The Nucleocapsid Protein of SARS-CoV-2, Combined with ODN-39M, Is a Potential Component for an Intranasal Bivalent Vaccine with Broader Functionality
Abstract
1. Introduction
2. Materials and Methods
2.1. Recombinant Proteins, Peptide and ODN-39M
2.2. In Vitro Aggregation Procedure of Nucleocapsid Protein with ODN-39M
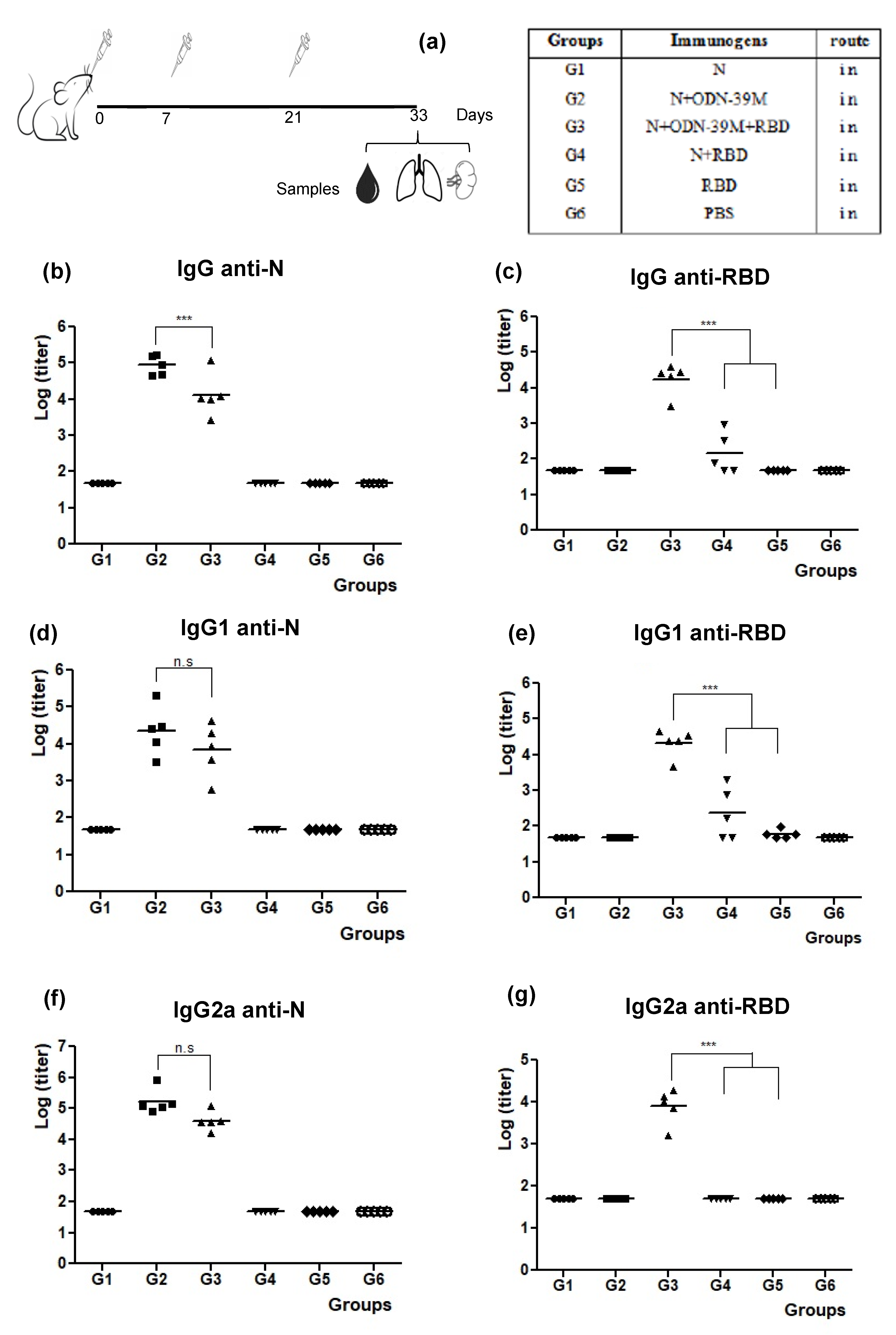
2.3. Immunization Experiments
2.4. Assessment of Humoral Immune Response by ELISA and a Surrogate Virus Neutralization Test
2.5. Assessment of Cellular Immune Response by IFN-γ ELISPOT
2.6. Pseudotyped VSV-Based Neutralization Assay
2.7. Statistical Analysis
3. Results
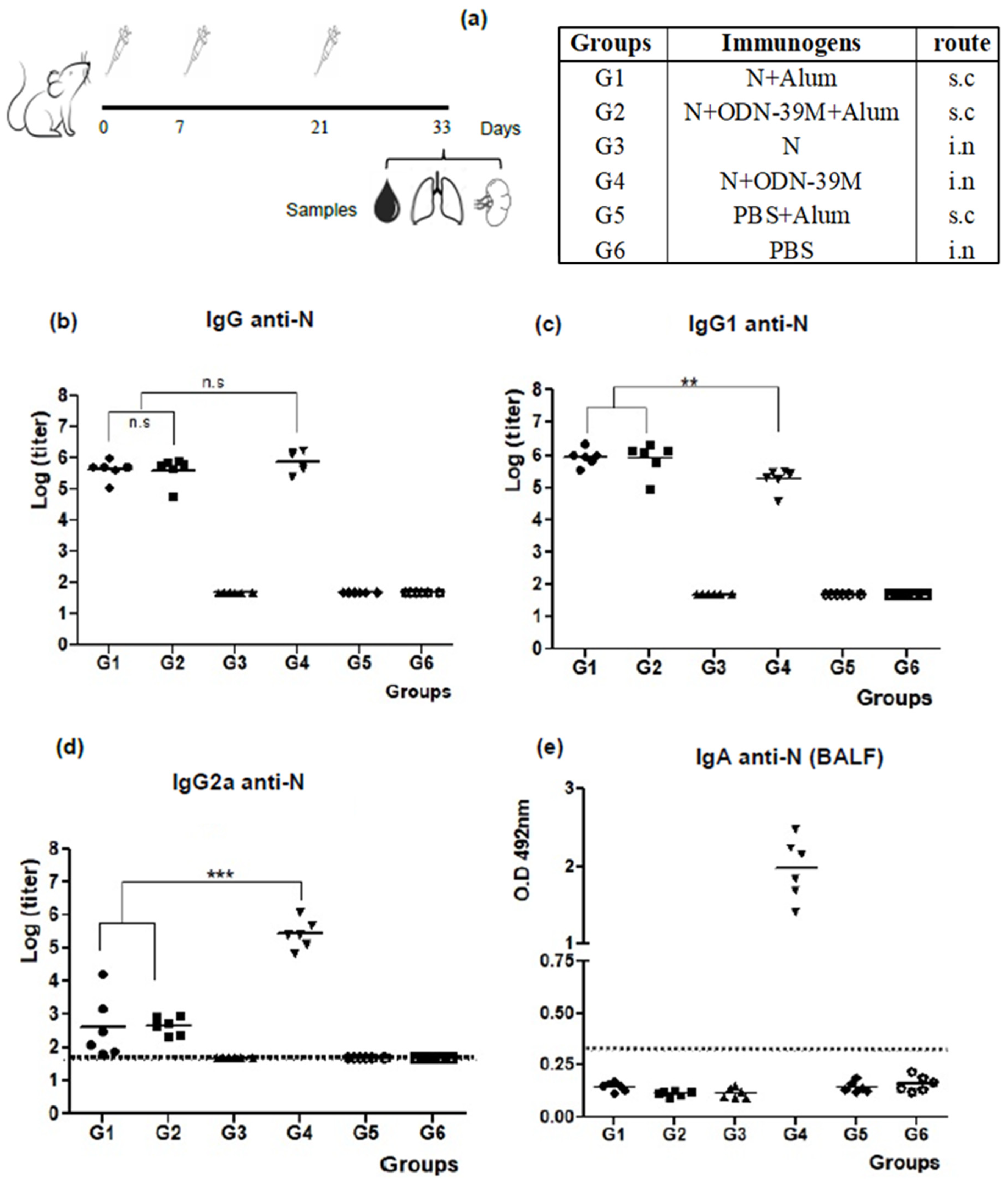
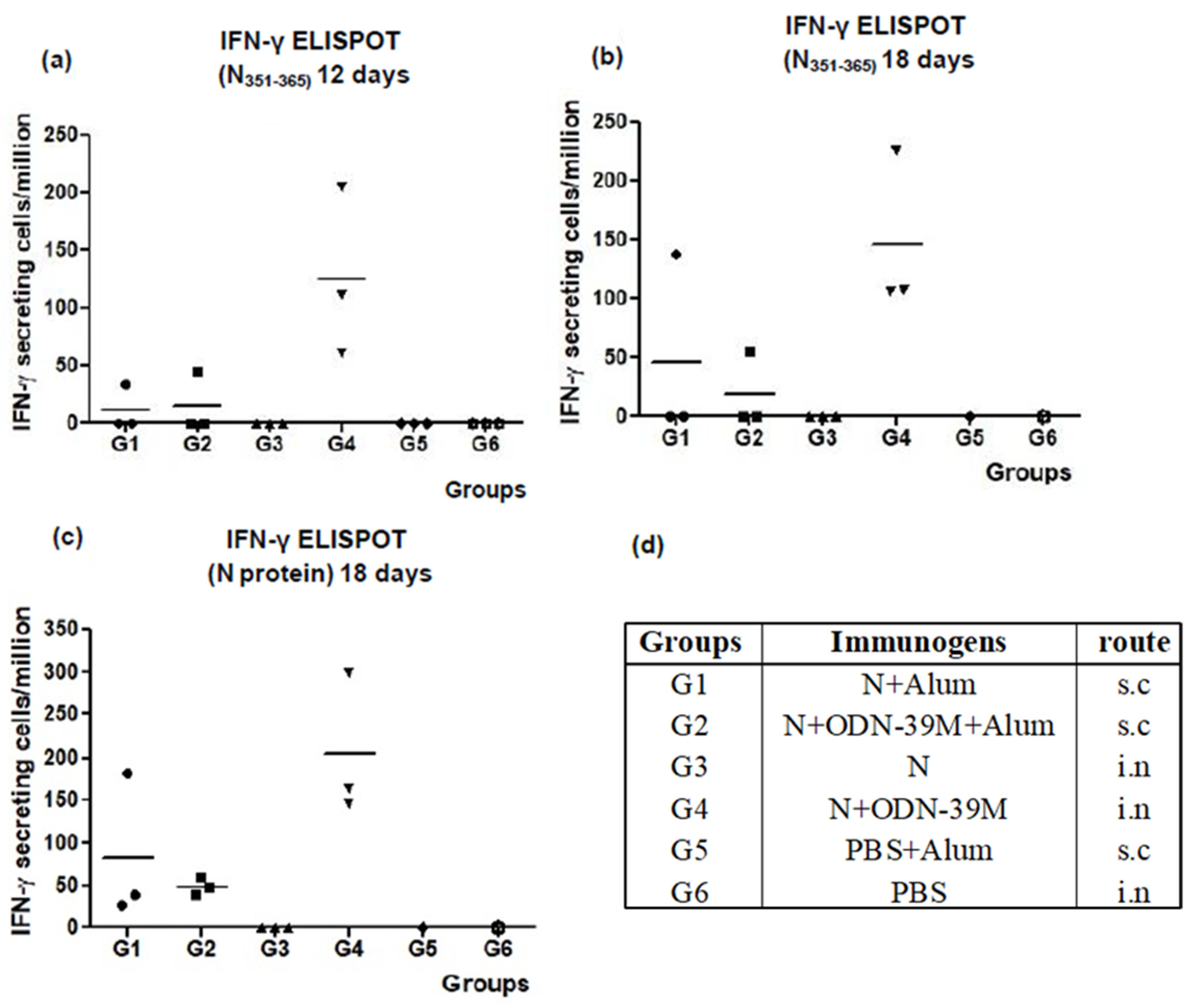
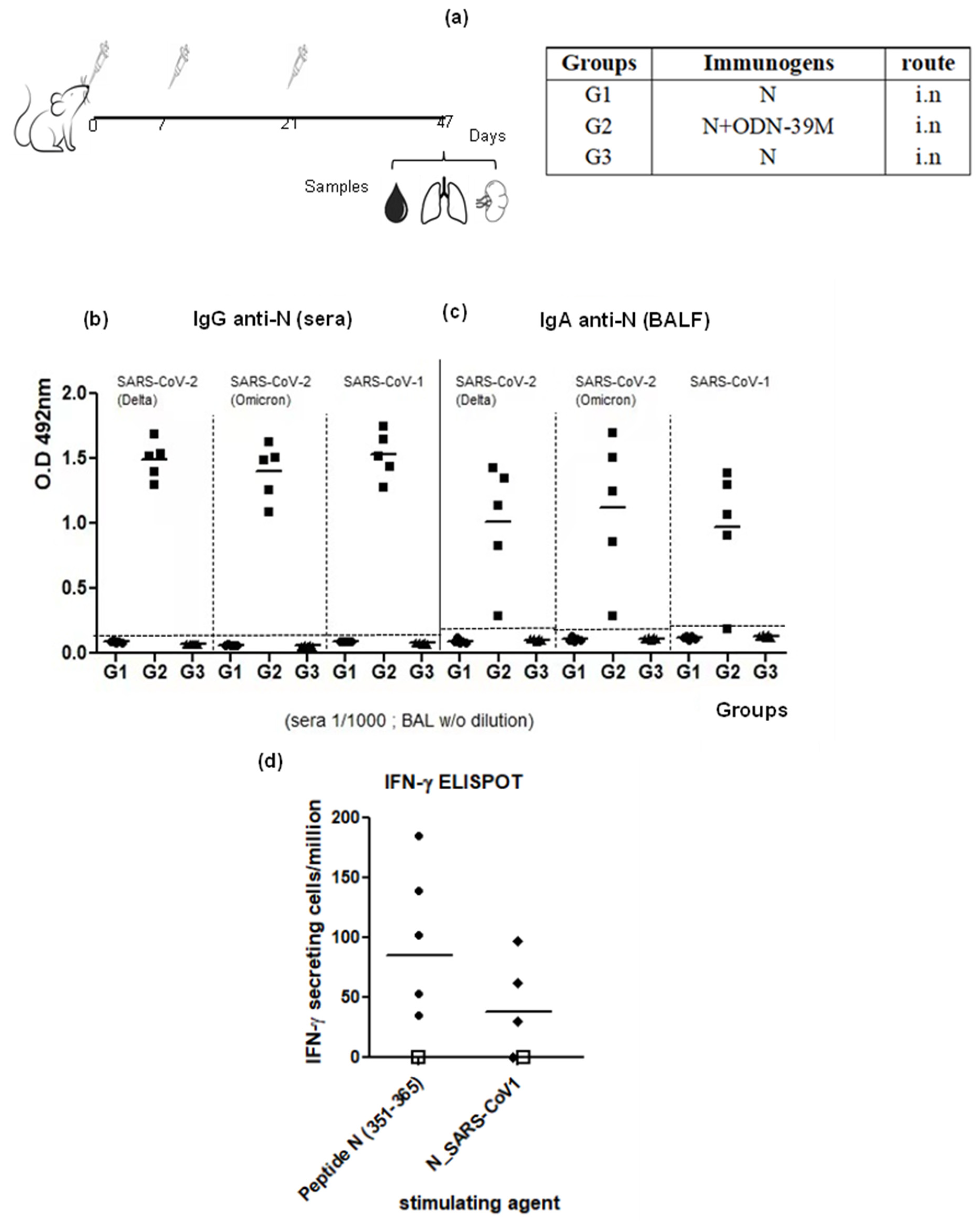
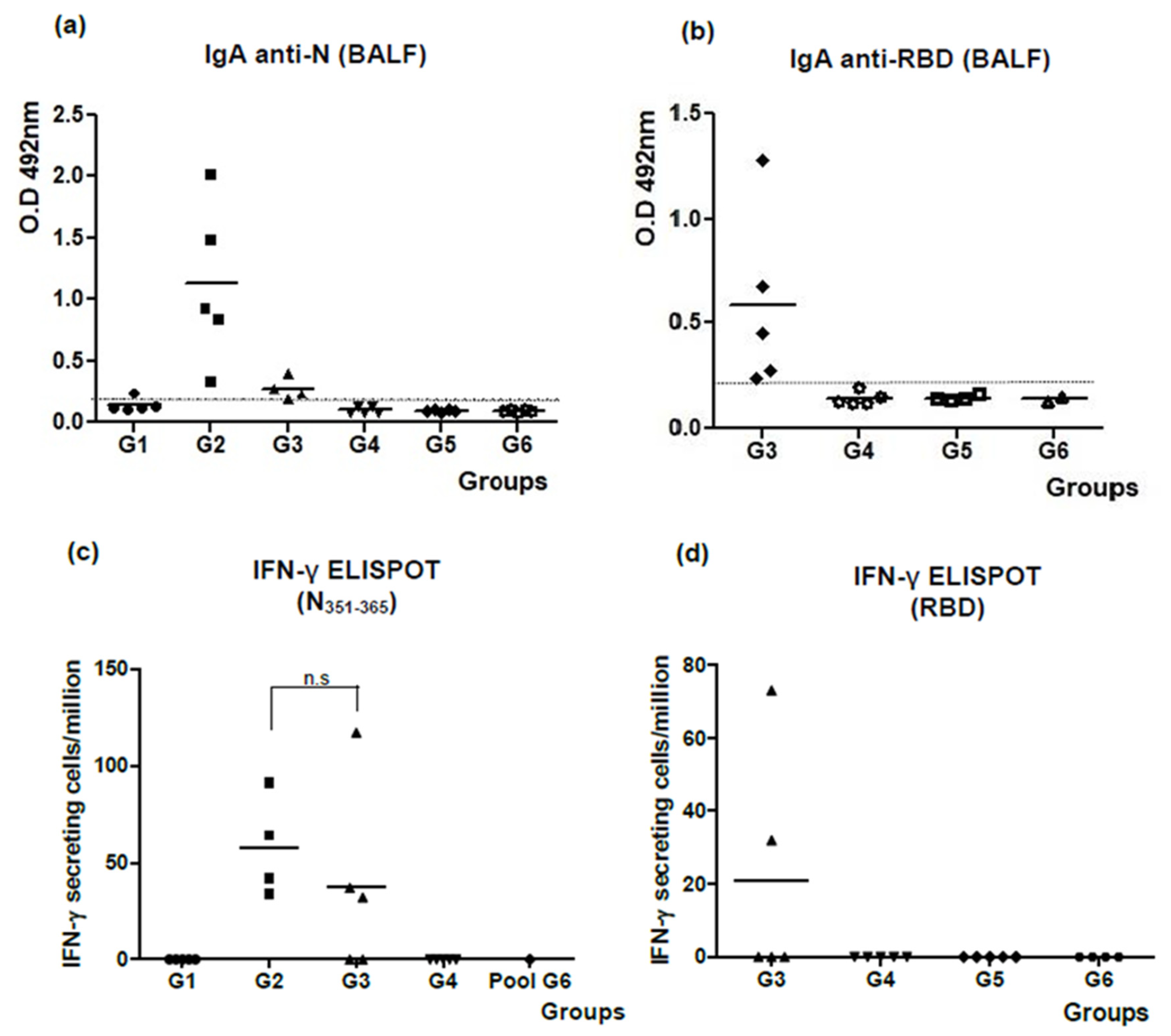
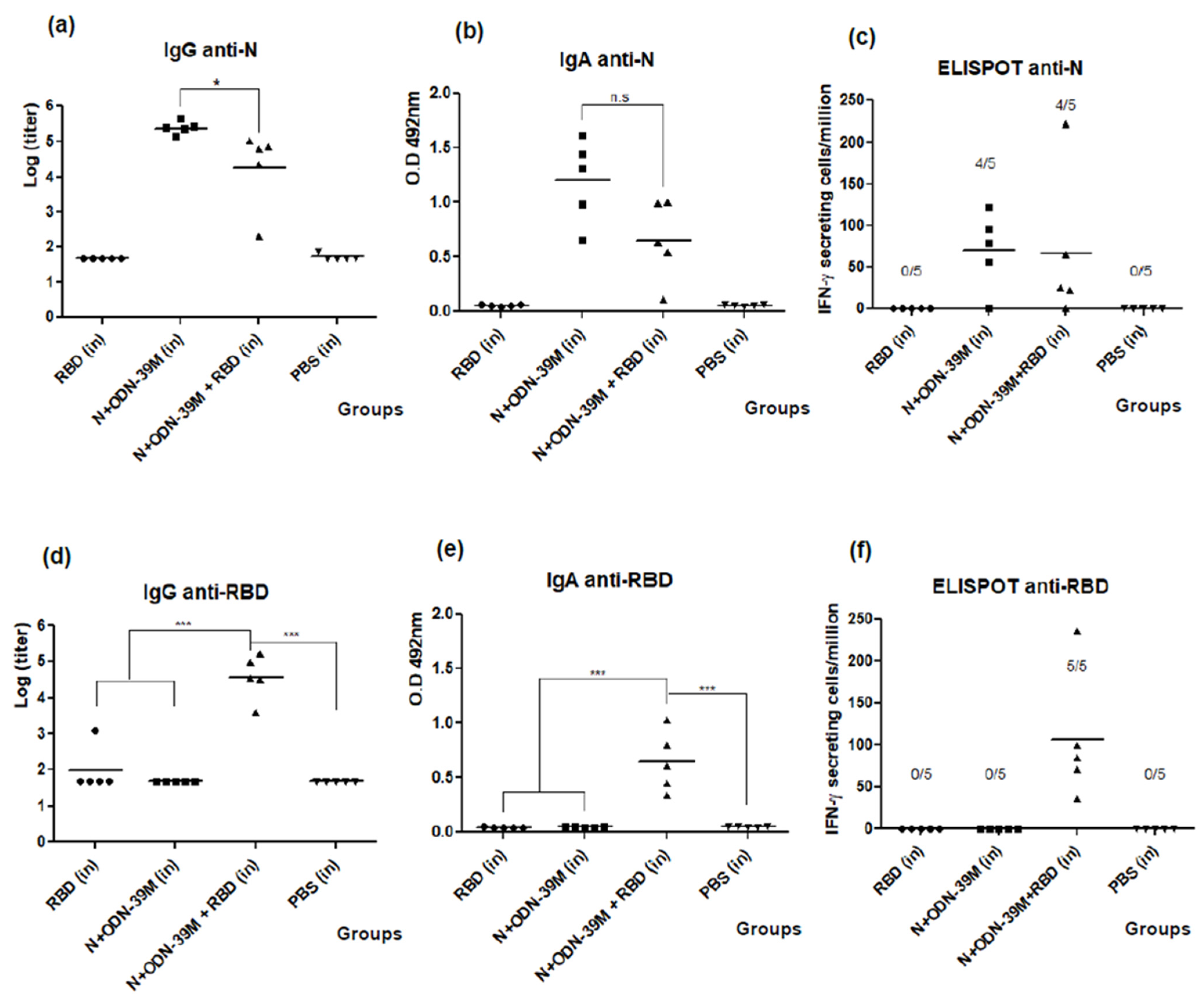
3.1. The N + ODN-39M Combination, Administered by Intranasal Route, Is Immunogenic in Balb/C Mice
3.2. Intranasally Administered N + ODN-39M Preparation Induces a Cross-Reactive Immune Response against N Protein until the Sarbecovirus Level
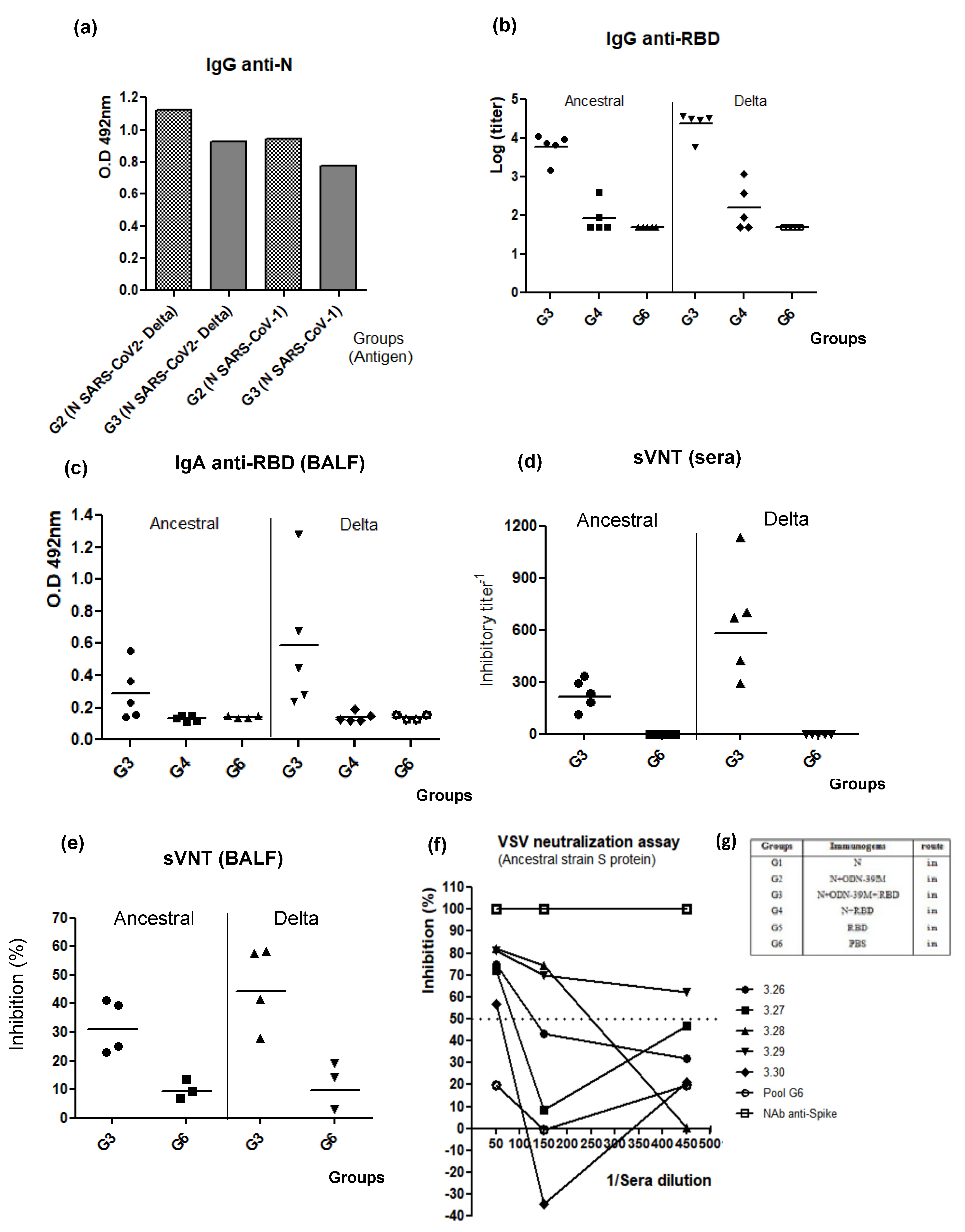
3.3. The N + ODN-39M Preparation Exerts an Adjuvant Effect on RBD Protein When Both Components Are Intranasally Co-Administered
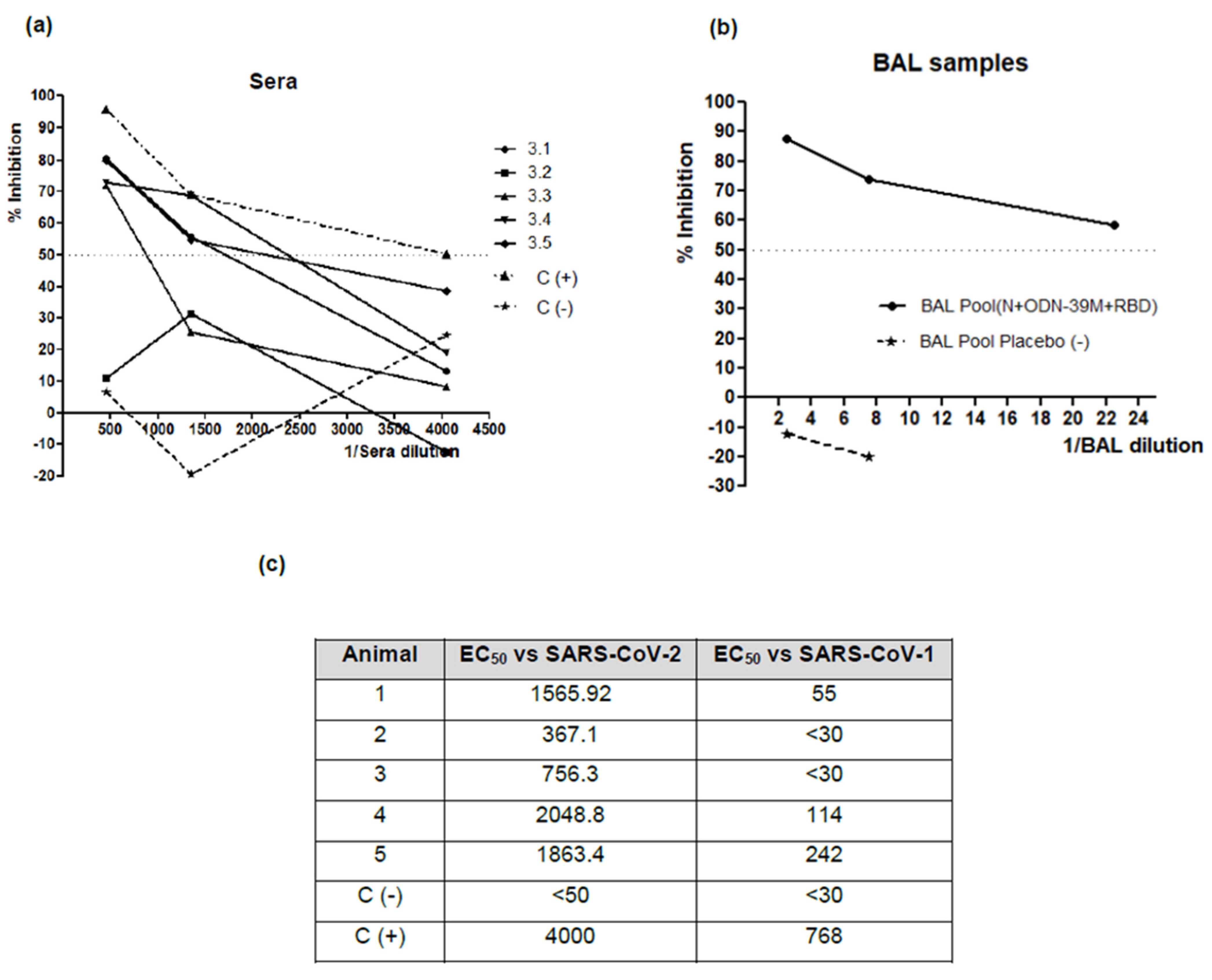
3.4. The Bivalent Formulation, N + ODN-39M + RBD, Induces Neutralizing Abs in Sera and Mucosal Samples, and Anti-RBD CMI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J.; et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults: A Randomized Clinical Trial. JAMA 2021, 326, 35. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-N.; Huang, Y.; Wang, W.; Jing, Q.-L.; Zhang, C.-H.; Qin, P.-Z.; Guan, W.-J.; Gan, L.; Li, Y.-L.; Liu, W.-H.; et al. Effectiveness of Inactivated SARS-CoV-2 Vaccines against the Delta Variant Infection in Guangzhou: A Test-Negative Case–Control Real-World Study. Emerg. Microbes Infect. 2021, 10, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Yokoyama, A.; Shichida, A.; Sasuga, K.; Maekawa, T.; Moriyama, T. Impact of sex and age on vaccine-related side effects and their progression after booster mRNA COVID-19 vaccine. Sci. Rep. 2023, 13, 19328. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.V.; Kumar, A. Increases in COVID-19 are unrelated to levels of vaccination across 68 countries and 2947 counties in the United States. Eur. J. Epidemiol. 2021, 36, 1237–1240. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.-M.; et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.W.; Moldoveanu, Z.; Ogra, P.L.; Mestecky, J. Mucosal Immunity in COVID-19: A Neglected but Critical Aspect of SARS-CoV-2 Infection. Front. Immunol. 2020, 11, 611337. [Google Scholar] [CrossRef]
- Steele, E.J.; Gorczynski, R.M.; Carnegie, P.; Tokoro, G.; Wallis, D.H.; Temple, R.; Wainwright, M.; Wickramasinghe, N.C. COVID-19 Sudden Outbreak of Mystery Case Transmissions in Victoria, Australia, May–June 2021: Strong Evidence of Tropospheric Transport of Human Passaged Infective Virions from the Indian Epidemic. Infect. Dis. Ther. 2021, 2, 1–28. [Google Scholar] [CrossRef]
- Steele, E.J.; Gorczynski, R.M.; Rebhan, H.; Carnegie, P.; Temple, R.; Tokoro, G.; Kondakon, A.; Coulson, S.G.; Wickramasinghe, D.T.; Wickramasinghe, N.C. Implications of haplotype switching for the origin and global spread of COVID-19. Virol. Curr. Res. 2020, 4, 1–13. [Google Scholar]
- Hammond, G.W.; Raddatz, R.L.; Gelskey, D.E. Impact of atmospheric dispersion and transport of viral aerosols on the epidemiology of Influenza. Rev. Infect. Dis. 1989, 11, 494–497. [Google Scholar] [CrossRef]
- Lindley, R.A.; Steele, E.J. 2021 Analysis of SARS-CoV-2 haplotypes and genomic sequences during 2020 in Victoria, Australia, in the context of putative deficits in innate immune deaminase anti-viral responses. Scand. J. Immunol. 2021, 94, e13100. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 1–20. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses re- veal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Xiao, Y.; Lidsky, P.V.; Shirogane, Y.; Aviner, R.; Wu, C.-T.; Li, W.; Zheng, W.; Talbot, D.; Catching, A.; Doitsh, G.; et al. A defective viral genome strategy elicits broad protective immunity against respiratory viruses. Cell 2021, 184, 6037–6051. [Google Scholar] [CrossRef]
- Oh, J.E.; Song, E.; Moriyama, M.; Wong, P.; Zhang, S. Intranasal priming induces local lung-resident B cell populations that secrete protective mucosal antiviral IgA. Sci. Immunol. 2021, 6, eabj5129. [Google Scholar] [CrossRef]
- Afkhami, S.; D’agostino, M.R.; Zhang, A.; Stacey, H.D.; Marzok, A.; Kang, A.; Singh, R.; Bavananthasivam, J.; Ye, G.; Luo, X.; et al. Respiratory mucosal delivery of next-generation COVID-19 vaccine provides robust protection against both ancestral and variant strains of SARS-CoV-2. Cell 2022, 185, 896–915. [Google Scholar] [CrossRef]
- Pagani, I.; Ghezzi, S.; Alberti, S.; Poli, G.; Vicenzi, E. Origin and evolution of SARS-CoV-2. Eur. Phys. J. Plus 2023, 138, 157. [Google Scholar] [CrossRef]
- Wickramasinghe, C.; Gorczynski, R.M.; Steele, E.J. (Eds.) Understanding the Origin and Global Spread of COVID-19; World Scientific Publishers: Singapore, 2022; Available online: https://www.amazon.com.au/UNDERSTANDING-ORIGIN-GLOBAL-SPREAD-COVID-19/dp/9811259070 (accessed on 13 February 2024).
- Graham, R.L.; Baric, R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef]
- Rubin, R. The Search for a Single Vaccine Against Coronaviruses Yet to Come. JAMA 2021, 326, 118–120. [Google Scholar] [CrossRef]
- Morens, D.M.; Taubenberger, J.K.; Fauci, A.S. Universal Coronavirus Vaccines—An Urgent Need. N. Engl. J. Med. 2022, 386, 297–299. [Google Scholar] [CrossRef]
- Cohen, A.A.; Gnanapragasam, P.N.P.; Lee, Y.E.; Hoffman, P.R.; Ou, S.; Kakutani, L.M.; Keeffe, J.R.; Wu, H.-J.; Howarth, M.; West, A.P.; et al. Mosaic Nanoparticles Elicit Cross-Reactive Immune Responses to Zoonotic Coronaviruses in Mice. Science 2021, 371, 735–741. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, J.; Yuan, R.Y.; Wang, M.Y.; He, P.; Su, J.G.; Han, Z.B.; Jin, Y.Q.; Hou, J.W.; Zhang, H.; et al. Structure and Computation-Guided Design of a Mutation-Integrated Trimeric RBD Candidate Vaccine with Broad Neutralization against SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wang, R.; Sun, C.; Ma, J.; Yu, C.; Kong, D.; Chen, M.; Liu, X.; Zhao, D.; Gao, S.; Kou, S.; et al. A Bivalent COVID-19 Vaccine Based on Alpha and Beta Variants Elicits Potent and Broad Immune Responses in Mice against SARS-CoV-2 Variants. Vaccines 2022, 10, 702. [Google Scholar] [CrossRef]
- Chiuppesi, F.; Salazar, M.D.; Contreras, H.; Nguyen, V.H.; Martinez, J.; Park, Y.; Nguyen, J.; Kha, M.; Iniguez, A.; Zhou, Q.; et al. Development of a Multi-Antigenic SARS-CoV-2 Vaccine Candidate Using a Synthetic Poxvirus Platform. Nat. Commun. 2020, 11, 6121. [Google Scholar] [CrossRef]
- Ahn, J.Y.; Lee, J.; Suh, Y.S.; Song, Y.G.; Choi, Y.-J.; Lee, K.H.; Seo, S.H.; Song, M.; Oh, J.-W.; Kim, M.; et al. Safety and Immunogenicity of a Recombinant DNA COVID-19 Vaccine Containing the Coding Regions of the Spike and Nucleocapsid Proteins: Preliminary Results from an Open-Label, Phase 1 Trial in Healthy Adults Aged 19–55 Years. medRxiv 2021. [Google Scholar] [CrossRef]
- Thura, M.; Sng, J.X.E.; Ang, K.H.; Li, J.; Gupta, A.; Hong, J.M.; Hong, C.W.; Zeng, Q. Targeting Intra-Viral Conserved Nucleocapsid (N) Proteins as Novel Vaccines against SARS-CoVs. Biosci. Rep. 2021, 41, BSR20211491. [Google Scholar] [CrossRef]
- Ahlen, G.; Frelin, L.; Nikouyan, N.; Weber, F.; Hoglund, U.; Larsson, O.; Westman, M.; Tuvesson, O.; Gidlund, E.K.; Cadossi, M.; et al. The SARS-CoV-2 N Protein Is a Good Component in a Vaccine. J. Virol. 2020, 94, e01279-20. [Google Scholar] [CrossRef]
- Smits, V.A.J.; Hernández-Carralero, E.; Paz-Cabrera, M.C.; Cabrera, E.; Hernández-Reyes, Y.; Hernández-Fernaud, J.R.; Gillespie, D.A.; Salido, E.; Hernández-Porto, M.; Freire, R. The Nucleocapsid protein triggers the main humoral immune response in COVID-19 patients. Biochem. Biophys. Res. Commun. 2021, 543, 45–49. [Google Scholar] [CrossRef]
- Hevesi, Z.; Gerges, D.; Kapps, S.; Freire, R.; Schmidt, S.; Pollak, D.; Schmetterer, K.; Freyn, T.; Lang, R.; Winnicki, W.; et al. Preclinical Establishment of a Divalent Vaccine against SARS-CoV-2. Vaccines 2022, 10, 516. [Google Scholar] [CrossRef]
- Scheiermann, J.; Klinman, D.M. Clinical evaluation of CpG oligonucleotides as adjuvants for Vaccines Targeting Infectious Diseases and Cancer. Vaccine 2014, 32, 6377–6389. [Google Scholar] [CrossRef] [PubMed]
- Gil, L.; Marcos, E.; Izquierdo, A.; Lazo, L.; Valdés, I.; Ambala, P.; Ochola, L.; Hitler, R.; Suzarte, E.; Álvarez, M.; et al. The Protein DIIIC-2, Aggregated with a Specific Oligodeoxynucleotide and Adjuvanted in Alum, Protects Mice and Monkeys against DENV-2. Immunol. Cell Biol. 2015, 93, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Gil, L.; Cobas, K.; Lazo, L.; Marcos, E.; Hernández, L.; Suzarte, E.; Izquierdo, A.; Valdés, I.; Blanco, A.; Puentes, P.; et al. A Tetravalent Formulation Based on Recombinant Nucleocapsid-like Particles from Dengue Viruses Induces a Functional Immune Response in Mice and Monkeys. J. Immunol. 2016, 197, 3597–3606. [Google Scholar] [CrossRef] [PubMed]
- Olivera, S.; Perez, A.; Falcon, V.; Urquiza, D.; Pichardo, D.; Martinez-Donato, G. Protective Cellular Immune Response against Hepatitis C Virus Elicited by Chimeric Protein Formulations in BALB/c Mice. Arch. Virol. 2020, 165, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Lobaina, Y.; Trujillo, H.; García, D.; Gambe, A.; Chacon, Y.; Blanco, A.; Aguilar, J.C. The Effect of the Parenteral Route of Administration on the Immune Response to Simultaneous Nasal and Parenteral Immunizations Using a New HBV Therapeutic Vaccine Candidate. Viral Immunol. 2010, 23, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Nie, J. Quantification of SARS-CoV-2 Neutralizing Antibody by a Pseudotyped Virus-Based Assay. Nat. Protoc. 2020, 15, 19. [Google Scholar] [CrossRef]
- Kundu, R.; Narean, J.S.; Wang, L.; Fenn, J.; Pillay, T.; Fernandez, N.D.; Conibear, E.; Koycheva, A.; Davies, M.; Tolosa-Wright, M.; et al. Cross-Reactive Memory T Cells Associate with Protection against SARS-CoV-2 Infection in COVID-19 Contacts. Nat. Commun. 2022, 13, 80. [Google Scholar] [CrossRef]
- Matchett, W.E.; Joag, V.; Stolley, J.M.; Shepherd, F.K.; Quarnstrom, C.F.; Mickelson, C.K.; Wijeyesinghe, S.; Soerens, A.G.; Becker, S.; Thiede, J.M.; et al. Cutting Edge: Nucleocapsid Vaccine Elicits Spike-Independent SARS-CoV-2 Protective Immunity. J. Immunol. 2021, 207, 376–379. [Google Scholar] [CrossRef]
- Dangi, T.; Class, J.; Palacio, N.; Richner, J.M.; Penaloza MacMaster, P. Combining Spike- and Nucleocapsid-Based Vaccines Improves Distal Control of SARS-CoV-2. Cell Rep. 2021, 36, 109664. [Google Scholar] [CrossRef] [PubMed]
- Dangi, T.; Sanchez, S.; Park, M.; Class, J.; Richner, M.; Richner, J.M.; Penaloza-MacMaster, P. Nucleocapsid-Specific Humoral Responses Improve the Control of SARS-CoV-2. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T. The Nucleocapsid Protein of SARS–CoV-2: A Target for Vaccine Development. J. Virol. 2020, 94, e00647-20. [Google Scholar] [CrossRef]
- Suzarte, E.; Gil, L.; Valdés, I.; Marcos, E.; Lazo, L.; Izquierdo, A.; García, A.; López, L.; Álvarez, M.; Pérez, Y.; et al. A Novel Tetravalent Formulation Combining the Four Aggregated Domain III-Capsid Proteins from Dengue Viruses Induces a Functional Immune Response in Mice and Monkeys. Int. Immunol. 2015, 27, 367–379. [Google Scholar] [CrossRef]
- Jack, A.; Ferro, L.S.; Trnka, M.J.; Wehri, E.; Nadgir, A.; Nguyenla, X.; Fox, D.; Costa, K.; Stanley, S.; Schaletzky, J.; et al. SARS-CoV-2 Nucleocapsid Protein Forms Condensates with Viral Genomic RNA. PLoS Biol. 2021, 19, e3001425. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Lai, X.; Sun, J.; Chen, Z.; Zhang, Z.; Dai, J.; Liu, D.; Li, Y.; Li, F.; Wang, Y.; et al. Mapping and Role of T Cell Response in SARS-CoV-2–Infected Mice. J. Exp. Med. 2021, 218, e20202187. [Google Scholar] [CrossRef]
- Bode, C.; Zhao, G.; Steinhagen, F.; Kinjo, T.; Klinman, D.M. CpG DNA as a Vaccine Adjuvant. Expert Rev. Vaccines 2011, 10, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Maeyama, J.; Takatsuka, H.; Suzuki, F.; Kubota, A.; Horiguchi, S.; Komiya, T.; Shimada, I.; Murata, E.; Osawa, Y.; Kitagawa, H.; et al. A Palindromic CpG-Containing Phosphodiester Oligodeoxynucleotide as a Mucosal Adjuvant Stimulates Plasmacytoid Dendritic Cell-Mediated TH1 Immunity. PLoS ONE 2014, 9, e88846. [Google Scholar] [CrossRef]
- Rothenfusser, S.; Tuma, E.; Endres, S.; Hartmann, G. Plasmacytoid Dendritic Cells: The Key to CpG. Hum. Immunol. 2002, 63, 1111–1119. [Google Scholar] [CrossRef]
- van Beek, L.F.; Langereis, J.D.; van den Berg van Saparoea, H.B.; Gillard, J.; Jong, W.S.P.; van Opzeeland, F.J.; Mesman, R.; van Niftrik, L.; Joosten, I.; Diavatopoulos, D.A.; et al. Intranasal Vaccination with Protein Bodies Elicit Strong Protection against Streptococcus Pneumoniae Colonization. Vaccine 2021, 39, 6920–6929. [Google Scholar] [CrossRef]
- Tilocca, B.; Soggiu, A.; Sanguinetti, M.; Musella, V.; Britti, D.; Bonizzi, L.; Urbani, A.; Roncada, P. Comparative Computational Analysis of SARS-CoV-2 Nucleocapsid Protein Epitopes in Taxonomically Related Coronaviruses. Microbes Infect. 2020, 22, 188–194. [Google Scholar] [CrossRef]
- Ravelomanantsoa, N.A.F.; Guth, S.; Andrianiaina, A.; Andry, S.; Gentles, A.; Ranaivoson, H.C.; Brook, C.E. The Zoonotic Potential of Bat-Borne Coronaviruses. Emerg. Top. Life Sci. 2020, 4, 353–369. [Google Scholar] [CrossRef]
- Crook, J.M.; Murphy, I.; Carter, D.P.; Pullan, S.T.; Carroll, M.; Vipond, R.; Cunningham, A.A.; Bell, D. Metagenomic Identification of a New Sarbecovirus from Horseshoe Bats in Europe. Sci. Rep. 2021, 11, 14723. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Bernal, F.; Ricardo-Cobas, M.C.; Martín-Bauta, Y.; Navarro-Rodríguez, Z.; Piñera-Martínez, M.; Quintana-Guerra, J.; Urrutia-Pérez, K.; Urrutia-Pérez, K.; Chávez-Chong, C.O.; Azor-Hernández, J.L.; et al. Safety, Tolerability, and Immunogenicity of a SARS-CoV-2 Recombinant Spike RBD Protein Vaccine: A Randomised, Double-Blind, Placebo-Controlled, Phase 1-2 Clinical Trial (ABDALA Study). eClinicalMedicine 2022, 46, 101383. [Google Scholar] [CrossRef] [PubMed]
- Reed, G. Cuban COVID-19 Vaccines for Children: Rinaldo Puga MD MS Principal Investigator, Pediatric Clinical Trials for Soberana 02 and Soberana Plus. MEDICC Rev. 2022, 24, 14–18. [Google Scholar] [CrossRef]
- Cao, Y.; Hao, X.; Wang, X.; Wu, Q.; Song, R.; Zhao, D.; Song, W.; Wang, Y.; Yisimayi, A.; Wang, W.; et al. Humoral Immunogenicity and Reactogenicity of CoronaVac or ZF2001 Booster after Two Doses of Inactivated Vaccine. Cell Res. 2022, 32, 107–109. [Google Scholar] [CrossRef]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; McMahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA Vaccine Protection against SARS-CoV-2 in Rhesus Macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Huang, B.; Deng, W.; Quan, Y.; Wang, W.; Xu, W.; Zhao, Y.; Li, N.; Zhang, J.; et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 2020, 182, 713–721.e9. [Google Scholar] [CrossRef]
- Sholukh, A.M.; Fiore-Gartland, A.; Ford, E.S.; Miner, M.D.; Hou, Y.J.; Tse, L.V.; Kaiser, H.; Zhu, H.; Lu, J.; Madarampalli, B.; et al. Evaluation of cell-based and surrogate SARS-CoV-2 neutralization assays. J. Clin. Microbiol. 2021, 59, e00527-21. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, J.; Li, Q.; Hu, H.; Lu, J.; Chen, Z. Advances in Neutralization Assays for SARS-CoV-2. Scand. J. Immunol. 2021, 94, e13088. [Google Scholar] [CrossRef]
- He, Q.; Mao, Q.; An, C.; Zhang, J.; Gao, F.; Bian, L.; Li, C.; Liang, Z.; Xu, M.; Wang, J. Heterologous Prime-Boost: Breaking the Protective Immune Response Bottleneck of COVID-19 Vaccine Candidates. Emerg. Microbes Infect. 2021, 10, 629–637. [Google Scholar] [CrossRef]
- An, Y.; Li, S.; Jin, X.; Han, J.; Xu, K.; Xu, S.; Han, Y.; Liu, C.; Zheng, T.; Liu, M.; et al. A Tandem-Repeat Dimeric RBD Protein-Based Covid-19 Vaccine Zf2001 Protects Mice and Nonhuman Primates. Emerg. Microbes Infect. 2022, 11, 1058–1071. [Google Scholar] [CrossRef]
- Mäkitalo, B.; Lundholm, P.; Hinkula, J.; Nilsson, C.; Karlén, K.; Mörner, A.; Sutter, G.; Erfle, V.; Heeney, J.L.; Wahren, B.; et al. Enhanced Cellular Immunity and Systemic Control of SHIV Infection by Combined Parenteral and Mucosal Administration of a DNA Prime MVA Boost Vaccine Regimen. J. Gen. Virol. 2004, 85, 2407–2419. [Google Scholar] [CrossRef]
- Vajdy, M.; Singh, M.; Kazzaz, J.; Soenawan, E.; Ugozzoli, M.; Zhou, F.; Srivastava, I.; Bin, Q.; Barnett, S.; Donnelly, J.; et al. Mucosal and Systemic Anti-HIV Responses in Rhesus Macaques Following Combinations of Intranasal and Parenteral Immunizations. AIDS Res. Hum. Retroviruses 2004, 20, 1269–1281. [Google Scholar] [CrossRef]
- Cao, H.; Mai, J.; Zhou, Z.; Li, Z.; Duan, R.; Watt, J.; Chen, Z.; Bandara, R.A.; Li, M.; Ahn, S.K.; et al. Intranasal HD-Ad Vaccine Protects the Upper and Lower Respiratory Tracts of HACE2 Mice against SARS-CoV-2. Cell Biosci. 2021, 11, 202. [Google Scholar] [CrossRef]
- Du, Y.; Xu, Y.; Feng, J.; Hu, L.; Zhang, Y.; Zhang, B.; Guo, W.; Mai, R.; Chen, L.; Fang, J.; et al. Intranasal Administration of a Recombinant RBD Vaccine Induced Protective Immunity against SARS-CoV-2 in Mouse. Vaccine 2021, 9, 2280–2287. [Google Scholar] [CrossRef] [PubMed]
- Jearanaiwitayakul, T.; Seesen, M.; Chawengkirttikul, R.; Limthongkul, J.; Apichirapokey, S.; Sapsutthipas, S.; Phumiamorn, S.; Sunintaboon, P.; Ubol, S. Intranasal Administration of RBD Nanoparticles Confers Induction of Mucosal and Systemic Immunity against SARS-CoV-2. Vaccines 2021, 9, 768. [Google Scholar] [CrossRef] [PubMed]
- Lazo, L.; Bequet-Romero, M.; Lemos, G.; Musacchio, A.; Cabrales, A.; Bruno, A.J.; Ariel Espinosa, L.; Saloheimo, M.; Vitikainen, M.; Hernández, A.; et al. A Recombinant SARS-CoV-2 Receptor-Binding Domain Expressed in an Engineered Fungal Strain of Thermothelomyces Heterothallica Induces a Functional Immune Response in Mice. Vaccine 2022, 40, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Schild, S. An Intranasal Vaccine Based on Outer Membrane Vesicles Against SARS-CoV-2. Front. Microbiol. 2021, 12, 16. [Google Scholar]
- Lobaina, Y. Nasal route for vaccine and drug delivery: Features and current opportunities. Int. J. Pharm. 2019, 572, 118813. [Google Scholar] [CrossRef] [PubMed]
- FDA. Information Regarding FluMist Quadrivalent Vaccine. Available online: https://www.fda.gov/biologicsbloodvaccines/vaccines/approvedproducts/ucm508761.htm (accessed on 13 February 2024).
- Aguilar, J.C.; Aguiar, J.A.; Akbar, S.M.F. Action Mechanisms and Scientific Rationale of Using Nasal Vaccine (HeberNasvac) for the Treatment of Chronic Hepatitis B. Vaccines 2022, 10, 2087. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobaina, Y.; Chen, R.; Suzarte, E.; Ai, P.; Huerta, V.; Musacchio, A.; Silva, R.; Tan, C.; Martín, A.; Lazo, L.; et al. The Nucleocapsid Protein of SARS-CoV-2, Combined with ODN-39M, Is a Potential Component for an Intranasal Bivalent Vaccine with Broader Functionality. Viruses 2024, 16, 418. https://doi.org/10.3390/v16030418
Lobaina Y, Chen R, Suzarte E, Ai P, Huerta V, Musacchio A, Silva R, Tan C, Martín A, Lazo L, et al. The Nucleocapsid Protein of SARS-CoV-2, Combined with ODN-39M, Is a Potential Component for an Intranasal Bivalent Vaccine with Broader Functionality. Viruses. 2024; 16(3):418. https://doi.org/10.3390/v16030418
Chicago/Turabian StyleLobaina, Yadira, Rong Chen, Edith Suzarte, Panchao Ai, Vivian Huerta, Alexis Musacchio, Ricardo Silva, Changyuan Tan, Alejandro Martín, Laura Lazo, and et al. 2024. "The Nucleocapsid Protein of SARS-CoV-2, Combined with ODN-39M, Is a Potential Component for an Intranasal Bivalent Vaccine with Broader Functionality" Viruses 16, no. 3: 418. https://doi.org/10.3390/v16030418
APA StyleLobaina, Y., Chen, R., Suzarte, E., Ai, P., Huerta, V., Musacchio, A., Silva, R., Tan, C., Martín, A., Lazo, L., Guillén-Nieto, G., Yang, K., Perera, Y., & Hermida, L. (2024). The Nucleocapsid Protein of SARS-CoV-2, Combined with ODN-39M, Is a Potential Component for an Intranasal Bivalent Vaccine with Broader Functionality. Viruses, 16(3), 418. https://doi.org/10.3390/v16030418






