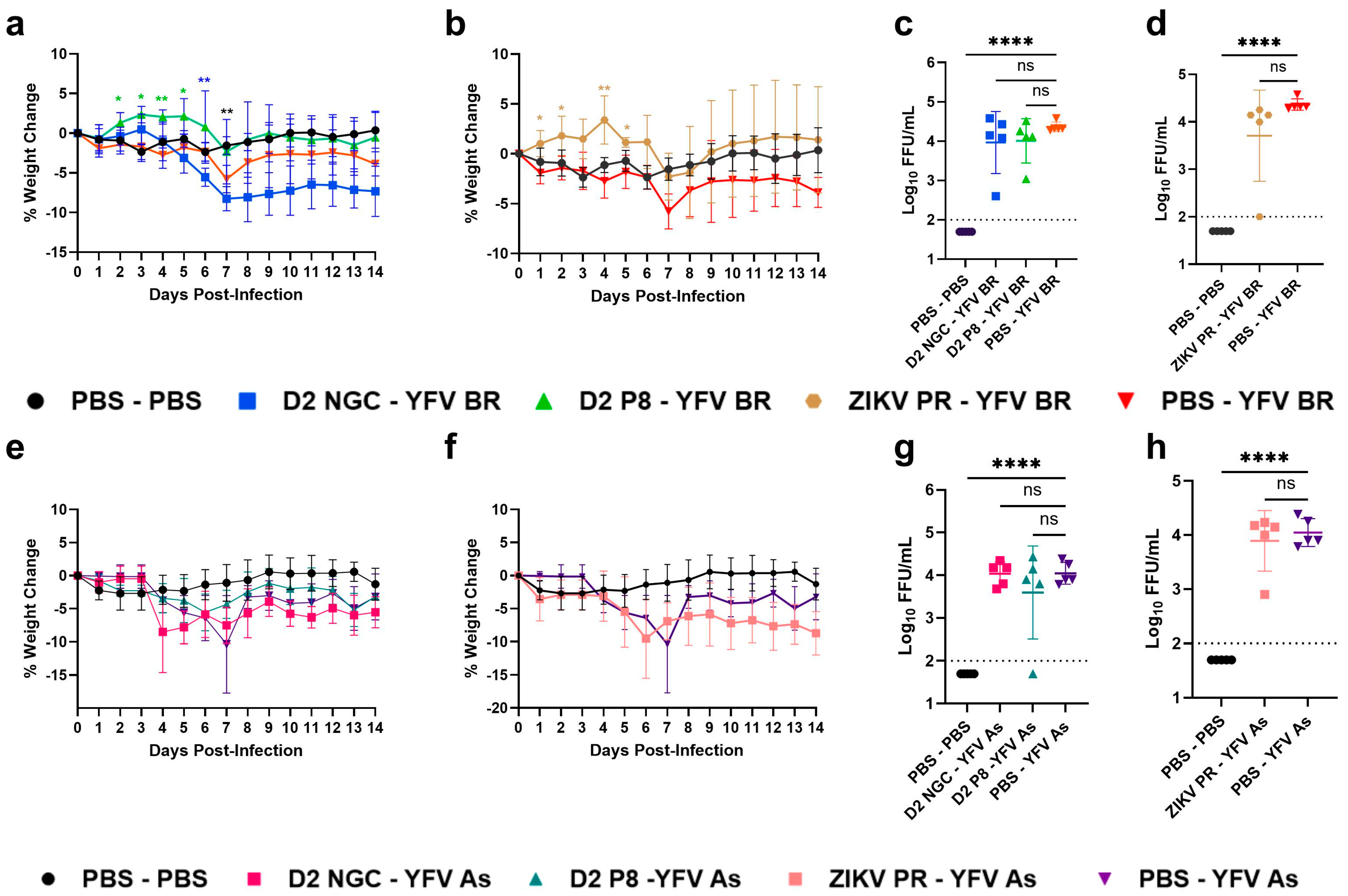
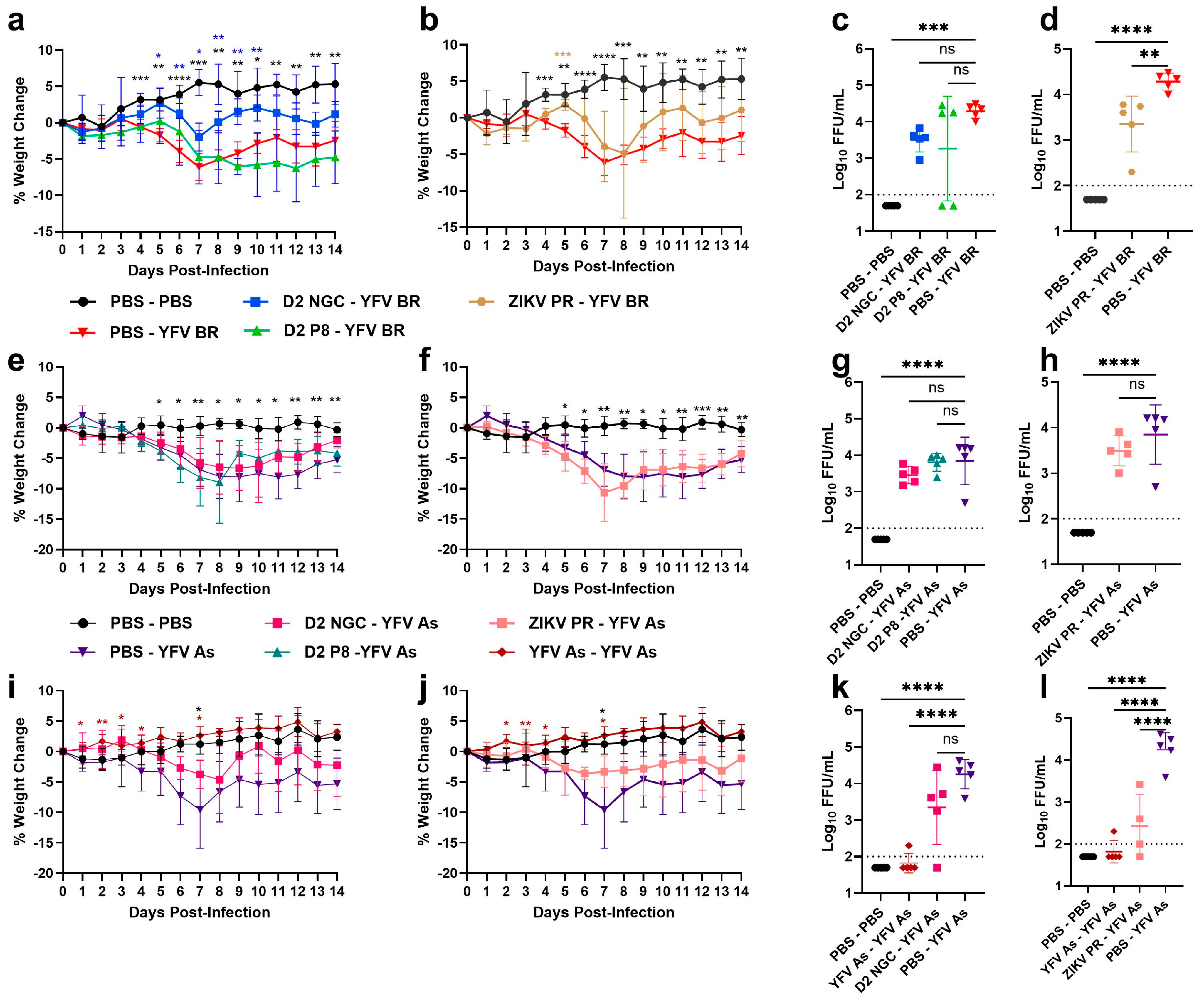
3.1. Prior Dengue-2 and Zika Virus Immunity Generally Suppresses Yellow Fever Viremia and Viral Loads in A129 Mice
3.2. CD4+ T Cells in DENV-2/ZIKV-Immune Mice Play Minimal Roles in Cross-Protection against YFV Infection
3.3. CD8+ T Cells in DENV-2/ZIKV-Immune Mice Play Little to No Role in Suppressing YFV Viremia
3.4. Antibodies Contribute to Cross-Protection in Sequential DENV-2/ZIKV and YFV Infections
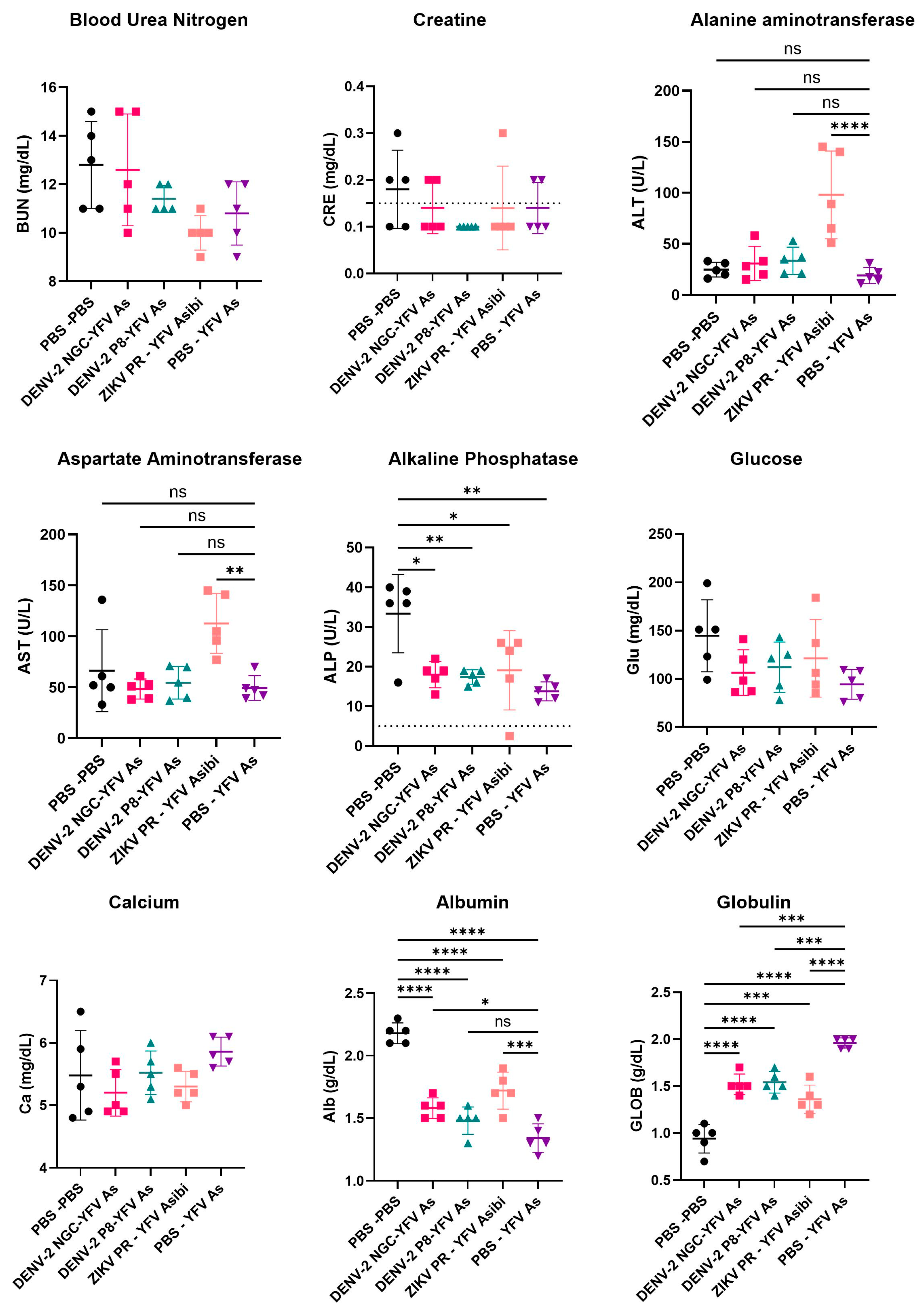
3.5. Serum Chemistries of YFV Asibi-Infected Mice with Varying Flavivirus Immune Profiles Generally Demonstrate No Significant Differences
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leta, S.; Beyene, T.J.; De Clercq, E.M.; Amenu, K.; Revie, C.W.; Kraemer, M.U.G. Global risk mapping for major diseases transmitted by Aedes aegypti and Aedes albopictus. Int. J. Infect. Dis. 2018, 67, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Flamand, C.; Bailly, S.; Fritzell, C.; Berthelot, L.; Vanhomwegen, J.; Salje, H.; Paireau, J.; Matheus, S.; Enfissi, A.; Fernandes-Pellerin, S.; et al. Impact of Zika Virus Emergence in French Guiana: A Large General Population Seroprevalence Survey. J. Infect. Dis. 2019, 220, 1915–1925. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, P.M.S.; Nurtop, E.; Pastorino, B.; Roca, Y.; Drexler, J.F.; Gallian, P.; Jaenisch, T.; Leparc-Goffart, I.; Priet, S.; Ninove, L.; et al. Zika virus epidemiology in Bolivia: A seroprevalence study in volunteer blood donors. PLoS Negl. Trop. Dis. 2018, 12, e0006239. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.K.; Rossi, S.L.; Tesh, R.B.; Weaver, S.C. Zoonotic mosquito-borne arboviruses: Spillover, spillback, and realistic mitigation strategies. Sci. Transl. Med. 2023, 15, eadj2166. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.; Koch, R.T.; Bialosuknia, S.M.; Ngo, K.; Zink, S.D.; Koetzner, C.A.; Maffei, J.G.; Dupuis, A.P.; Backenson, P.B.; Oliver, J.; et al. Dynamics of eastern equine encephalitis virus during the 2019 outbreak in the Northeast United States. Curr. Biol. 2023, 33, 2515–2527.e6. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Forrester, N.L. Chikungunya: Evolutionary history and recent epidemic spread. Antivir. Res. 2015, 120, 32–39. [Google Scholar] [CrossRef]
- Weaver, S.C.; Charlier, C.; Vasilakis, N.; Lecuit, M. Zika, Chikungunya, and Other Emerging Vector-Borne Viral Diseases. Annu. Rev. Med. 2018, 69, 395. [Google Scholar] [CrossRef] [PubMed]
- de Souza, W.M.; Ribeiro, G.S.; de Lima, S.T.S.; de Jesus, R.; Moreira, F.R.R.; Whittaker, C.; Sallum, M.A.M.; Carrington, C.V.F.; Sabino, E.C.; Kitron, U.; et al. Chikungunya: A decade of burden in the Americas. Lancet Reg. Health Am. 2024, 30, 100673. [Google Scholar] [CrossRef]
- Ceconi, M.; Ariën, K.K.; Delputte, P. Diagnosing Arthropod-Borne Flaviviruses: Non-Structural Protein 1 (NS1) as a Biomarker. Trends in Microbiology. 2023. Available online: https://www.sciencedirect.com/science/article/pii/S0966842X23003347 (accessed on 11 March 2024).
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef]
- Monath, T.P. The Arboviruses: Epidemiology and Ecology; CRC Press: Boca Raton, FL, USA, 2021; Volume V. [Google Scholar]
- Shinde, D.P.; Plante, J.A.; Plante, K.S.; Weaver, S.C. Yellow Fever: Roles of Animal Models and Arthropod Vector Studies in Understanding Epidemic Emergence. Microorganisms 2022, 10, 1578. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.N.; Harbach, R.E. History of the discovery of the mode of transmission of yellow fever virus. J. Vector Ecol. J. Soc. Vector Ecol. 2017, 42, 208–222. [Google Scholar] [CrossRef]
- Mutebi, J.-P.; Barrett, A.D.T. The epidemiology of yellow fever in Africa. Microbes Infect. 2002, 4, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.E.; Holmes, E.C.; Barrett, A.D.T. Out of Africa: A molecular perspective on the introduction of yellow fever virus into the Americas. PLOS Pathog. 2007, 3, e75. [Google Scholar] [CrossRef]
- Lwande, O.W.; Obanda, V.; Lindström, A.; Ahlm, C.; Evander, M.; Näslund, J.; Bucht, G. Globe-Trotting Aedes aegypti and Aedes albopictus: Risk Factors for Arbovirus Pandemics. Vector-Borne Zoonotic Dis. 2020, 20, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G. The absence of yellow fever in Asia: History, hypotheses, vector dispersal, possibility of YF in Asia, and other enigmas. Viruses 2020, 12, 1349. [Google Scholar] [CrossRef]
- Wasserman, S.; Tambyah, P.A.; Lim, P.L. Yellow fever cases in Asia: Primed for an epidemic. Int. J. Infect. Dis. 2016, 48, 98–103. [Google Scholar] [CrossRef]
- Agampodi, S.B.; Wickramage, K. Is there a risk of yellow fever virus transmission in South Asian countries with hyperendemic dengue? BioMed Res. Int. 2013, 2013, e905043. [Google Scholar] [CrossRef] [PubMed]
- de Abreu, F.V.S.; Ribeiro, I.P.; Ferreira-de-Brito, A.; dos Santos, A.A.C.; de Miranda, R.M.; Bonelly, I.D.S.; Neves, M.S.A.S.; Bersot, M.I.; dos Santos, T.P.; Gomes, M.Q.; et al. Haemagogus leucocelaenus and Haemagogus janthinomys are the primary vectors in the major yellow fever outbreak in Brazil, 2016–2018. Emerg. Microbes Infect. 2019, 8, 218–231. [Google Scholar] [CrossRef]
- Couto-Lima, D.; Madec, Y.; Bersot, M.I.; Campos, S.S.; de Albuquerque Motta, M.; dos Santos, F.B.; Vazeille, M.; Vasconcelos, P.F.D.C.; Lourenço-de-Oliveira, R.; Failloux, A.-B. Potential risk of re-emergence of urban transmission of Yellow Fever virus in Brazil facilitated by competent Aedes populations. Sci. Rep. 2017, 7, 4848. [Google Scholar] [CrossRef]
- Silva, N.I.O.; Sacchetto, L.; de Rezende, I.M.; Trindade, G.D.S.; LaBeaud, A.D.; de Thoisy, B.; Drumond, B.P. Recent sylvatic yellow fever virus transmission in Brazil: The news from an old disease. Virol. J. 2020, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Possas, C.; Lourenço-de-Oliveira, R.; Tauil, P.L.; Pinheiro, F.D.P.; Pissinatti, A.; Cunha, R.V.D.; Freire, M.; Martins, R.M.; Homma, A. Yellow fever outbreak in Brazil: The puzzle of rapid viral spread and challenges for immunisation. Mem. Inst. Oswaldo Cruz 2018, 113, e180278. [Google Scholar] [CrossRef] [PubMed]
- Theiler, M.; Anderson, C.R. The Relative Resistance of Dengue-Immune Monkeys to Yellow Fever Virus. Am. J. Trop. Med. Hyg. 1975, 24, 115–117. [Google Scholar] [CrossRef] [PubMed]
- Lataillade, L.D.G.D.; Vazeille, M.; Obadia, T.; Madec, Y.; Mousson, L.; Kamgang, B.; Chen, C.-H.; Failloux, A.-B.; Yen, P.-S. Risk of yellow fever virus transmission in the Asia-Pacific region. Nat. Commun. 2020, 11, 5801. [Google Scholar] [CrossRef] [PubMed]
- Duong, V.; Dussart, P.; Buchy, P. Zika virus in Asia. Int. J. Infect. Dis. 2017, 54, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Dick, G.W.A.; Kitchen, S.F.; Haddow, A.J. Zika Virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Ikejezie, J.; Shapiro, C.N.; Kim, J.; Chiu, M.; Almiron, M.; Ugarte, C.; Espinal, M.A.; Aldighieri, S. Zika Virus Transmission—Region of the Americas, May 15, 2015–December 15, 2016. Morb. Mortal. Wkly. Rep. 2017, 66, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Aliota, M.T.; Bassit, L.; Bradrick, S.S.; Cox, B.; Garcia-Blanco, M.A.; Gavegnano, C.; Friedrich, T.C.; Golos, T.G.; Griffin, D.E.; Haddow, A.D.; et al. Zika in the Americas, year 2: What have we learned? What gaps remain? A report from the Global Virus Network. Antivir. Res. 2017, 144, 223–246. [Google Scholar] [CrossRef] [PubMed]
- Shinde, D.P.; Plante, J.A.; Scharton, D.; Mitchell, B.; Walker, J.; Azar, S.R.; Campos, R.K.; Sacchetto, L.; Drumond, B.P.; Vasilakis, N.; et al. Yellow Fever Emergence: Role of Heterologous Flavivirus Immunity in Preventing Urban Transmission. 2024. Available online: https://www.biorxiv.org/content/10.1101/2024.03.03.583168v1 (accessed on 11 March 2024).
- Dejnirattisai, W.; Supasa, P.; Wongwiwat, W.; Rouvinski, A.; Barba-Spaeth, G.; Duangchinda, T.; Sakuntabhai, A.; Cao-Lormeau, V.-M.; Malasit, P.; Rey, F.A.; et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 2016, 17, 1102–1108. [Google Scholar] [CrossRef]
- Piauilino, I.C.R.; Souza, R.K.D.S.; Lima, M.T.; Rodrigues, Y.K.B.; da Silva, L.F.A.; Gouveia, A.S.; Neto, A.V.D.S.; Chaves, B.A.; Alecrim, M.D.G.C.; de Menezes, C.H.A.B.; et al. Does the Presence or a High Titer of Yellow Fever Virus Antibodies Interfere with Pregnancy Outcomes in Women with Zika Virus Infection? Viruses 2023, 15, 2244. [Google Scholar] [CrossRef]
- Rodriguez-Barraquer, I.; Costa, F.; Nascimento, E.J.M.; Júnior, N.N.; Castanha, P.M.S.; Sacramento, G.A.; Cruz, J.; Carvalho, M.; De Olivera, D.; Hagan, J.E.; et al. Impact of preexisting dengue immunity on Zika virus emergence in a dengue endemic region. Science 2019, 363, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Regla-Nava, J.A.; Wang, Y.-T.; Fontes-Garfias, C.R.; Liu, Y.; Syed, T.; Susantono, M.; Gonzalez, A.; Viramontes, K.M.; Verma, S.K.; Kim, K.; et al. A Zika virus mutation enhances transmission potential and confers escape from protective dengue virus immunity. Cell Rep. 2022, 39, 110655. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Wang, Y.-T.; Valentine, K.M.; Dos Santos Alves, R.P.; Xu, Z.; Regla-Nava, J.A.; Ngono, A.E.; Young, M.P.; Ferreira, L.C.S.; Shresta, S. CD4+ T Cells Cross-Reactive with Dengue and Zika Viruses Protect against Zika Virus Infection. Cell Rep. 2020, 31, 107566. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Elong Ngono, A.; Regla-Nava, J.A.; Kim, K.; Gorman, M.J.; Diamond, M.S.; Shresta, S. Dengue virus-reactive CD8+ T cells mediate cross-protection against subsequent Zika virus challenge. Nat. Commun. 2017, 8, 1459. [Google Scholar] [CrossRef] [PubMed]
- Turtle, L.; Tatullo, F.; Bali, T.; Ravi, V.; Soni, M.; Chan, S.; Chib, S.; Venkataswamy, M.M.; Fadnis, P.; Yaïch, M.; et al. Cellular Immune Responses to Live Attenuated Japanese Encephalitis (JE) Vaccine SA14-14-2 in Adults in a JE/Dengue Co-Endemic Area. PLoS Negl. Trop. Dis. 2017, 11, e0005263. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Rothman, A.L.; Potts, J.; Guirakhoo, F.; Ennis, F.A.; Green, S. Sequential Immunization with Heterologous Chimeric Flaviviruses Induces Broad-Spectrum Cross-Reactive CD8+ T Cell Responses. J. Infect. Dis. 2010, 202, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhen, Z.; Turtle, L.; Hou, B.; Li, Y.; Wu, N.; Gao, N.; Fan, D.; Chen, H.; An, J. T cell immunity rather than antibody mediates cross-protection against Zika virus infection conferred by a live attenuated Japanese encephalitis SA14-14-2 vaccine. Appl. Microbiol. Biotechnol. 2020, 104, 6779–6789. [Google Scholar] [CrossRef]
- Subramaniam, K.S.; Lant, S.; Goodwin, L.; Grifoni, A.; Weiskopf, D.; Turtle, L. Two Is Better Than One: Evidence for T-Cell Cross-Protection between Dengue and Zika and Implications on Vaccine Design. Front. Immunol. 2020, 11, 517. [Google Scholar] [CrossRef]
- Meier, K.C.; Gardner, C.L.; Khoretonenko, M.V.; Klimstra, W.B.; Ryman, K.D. A mouse model for studying viscerotropic disease caused by yellow fever virus infection. PLoS Pathog. 2009, 5, e1000614. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shinde, D.P.; Walker, J.; Reyna, R.A.; Scharton, D.; Mitchell, B.; Dulaney, E.; Bonam, S.R.; Hu, H.; Plante, J.A.; Plante, K.S.; et al. Mechanisms of Flavivirus Cross-Protection against Yellow Fever in a Mouse Model. Viruses 2024, 16, 836. https://doi.org/10.3390/v16060836
Shinde DP, Walker J, Reyna RA, Scharton D, Mitchell B, Dulaney E, Bonam SR, Hu H, Plante JA, Plante KS, et al. Mechanisms of Flavivirus Cross-Protection against Yellow Fever in a Mouse Model. Viruses. 2024; 16(6):836. https://doi.org/10.3390/v16060836
Chicago/Turabian StyleShinde, Divya P., Jordyn Walker, Rachel A. Reyna, Dionna Scharton, Brooke Mitchell, Ennid Dulaney, Srinivisa Reddy Bonam, Haitao Hu, Jessica A. Plante, Kenneth S. Plante, and et al. 2024. "Mechanisms of Flavivirus Cross-Protection against Yellow Fever in a Mouse Model" Viruses 16, no. 6: 836. https://doi.org/10.3390/v16060836
APA StyleShinde, D. P., Walker, J., Reyna, R. A., Scharton, D., Mitchell, B., Dulaney, E., Bonam, S. R., Hu, H., Plante, J. A., Plante, K. S., & Weaver, S. C. (2024). Mechanisms of Flavivirus Cross-Protection against Yellow Fever in a Mouse Model. Viruses, 16(6), 836. https://doi.org/10.3390/v16060836






