Detection and Characterization of Influenza A Virus Endemic Circulation in Suckling and Nursery Pigs Originating from Vaccinated Farms in the Same Production System
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. Influenza A Virus RNA Detection and Subtyping
2.3. Detection of Pandemic or North American TRIG Matrix Genes
2.4. Influenza A Virus Isolation
2.5. Influenza A Virus Complete Genome Sequencing
2.6. Influenza A Virus Phylogenetic Analysis
3. Results
3.1. Influenza A Virus Respiratory Clinical Signs
3.2. Detection of Influenza A Virus RNA in Suckling Pig Nasal Swabs
3.3. Detection of Influenza A Virus RNA in Nursery Pig Oral Fluids
3.4. Detection of Pandemic or North American TRIG Matrix
3.5. Influenza A Virus Isolation
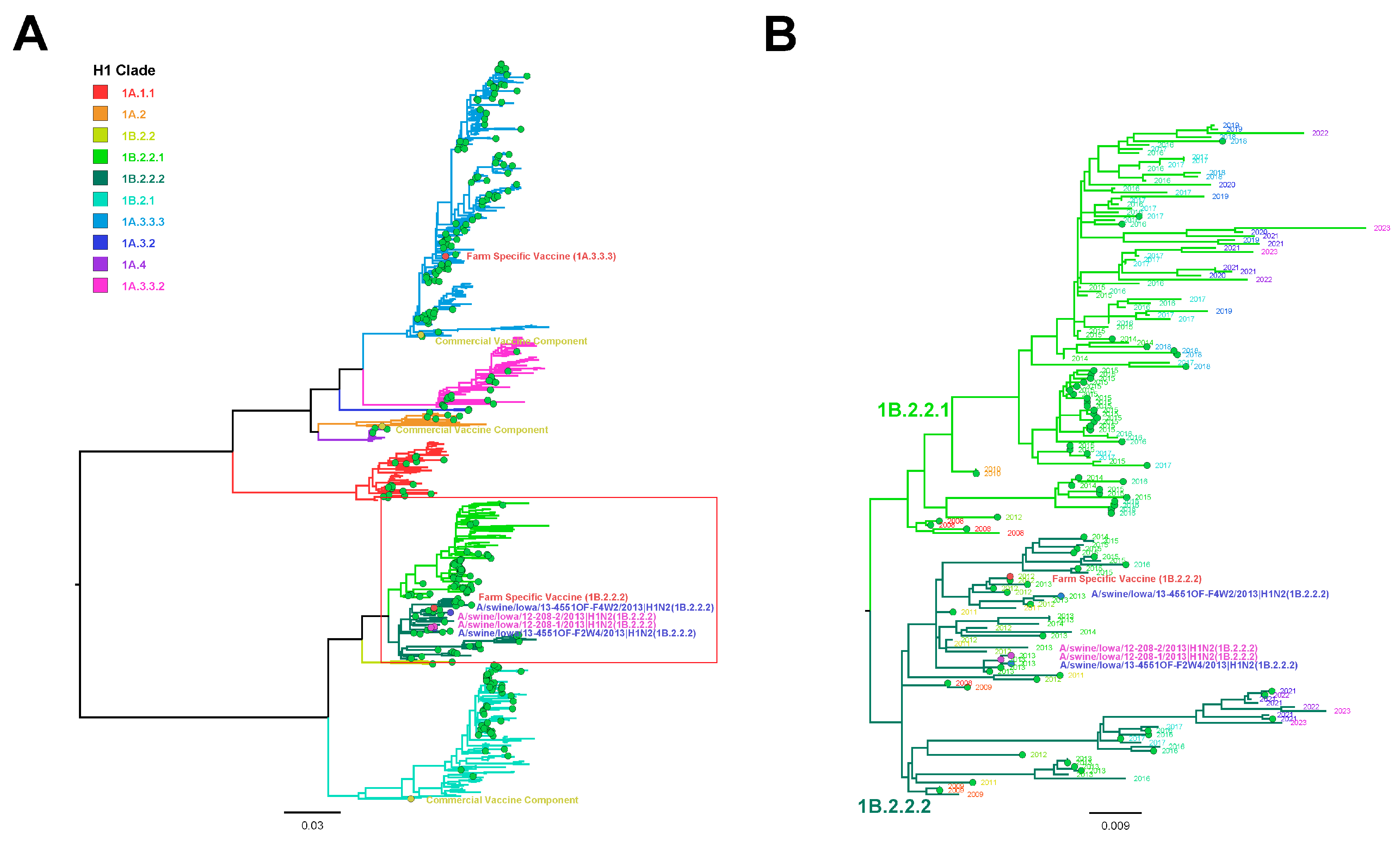
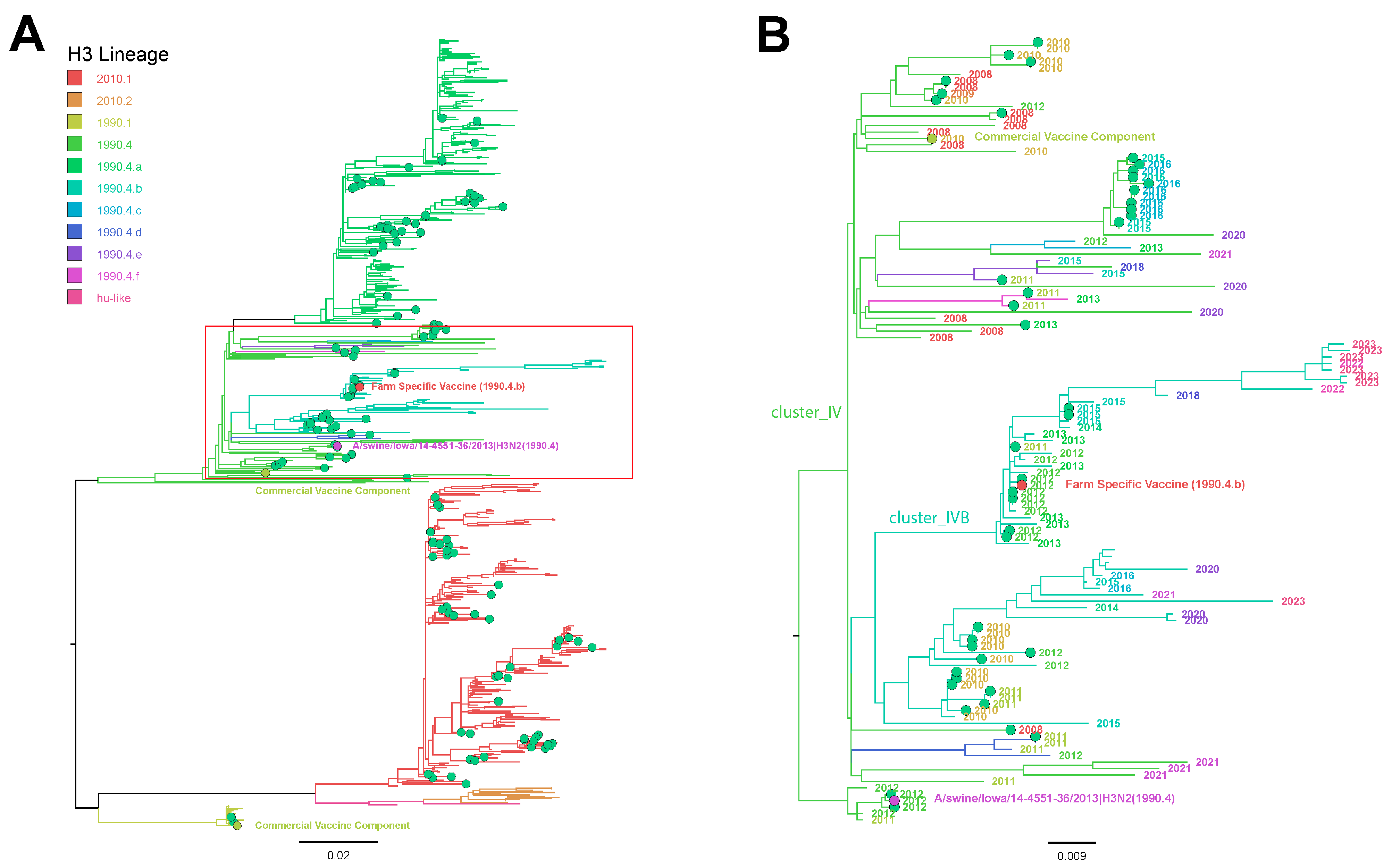
3.6. Influenza A Virus Gene Sequencing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ellis, J.; Clark, E.; Haines, D.; West, K.; Krakowka, S.; Kennedy, S.; Allan, G.M. Porcine circovirus-2 and concurrent infections in the field. Vet. Microbiol. 2004, 98, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.J.; Brown, I.H. Recent zoonoses caused by influenza A viruses. Rev. Sci. Tech. 2000, 19, 197–225. [Google Scholar] [CrossRef]
- Richt, J.A.; Lager, K.M.; Janke, B.H.; Woods, R.D.; Webster, R.G.; Webby, R.J. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J. Clin. Microbiol. 2003, 41, 3198–3205. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Richt, J.A. Swine influenza viruses a North American perspective. Adv. Virus Res. 2008, 72, 127–154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.N.; Senne, D.A.; Landgraf, J.S.; Swenson, S.L.; Erickson, G.; Rossow, K.; Liu, L.; Yoon, K.; Krauss, S.; Webster, R.G. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 1999, 73, 8851–8856. [Google Scholar] [CrossRef]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Gramer, M.R.; Richt, J.A.; Janke, B.H. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes. 2009, 39, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Anderson, T.K.; Chang, J.; Arendsee, Z.W.; Venkatesh, D.; Souza, C.K.; Kimble, J.B.; Lewis, N.S.; Davis, C.T.; Vincent, A.L. Swine Influenza A Viruses and the Tangled Relationship with Humans. Cold Spring Harb. Perspect. Med. 2021, 11, a038737. [Google Scholar] [CrossRef] [PubMed]
- Kitikoon, P.; Nelson, M.I.; Killian, M.L.; Anderson, T.K.; Koster, L.; Culhane, M.R.; Vincent, A.L. Genotype patterns of contemporary reassorted H3N2 virus in US swine. J. Gen. Virol. 2013, 94, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Rajao, D.S.; Vincent, A.L.; Perez, D.R. Adaptation of Human Influenza Viruses to Swine. Front. Vet. Sci. 2018, 5, 347. [Google Scholar] [CrossRef]
- Vincent, A.L.; Anderson, T.K.; Lager, K.M. A Brief Introduction to Influenza A Virus in Swine. Methods Mol. Biol. 2020, 2123, 249–271. [Google Scholar] [CrossRef]
- Walia, R.R.; Anderson, T.K.; Vincent, A.L. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses 2019, 13, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Chamba Pardo, F.O.; Schelkopf, A.; Allerson, M.; Morrison, R.; Culhane, M.; Perez, A.; Torremorell, M. Breed-to-wean farm factors associated with influenza A virus infection in piglets at weaning. Prev. Vet. Med. 2018, 161, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Perez, D.R.; Rajao, D.; Anderson, T.K.; Abente, E.J.; Walia, R.R.; Lewis, N.S. Influenza A virus vaccines for swine. Vet. Microbiol. 2017, 206, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Wymore Brand, M.; Anderson, T.K.; Kitikoon, P.; Brian Kimble, J.; Otis, N.; Gauger, P.C.; Souza, C.K.; Kaplan, B.; Mogler, M.; Strait, E.; et al. Bivalent hemagglutinin and neuraminidase influenza replicon particle vaccines protect pigs against influenza a virus without causing vaccine associated enhanced respiratory disease. Vaccine 2022, 40, 5569–5578. [Google Scholar] [CrossRef] [PubMed]
- Vander Veen, R.L.; Loynachan, A.T.; Mogler, M.A.; Russell, B.J.; Harris, D.L.; Kamrud, K.I. Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine 2012, 30, 1944–1950. [Google Scholar] [CrossRef] [PubMed]
- Vander Veen, R.L.; Harris, D.L.; Kamrud, K.I. Alphavirus replicon vaccines. Anim. Health Res. Rev. 2012, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chase, C.C. Autogenous vaccines: Current use in the field in the U.S. cattle and hog industry. Dev. Biol. 2004, 117, 69–71. [Google Scholar]
- Allerson, M.; Deen, J.; Detmer, S.E.; Gramer, M.R.; Joo, H.S.; Romagosa, A.; Torremorell, M. The impact of maternally derived immunity on influenza A virus transmission in neonatal pig populations. Vaccine 2013, 31, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Kitikoon, P.; Nilubol, D.; Erickson, B.J.; Janke, B.H.; Hoover, T.C.; Sornsen, S.A.; Thacker, E.L. The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet. Immunol. Immunopathol. 2006, 112, 117–128. [Google Scholar] [CrossRef]
- Loeffen, W.L.; Heinen, P.P.; Bianchi, A.T.; Hunneman, W.A.; Verheijden, J.H. Effect of maternally derived antibodies on the clinical signs and immune response in pigs after primary and secondary infection with an influenza H1N1 virus. Vet. Immunol. Immunopathol. 2003, 92, 23–35. [Google Scholar] [CrossRef]
- Rajao, D.S.; Sandbulte, M.R.; Gauger, P.C.; Kitikoon, P.; Platt, R.; Roth, J.A.; Perez, D.R.; Loving, C.L.; Vincent, A.L. Heterologous challenge in the presence of maternally-derived antibodies results in vaccine-associated enhanced respiratory disease in weaned piglets. Virology 2016, 491, 79–88. [Google Scholar] [CrossRef]
- White, L.A.; Torremorell, M.; Craft, M.E. Influenza A virus in swine breeding herds: Combination of vaccination and biosecurity practices can reduce likelihood of endemic piglet reservoir. Prev. Vet. Med. 2017, 138, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Corzo, C.A.; Culhane, M.; Dee, S.; Morrison, R.B.; Torremorell, M. Airborne detection and quantification of swine influenza a virus in air samples collected inside, outside and downwind from swine barns. PLoS ONE 2013, 8, e71444. [Google Scholar] [CrossRef]
- Diaz, A.; Perez, A.; Sreevatsan, S.; Davies, P.; Culhane, M.; Torremorell, M. Association between Influenza A Virus Infection and Pigs Subpopulations in Endemically Infected Breeding Herds. PLoS ONE 2015, 10, e0129213. [Google Scholar] [CrossRef]
- Allerson, M.W.; Davies, P.R.; Gramer, M.R.; Torremorell, M. Infection dynamics of pandemic 2009 H1N1 influenza virus in a two-site swine herd. Transbound. Emerg. Dis. 2014, 61, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Marthaler, D.; Culhane, M.; Sreevatsan, S.; Alkhamis, M.; Torremorell, M. Complete Genome Sequencing of Influenza A Viruses within Swine Farrow-to-Wean Farms Reveals the Emergence, Persistence, and Subsidence of Diverse Viral Genotypes. J. Virol. 2017, 91, e00745-17. [Google Scholar] [CrossRef]
- Nelson, M.I.; Viboud, C.; Vincent, A.L.; Culhane, M.R.; Detmer, S.E.; Wentworth, D.E.; Rambaut, A.; Suchard, M.A.; Holmes, E.C.; Lemey, P. Global migration of influenza A viruses in swine. Nat. Commun. 2015, 6, 6696. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.H. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 2000, 74, 29–46. [Google Scholar] [CrossRef]
- Chamba Pardo, F.O.; Wayne, S.; Culhane, M.R.; Perez, A.; Allerson, M.; Torremorell, M. Effect of strain-specific maternally-derived antibodies on influenza A virus infection dynamics in nursery pigs. PLoS ONE 2019, 14, e0210700. [Google Scholar] [CrossRef]
- CPardo, F.O.C.; Allerson, M.W.; Culhane, M.R.; Morrison, R.B.; Davies, P.R.; Perez, A.; Torremorell, M. Effect of influenza A virus sow vaccination on infection in pigs at weaning: A prospective longitudinal study. Transbound. Emerg. Dis. 2021, 68, 183–193. [Google Scholar] [CrossRef]
- Prickett, J.R.; Zimmerman, J.J. The development of oral fluid-based diagnostics and applications in veterinary medicine. Anim. Health Res. Rev. 2010, 11, 207–216. [Google Scholar] [CrossRef]
- Harmon, K.; Bower, L.; Kim, W.I.; Pentella, M.; Yoon, K.J. A matrix gene-based multiplex real-time RT-PCR for detection and differentiation of 2009 pandemic H1N1 and other influenza A viruses in North America. Influenza Other Respir Viruses 2010, 4, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gauger, P.C. Isolation of swine influenza virus in cell cultures and embryonated chicken eggs. Methods Mol. Biol. 2014, 1161, 265–276. [Google Scholar] [CrossRef]
- Bowman, A.S.; Sreevatsan, S.; Killian, M.L.; Page, S.L.; Nelson, S.W.; Nolting, J.M.; Cardona, C.; Slemons, R.D. Molecular evidence for interspecies transmission of H3N2pM/H3N2v influenza A viruses at an Ohio agricultural fair, July 2012. Emerg. Microbes Infect. 2012, 1, e33. [Google Scholar] [CrossRef]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 475, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Anderson, T.K.; Nelson, M.I.; Kitikoon, P.; Swenson, S.L.; Korslund, J.A.; Vincent, A.L. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 2013, 7 (Suppl. S4), 42–51. [Google Scholar] [CrossRef]
- Beaudoin, A.; Johnson, S.; Davies, P.; Bender, J.; Gramer, M. Characterization of influenza a outbreaks in Minnesota swine herds and measures taken to reduce the risk of zoonotic transmission. Zoonoses Public. Health 2012, 59, 96–106. [Google Scholar] [CrossRef]
- Lopez-Moreno, G.; Culhane, M.R.; Davies, P.; Corzo, C.; Allerson, M.W.; Torremorell, M. Farm management practices associated with influenza A virus contamination of people working in Midwestern United States swine farms. Porc. Health Manag. 2023, 9, 13. [Google Scholar] [CrossRef]
- Pitzer, V.E.; Aguas, R.; Riley, S.; Loeffen, W.L.; Wood, J.L.; Grenfell, B.T. High turnover drives prolonged persistence of influenza in managed pig herds. J. R. Soc. Interface 2016, 13, 20160138. [Google Scholar] [CrossRef]
- Reynolds, J.J.; Torremorell, M.; Craft, M.E. Mathematical modeling of influenza A virus dynamics within swine farms and the effects of vaccination. PLoS ONE 2014, 9, e106177. [Google Scholar] [CrossRef]
- Simon-Grifé, M.; Martín-Valls, G.E.; Vilar, M.J.; Busquets, N.; Mora-Salvatierra, M.; Bestebroer, T.M.; Fouchier, R.A.; Martín, M.; Mateu, E.; Casal, J. Swine influenza virus infection dynamics in two pig farms; results of a longitudinal assessment. Vet. Res. 2012, 43, 24. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.N.; Zhang, M.; Zimmerman, J.J.; Holtkamp, D.J.; Linhares, D.C.L. Finding PRRSV in sow herds: Family oral fluids vs. serum samples from due-to-wean pigs. Prev. Vet. Med. 2021, 193, 105397. [Google Scholar] [CrossRef]
- Garrido-Mantilla, J.; Alvarez, J.; Culhane, M.; Nirmala, J.; Cano, J.P.; Torremorell, M. Comparison of individual, group and environmental sampling strategies to conduct influenza surveillance in pigs. BMC Vet. Res. 2019, 15, 61. [Google Scholar] [CrossRef]
- Rose, N.; Herve, S.; Eveno, E.; Barbier, N.; Eono, F.; Dorenlor, V.; Andraud, M.; Camsusou, C.; Madec, F.; Simon, G. Dynamics of influenza A virus infections in permanently infected pig farms: Evidence of recurrent infections, circulation of several swine influenza viruses and reassortment events. Vet. Res. 2013, 44, 72. [Google Scholar] [CrossRef]
- Sandbulte, M.R.; Spickler, A.R.; Zaabel, P.K.; Roth, J.A. Optimal Use of Vaccines for Control of Influenza A Virus in Swine. Vaccines 2015, 3, 22–73. [Google Scholar] [CrossRef] [PubMed]
- Meiners, C.; Loesken, S.; Doehring, S.; Starick, E.; Pesch, S.; Maas, A.; Noe, T.; Beer, M.; Harder, T.; Grosse Beilage, E. Field study on swine influenza virus (SIV) infection in weaner pigs and sows. Tierarztl. Prax. Ausg. G. Grosstiere Nutztiere 2014, 42, 351–359. [Google Scholar] [CrossRef]
- Janke, B.H. Clinicopathological features of Swine influenza. Curr. Top. Microbiol. Immunol. 2013, 370, 69–83. [Google Scholar] [CrossRef]
- Prickett, J.R.; Kim, W.; Simer, R.; Yoon, K.J.; Zimmerman, J. Oral-fluid samples for surveillance of commercial growing pigs for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 infections. J. Swine Health Prod. 2008, 16, 6. [Google Scholar]
- Wills, R.W.; Zimmerman, J.J.; Yoon, K.J.; Swenson, S.L.; Hoffman, L.J.; McGinley, M.J.; Hill, H.T.; Platt, K.B. Porcine reproductive and respiratory syndrome virus: Routes of excretion. Vet. Microbiol. 1997, 57, 69–81. [Google Scholar] [CrossRef]
- Detmer, S.E.; Patnayak, D.P.; Jiang, Y.; Gramer, M.R.; Goyal, S.M. Detection of Influenza A virus in porcine oral fluid samples. J. Vet. Diagn. Invest. 2011, 23, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Vander Veen, R.; Kamrud, K.; Mogler, M.; Loynachan, A.T.; McVicker, J.; Berglund, P.; Owens, G.; Timberlake, S.; Lewis, W.; Smith, J.; et al. Rapid development of an efficacious swine vaccine for novel H1N1. PLoS Curr. 2009, 1, Rrn1123. [Google Scholar] [CrossRef]
- Vander Veen, R.L.; Mogler, M.A.; Russell, B.J.; Loynachan, A.T.; Harris, D.L.; Kamrud, K.I. Haemagglutinin and nucleoprotein replicon particle vaccination of swine protects against the pandemic H1N1 2009 virus. Vet. Rec. 2013, 173, 344. [Google Scholar] [CrossRef] [PubMed]
- Ducatez, M.F.; Hause, B.; Stigger-Rosser, E.; Darnell, D.; Corzo, C.; Juleen, K.; Simonson, R.; Brockwell-Staats, C.; Rubrum, A.; Wang, D.; et al. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg. Infect. Dis. 2011, 17, 1624–1629. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Anderson, T.K.; Walia, R.R.; Dorman, K.S.; Janas-Martindale, A.; Vincent, A.L. The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol 2017, 98, 2001–2010. [Google Scholar] [CrossRef]
- Nelson, M.I.; Gramer, M.R.; Vincent, A.L.; Holmes, E.C. Global transmission of influenza viruses from humans to swine. J. Gen. Virol. 2012, 93, 2195–2203. [Google Scholar] [CrossRef]
- Nelson, M.I.; Vincent, A.L. Reverse zoonosis of influenza to swine: New perspectives on the human-animal interface. Trends Microbiol. 2015, 23, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Epperson, S.; Jhung, M.; Richards, S.; Quinlisk, P.; Ball, L.; Moll, M.; Boulton, R.; Haddy, L.; Biggerstaff, M.; Brammer, L.; et al. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin. Infect. Dis. 2013, 57 (Suppl. S1), S4–S11. [Google Scholar] [CrossRef]
- Schicker, R.S.; Rossow, J.; Eckel, S.; Fisher, N.; Bidol, S.; Tatham, L.; Matthews-Greer, J.; Sohner, K.; Bowman, A.S.; Avrill, J.; et al. Outbreak of Influenza A(H3N2) Variant Virus Infections Among Persons Attending Agricultural Fairs Housing Infected Swine—Michigan and Ohio, July–August 2016. Morb. Mortal. Wkly. Rep. 2016, 65, 1157–1160. [Google Scholar] [CrossRef]
| Sample Collection | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Collection Date | 11–15 March | 25–29 March | 8–12 April | 22–26 April | 6–9 May | 8–12 July | 22–26 July | 5–9 August |
| Vaccine Dose | Prime dose | Prime dose | Boost dose | Boost dose | ||||
| Administration Date | March 4 | March 25 | April 1 | April 15 | ||||
| Farms Vaccinated | F1, F2, F3 | F4 * | F1, F3 | F2 | ||||
| Group | Location | Age (Days) | Sample Type | Samples/Farm | # of Farms | # of Samplings | Total Samples |
|---|---|---|---|---|---|---|---|
| Suckling Pigs | Farrowing | 12–17 | Nasal Swabs | 135 | 4 | 8 | 4320 |
| Nursery Pigs | Nursery | 28–35 | Oral Fluids | 3–8 | 4 | 8 | 158 |
| Farm 1 | Farm 2 | Farm 3 | Farm 4 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H3 | H1/H3 | H1 | H3 | H1/H3 | H1 | H3 | H1/H3 | H1 | H3 | H1/H3 | |||||||||
| Time | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) |
| 1 | - | - | - | - | - | 38/135 (28.1) 5 * | 29.6 [0.6] | 3/135 (2.2) 29 * | 34.4 [0.4] | 11/135 (8.1) 2 * | 1/135 (0.7) 1 * | 31.7 NA | - | - | - | 2/135 (1.5) | 33.8 [1.1] | - | - | - |
| 2 | 6/135 (4.4) | 32.6 [0.6] | - | - | - | 8/135 (6.0) 6 * | 33.3 [0.6] | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 6 | 2/135 (1.5) 1 * | 33.3 [0.1] | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 * | NA | - |
| 7 | 2 * | NA | 1 * | NA | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Pos/ Farm | 8/1080 (0.7) | 32.8 [0.4] | - | - | - | 46/1080 (4.2) | 30.2 [0.5] | 3/1080 (0.3) | 34.4 [0.8] | 11/1080 (1.0) | 1/1080 (0.1) | 31.7 NA | - | - | - | 2/1080 (0.2) | 33.8 [1.1] | - | - | - |
| Farm 1 | Farm 2 | Farm 3 | Farm 4 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | N1/N2 | N1 | N2 | N1/N2 | N1 | N2 | N1/N2 | N1 | N2 | N1/N2 | |||||||||
| Time | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) | Mean Ct [SEM] | #/# Pos. (%) |
| 1 | - | - | - | - | - | 15 * | NA | 54/135 (28.1) | 29.7 [0.5] | - | - | - | 2/135 (1.5) | 32.8 [1.9] | - | - | - | 2/135 (1.5) | 33.4 [0.9] | - |
| 2 | - | - | 6/135 (4.4) | 32.0 [0.5] | - | 1 * | NA | 10/135 (7.4) 6 * | 32.9 [0.4] | - | - | - | - | - | - | - | - | - | - | - |
| 3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 6 | - | - | 2/135 (1.5) 1 * | 32.9 [0.5] | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Pos/ Farm | - | - | 8/1080 (0.7) | 30.2 [0.4] | - | - | - | 64/1080 (5.9) | 30.2 [0.4] | - | - | - | 2/1080 (0.2) | 32.8 [1.9] | - | - | - | 2/1080 (0.2) | 33.4 [0.9] | - |
| Farm 1 | Farm 2 | Farm 3 | Farm 4 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H3 | H1/H3 | H1 | H3 | H1/H3 | H1 | H3 | H1/H3 | H1 | H3 | H1/H3 | |||||||||
| Time | #/# Pos. (%) | Mean Ct (SEM) | #/# Pos. (%) | Mean Ct (SEM) | #/# Pos. (%) | #/# Pos. (%) | Mean Ct (SEM) | #/# Pos. (%) | Mean Ct (SEM) | #/# Pos. (%) | #/# Pos. (%) | Mean Ct (SEM) | #/# Pos. (%) | Mean Ct (SEM) | #/# Pos. (%) | #/# Pos. (%) | Mean Ct (SEM) | #/# Pos. (%) | Mean Ct (SEM) | #/# Pos. (%) |
| 1 | - | - | 1 * | NA | - | 4/8 (50.0) 1 * | 34.0 [0.3] | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 2 | - | - | - | - | - | 1/8 (12.5) 3 * | 33.5 [0.0] | - | - | - | 1 * | NA | - | - | 1 * | 1/8 (12.5) 1 * | 31.9 [0.0] | - | - | - |
| 3 | 1 * | NA | - | - | - | 2/7 (28.6) 2 * | 32.5 [0.3] | - | - | - | - | - | - | - | 1 * | - | - | 3/49 (6.1) | 29.6 [1.2] | - |
| 4 | - | - | - | - | - | 6/8 (75.0) 1 * | 32.2 [0.8] | - | - | - | NT | NT | NT | NT | NT | - | - | - | - | 1 * |
| 5 | - | - | - | - | - | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT | NT |
| 6 | 1 * | NA | 1 * | NA | - | - | - | - | - | - | - | - | - | - | - | 1 * | NA | - | - | - |
| 7 | NT | NT | NT | NT | NT | - | - | - | - | - | NT | NT | NT | NT | NT | - | - | 1 * | NA | - |
| 8 | NT | NT | NT | NT | NT | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Farm Total | - | - | - | - | - | 13/49 (26.5) | 32.9 [0.4] | - | - | - | - | - | - | - | - | 1/49 (2.0) | 31.9 [0.0] | 3/49 (6.1) | 30.1 [1.8] | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias, A.S.; Baker, A.L.V.; Baker, R.B.; Zhang, J.; Zeller, M.A.; Kitikoon, P.; Gauger, P.C. Detection and Characterization of Influenza A Virus Endemic Circulation in Suckling and Nursery Pigs Originating from Vaccinated Farms in the Same Production System. Viruses 2024, 16, 626. https://doi.org/10.3390/v16040626
Dias AS, Baker ALV, Baker RB, Zhang J, Zeller MA, Kitikoon P, Gauger PC. Detection and Characterization of Influenza A Virus Endemic Circulation in Suckling and Nursery Pigs Originating from Vaccinated Farms in the Same Production System. Viruses. 2024; 16(4):626. https://doi.org/10.3390/v16040626
Chicago/Turabian StyleDias, Alessandra Silva, Amy L. Vincent Baker, Rodney B. Baker, Jianqiang Zhang, Michael A. Zeller, Pravina Kitikoon, and Phillip C. Gauger. 2024. "Detection and Characterization of Influenza A Virus Endemic Circulation in Suckling and Nursery Pigs Originating from Vaccinated Farms in the Same Production System" Viruses 16, no. 4: 626. https://doi.org/10.3390/v16040626
APA StyleDias, A. S., Baker, A. L. V., Baker, R. B., Zhang, J., Zeller, M. A., Kitikoon, P., & Gauger, P. C. (2024). Detection and Characterization of Influenza A Virus Endemic Circulation in Suckling and Nursery Pigs Originating from Vaccinated Farms in the Same Production System. Viruses, 16(4), 626. https://doi.org/10.3390/v16040626






