The Chemokine CXCL14 as a Potential Immunotherapeutic Agent for Cancer Therapy
Abstract
1. Tumor-Suppressive and Tumor-Promoting Roles of Chemokines in Cancer
2. Utilizing Chemokines to Augment Immunotherapy
3. CXCL14 and Cancer
4. Developing CXCL14 as an Immunotherapeutic Agent
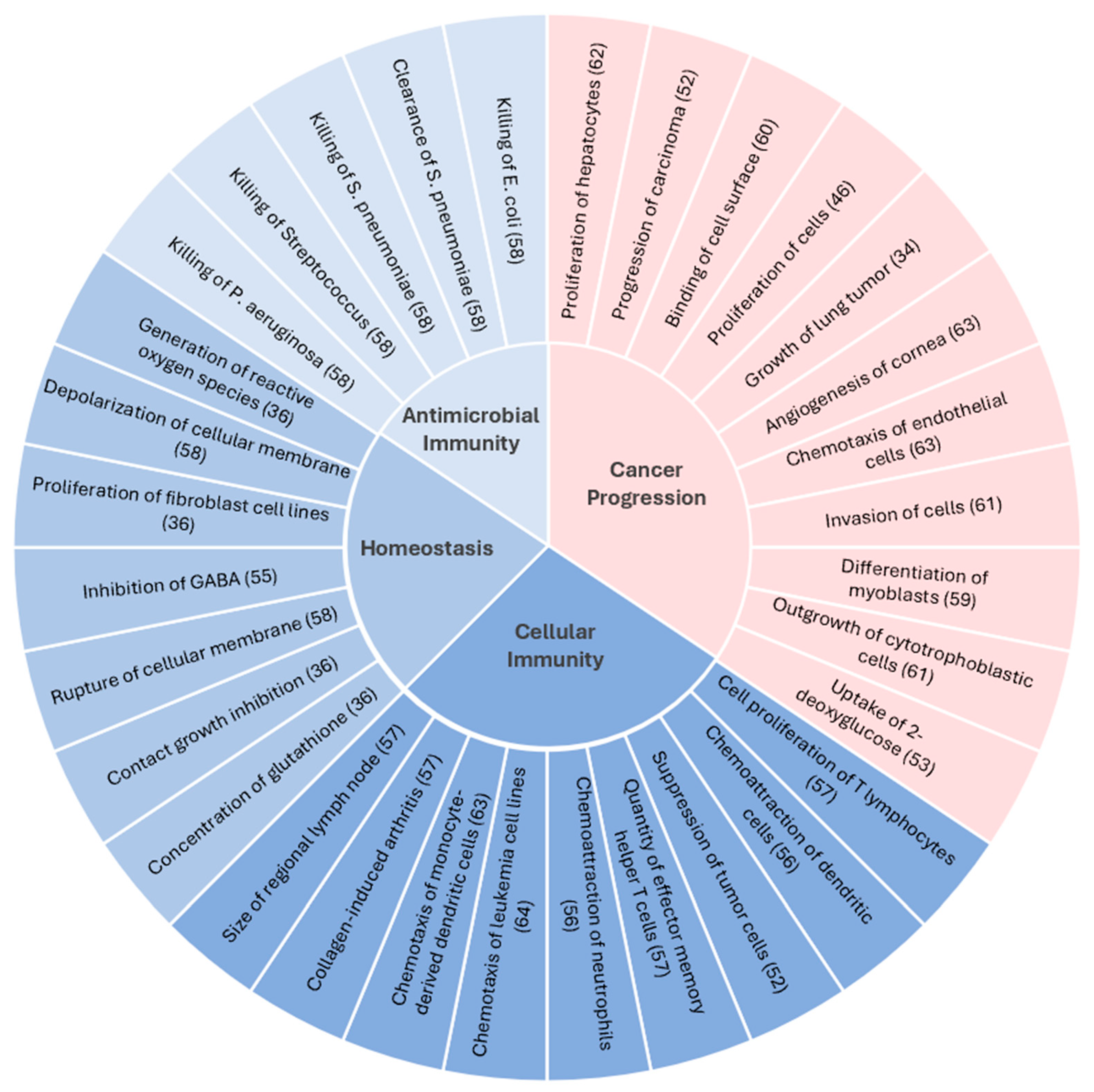
5. Understanding the Mechanism of CXCL14-Mediated Tumor Suppression
6. Developing Effective Delivery Tools for CXCL14
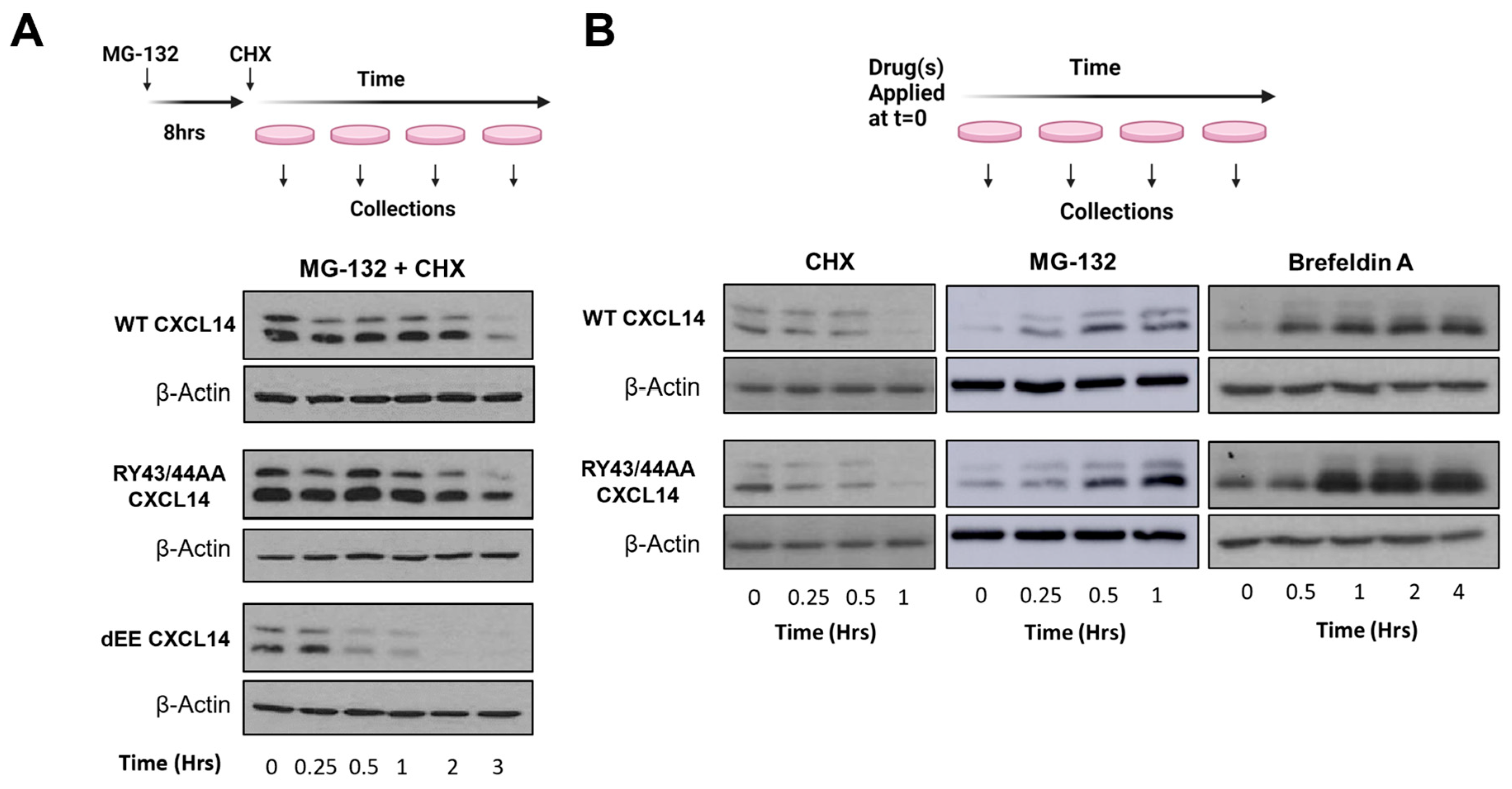
7. Optimizing CXCL14 Protein Stability
8. CXCL14 Therapeutic Dose and Combination Therapy
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Ozga, A.J.; Chow, M.T.; Luster, A.D. Chemokines and the immune response to cancer. Immunity 2021, 54, 859–874. [Google Scholar] [CrossRef]
- Drouillard, D.; Craig, B.T.; Dwinell, M.B. Physiology of chemokines in the cancer microenvironment. Am. J. Physiol. Cell Physiol. 2023, 324, C167–C182. [Google Scholar] [CrossRef]
- Märkl, F.; Huynh, D.; Endres, S.; Kobold, S. Utilizing chemokines in cancer immunotherapy. Trends Cancer 2022, 8, 670–682. [Google Scholar] [CrossRef]
- Hoch, T.; Schulz, D.; Eling, N.; Gómez, J.M.; Levesque, M.P.; Bodenmiller, B. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci. Immunol. 2022, 7, eabk1692. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Zumwalt, T.J.; Arnold, M.; Goel, A.; Boland, C.R. Active Secretion of CXCL10 and CCL5 from Colorectal Cancer Microenvironments Associates with GranzymeB+ CD8+ T-Cell Infiltration. Oncotarget 2015, 6, 2981–2991. [Google Scholar] [CrossRef]
- Mikucki, M.E.; Fisher, D.T.; Matsuzaki, J.; Skitzki, J.J.; Gaulin, N.B.; Muhitch, J.B.; Ku, A.W.; Frelinger, J.G.; Odunsi, K.; Gajewski, T.F.; et al. Non-Redundant Requirement for CXCR3 Signalling during Tumoricidal T-Cell Trafficking across Tumour Vascular Checkpoints. Nat. Commun. 2015, 6, 7458. [Google Scholar] [CrossRef]
- Karin, N. CXCR3 Ligands in Cancer and Autoimmunity, Chemoattraction of Effector T Cells, and Beyond. Front. Immunol. 2020, 11, 976. [Google Scholar] [CrossRef]
- Spranger, S.; Gajewski, T.F. Impact of Oncogenic Pathways on Evasion of Antitumour Immune Responses. Nat. Rev. Cancer 2018, 18, 139–147. [Google Scholar] [CrossRef]
- Harlin, H.; Meng, Y.; Peterson, A.C.; Zha, Y.; Tretiakova, M.; Slingluff, C.; McKee, M.; Gajewski, T.F. Chemokine Expression in Melanoma Metastases Associated with CD8+ T-Cell Recruitment. Cancer Res. 2009, 69, 3077–3085. [Google Scholar] [CrossRef]
- Rubio, A.J.; Porter, T.; Zhong, X. Duality of B Cell-CXCL13 Axis in Tumor Immunology. Front. Immunol. 2020, 11, 521110. [Google Scholar] [CrossRef]
- Ukita, M.; Hamanishi, J.; Yoshitomi, H.; Yamanoi, K.; Takamatsu, S.; Ueda, A.; Suzuki, H.; Hosoe, Y.; Furutake, Y.; Taki, M.; et al. CXCL13-Producing CD4+ T Cells Accumulate in the Early Phase of Tertiary Lymphoid Structures in Ovarian Cancer. JCI Insight 2022, 7, e157215. [Google Scholar] [CrossRef]
- Galeano Niño, J.L.; Pageon, S.V.; Tay, S.S.; Colakoglu, F.; Kempe, D.; Hywood, J.; Mazalo, J.K.; Cremasco, J.; Govendir, M.A.; Dagley, L.F.; et al. Cytotoxic T Cells Swarm by Homotypic Chemokine Signalling. eLife 2020, 9, e56554. [Google Scholar] [CrossRef]
- Li, F.; Kitajima, S.; Kohno, S.; Yoshida, A.; Tange, S.; Sasaki, S.; Okada, N.; Nishimoto, Y.; Muranaka, H.; Nagatani, N.; et al. Retinoblastoma Inactivation Induces a Protumoral Microenvironment via Enhanced CCL2 Secretion. Cancer Res. 2019, 79, 3903–3915. [Google Scholar] [CrossRef]
- Xu, W.; Wu, Y.; Liu, W.; Anwaier, A.; Tian, X.; Su, J.; Huang, H.; Wei, G.; Qu, Y.; Zhang, H.; et al. Tumor-Associated Macrophage-Derived Chemokine CCL5 Facilitates the Progression and Immunosuppressive Tumor Microenvironment of Clear Cell Renal Cell Carcinoma. Int. J. Biol. Sci. 2022, 18, 4884–4900. [Google Scholar] [CrossRef]
- Propper, D.J.; Balkwill, F.R. Harnessing Cytokines and Chemokines for Cancer Therapy. Nat. Rev. Clin. Oncol. 2022, 19, 237–253. [Google Scholar] [CrossRef]
- Homey, B.; Müller, A.; Zlotnik, A. Chemokines: Agents for the Immunotherapy of Cancer? Nat. Rev. Immunol. 2002, 2, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Chow, M.T.; Ozga, A.J.; Servis, R.L.; Frederick, D.T.; Lo, J.A.; Fisher, D.E.; Freeman, G.J.; Boland, G.M.; Luster, A.D. Intratumoral Activity of the CXCR3 Chemokine System Is Required for the Efficacy of Anti-PD-1 Therapy. Immunity 2019, 50, 1498–1512.e5. [Google Scholar] [CrossRef] [PubMed]
- House, I.G.; Savas, P.; Lai, J.; Chen, A.X.Y.; Oliver, A.J.; Teo, Z.L.; Todd, K.L.; Henderson, M.A.; Giuffrida, L.; Petley, E.V.; et al. Macrophage-Derived CXCL9 and CXCL10 Are Required for Antitumor Immune Responses Following Immune Checkpoint Blockade. Clin. Cancer Res. 2020, 26, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Dangaj, D.; Bruand, M.; Grimm, A.J.; Ronet, C.; Barras, D.; Duttagupta, P.A.; Lanitis, E.; Duraiswamy, J.; Tanyi, J.L.; Benencia, F.; et al. Cooperation between Constitutive and Inducible Chemokines Enables T Cell Engraftment and Immune Attack in Solid Tumors. Cancer Cell. 2019, 35, 885–900.e10. [Google Scholar] [CrossRef]
- Bockorny, B.; Semenisty, V.; Macarulla, T.; Borazanci, E.; Wolpin, B.M.; Stemmer, S.M.; Golan, T.; Geva, R.; Borad, M.J.; Pedersen, K.S.; et al. BL-8040, a CXCR4 Antagonist, in Combination with Pembrolizumab and Chemotherapy for Pancreatic Cancer: The COMBAT Trial. Nat. Med. 2020, 26, 878–885. [Google Scholar] [CrossRef]
- Gong, R.; Ren, H. Targeting Chemokines/Chemokine Receptors: A Promising Strategy for Enhancing the Immunotherapy of Pancreatic Ductal Adenocarcinoma. Signal Transduct. Target. Ther. 2020, 5, 149. [Google Scholar] [CrossRef]
- Sterner, R.C.; Sterner, R.M. CAR-T Cell Therapy: Current Limitations and Potential Strategies. Blood Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Q.; Ye, Y.; Sun, X.; Xie, S.; Zhan, Y.; Song, J.; Fan, X.; Zhang, B.; Yang, M.; et al. FGF-2 Signaling in Nasopharyngeal Carcinoma Modulates Pericyte-Macrophage Crosstalk and Metastasis. JCI Insight 2022, 7, e157874. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, Z.; Zhong, K.; Wang, Z.; Yang, N.; Tang, X.; Li, H.; Lu, Q.; Wu, Z.; Yuan, B.; et al. CXCL11-Armed Oncolytic Adenoviruses Enhance CAR-T Cell Therapeutic Efficacy and Reprogram Tumor Microenvironment in Glioblastoma. Mol. Ther. 2023, 31, 134–153. [Google Scholar] [CrossRef]
- Gao, Q.; Zhang, Y. CXCL11 Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2021, 1302, 41–50. [Google Scholar] [CrossRef]
- Hromas, R.; Broxmeyer, H.E.; Kim, C.; Nakshatri, H.; Christopherson, K.; Azam, M.; Hou, Y.H. Cloning of BRAK, a Novel Divergent CXC Chemokine Preferentially Expressed in Normal versus Malignant Cells. Biochem. Biophys. Res. Commun. 1999, 255, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Frederick, M.J.; Henderson, Y.; Xu, X.; Deavers, M.T.; Sahin, A.A.; Wu, H.; Lewis, D.E.; El-Naggar, A.K.; Clayman, G.L. In Vivo Expression of the Novel CXC Chemokine BRAK in Normal and Cancerous Human Tissue. Am. J. Pathol. 2000, 156, 1937–1950. [Google Scholar] [CrossRef] [PubMed]
- Westrich, J.A.; Vermeer, D.W.; Colbert, P.L.; Spanos, W.C.; Pyeon, D. The Multifarious Roles of the Chemokine CXCL14 in Cancer Progression and Immune Responses. Mol. Carcinog. 2020, 59, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Liu, Q.; Sun, Y.; Hu, X.; He, X.; Xu, C. CXCL14 Inhibits the Growth and Promotes Apoptosis of Hepatocellular Carcinoma Cells via Suppressing Akt/MTOR Pathway. J. Recept. Signal Transduct. 2021, 41, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mohamed, E.; Tong, S.; Chen, K.; Mukherjee, J.; Lim, Y.; Wong, C.M.; Boosalis, Z.; Shai, A.; Pieper, R.O.; et al. CXCL14 Promotes a Robust Brain Tumor-Associated Immune Response in Glioma. Clin. Cancer Res. 2022, 28, 2898–2910. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Won, B.H.; Choi, J.I.; Lee, I.; Lee, J.H.; Park, J.H.; Choi, Y.S.; Kim, J.-H.; Cho, S.; Lim, J.-B.; et al. BRAK and APRIL as Novel Biomarkers for Ovarian Tumors. Biomark. Med. 2022, 16, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Tessema, M.; Klinge, D.M.; Yingling, C.M.; Do, K.; Van Neste, L.; Belinsky, S.A. Re-Expression of CXCL14, a Common Target for Epigenetic Silencing in Lung Cancer, Induces Tumor Necrosis. Oncogene 2010, 29, 5159–5170. [Google Scholar] [CrossRef]
- Yang, X.-Y.; Ozawa, S.; Kato, Y.; Maehata, Y.; Izukuri, K.; Ikoma, T.; Kanamori, K.; Akasaka, T.; Suzuki, K.; Iwabuchi, H.; et al. C-X-C Motif Chemokine Ligand 14 Is a Unique Multifunctional Regulator of Tumor Progression. Int. J. Mol. Sci. 2019, 20, 1872. [Google Scholar] [CrossRef]
- Augsten, M.; Sjöberg, E.; Frings, O.; Vorrink, S.U.; Frijhoff, J.; Olsson, E.; Borg, Å.; Östman, A. Cancer-Associated Fibroblasts Expressing CXCL14 Rely upon NOS1-Derived Nitric Oxide Signaling for Their Tumor-Supporting Properties. Cancer Res. 2014, 74, 2999–3010. [Google Scholar] [CrossRef]
- Wei, S.-T.; Chiang, J.-Y.; Wang, H.-L.; Lei, F.-J.; Huang, Y.-C.; Wang, C.-C.; Cho, D.-Y.; Hsieh, C.-H. Hypoxia-Induced CXC Chemokine Ligand 14 Expression Drives Protumorigenic Effects through Activation of Insulin-like Growth Factor-1 Receptor Signaling in Glioblastoma. Cancer Sci. 2023, 114, 174–186. [Google Scholar] [CrossRef]
- Chang, T.-M.; Chiang, Y.-C.; Lee, C.-W.; Lin, C.-M.; Fang, M.-L.; Chi, M.-C.; Liu, J.-F.; Kou, Y.R. CXCL14 Promotes Metastasis of Non-Small Cell Lung Cancer through ACKR2-Depended Signaling Pathway. Int. J. Biol. Sci. 2023, 19, 1455–1470. [Google Scholar] [CrossRef]
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Maby, P.; Angelova, M.; Tougeron, D.; Church, S.E.; Lafontaine, L.; Fischer, M.; Fredriksen, T.; et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44, 698–711. [Google Scholar] [CrossRef]
- Cicchini, L.; Blumhagen, R.Z.; Westrich, J.A.; Myers, M.E.; Warren, C.J.; Siska, C.; Raben, D.; Kechris, K.J.; Pyeon, D. High-Risk Human Papillomavirus E7 Alters Host DNA Methylome and Represses HLA-E Expression in Human Keratinocytes. Sci. Rep. 2017, 7, 3633. [Google Scholar] [CrossRef] [PubMed]
- Westrich, J.A.; Vermeer, D.W.; Silva, A.; Bonney, S.; Berger, J.N.; Cicchini, L.; Greer, R.O.; Song, J.I.; Raben, D.; Slansky, J.E.; et al. CXCL14 Suppresses Human Papillomavirus-Associated Head and Neck Cancer through Antigen-Specific CD8+ T-Cell Responses by Upregulating MHC-I Expression. Oncogene 2019, 38, 7166–7180. [Google Scholar] [CrossRef]
- Cicchini, L.; Westrich, J.A.; Xu, T.; Vermeer, D.W.; Berger, J.N.; Clambey, E.T.; Lee, D.; Song, J.I.; Lambert, P.F.; Greer, R.O.; et al. Suppression of Antitumor Immune Responses by Human Papillomavirus through Epigenetic Downregulation of CXCL14. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Dolinska, M.; Cai, H.; Månsson, A.; Shen, J.; Xiao, P.; Bouderlique, T.; Li, X.; Leonard, E.; Chang, M.; Gao, Y.; et al. Characterization of the Bone Marrow Niche in Patients with Chronic Myeloid Leukemia Identifies CXCL14 as a New Therapeutic Option. Blood 2023, 142, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.; Shin, J.; Faquin, W.; Lin, D.T.; Tirosh, I.; Sunwoo, J.B.; Puram, S. V Malignant Cell-Specific CXCL14 Promotes Tumor Lymphocyte Infiltration in Oral Cavity Squamous Cell Carcinoma. J Immunother Cancer 2020, 8, e001048. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Yang, Y.; Pan, Y.; Jia, Y.; Brock, M.V.; Herman, J.G.; Guo, M. Epigenetic Silencing of CXCL14 Induced Colorectal Cancer Migration and Invasion. Discov. Med. 2013, 16, 137–147. [Google Scholar] [PubMed]
- Gu, X.-L.; Ou, Z.-L.; Lin, F.-J.; Yang, X.-L.; Luo, J.-M.; Shen, Z.-Z.; Shao, Z.-M. Expression of CXCL14 and Its Anticancer Role in Breast Cancer. Breast Cancer Res. Treat. 2012, 135, 725–735. [Google Scholar] [CrossRef]
- Sjöberg, E.; Meyrath, M.; Milde, L.; Herrera, M.; Lövrot, J.; Hägerstrand, D.; Frings, O.; Bartish, M.; Rolny, C.; Sonnhammer, E.; et al. A Novel ACKR2-Dependent Role of Fibroblast-Derived CXCL14 in Epithelial-to-Mesenchymal Transition and Metastasis of Breast Cancer. Clin. Cancer Res. 2019, 25, 3702–3717. [Google Scholar] [CrossRef]
- Wente, M.N.; Mayer, C.; Gaida, M.M.; Michalski, C.W.; Giese, T.; Bergmann, F.; Giese, N.A.; Büchler, M.W.; Friess, H. CXCL14 Expression and Potential Function in Pancreatic Cancer. Cancer Lett. 2008, 259, 209–217. [Google Scholar] [CrossRef]
- Horny, K.; Sproll, C.; Peiffer, L.; Furtmann, F.; Gerhardt, P.; Gravemeyer, J.; Stoecklein, N.H.; Spassova, I.; Becker, J.C. Mesenchymal-Epithelial Transition in Lymph Node Metastases of Oral Squamous Cell Carcinoma Is Accompanied by ZEB1 Expression. J. Transl. Med. 2023, 21, 267. [Google Scholar] [CrossRef]
- Gowhari Shabgah, A.; Haleem Al-Qaim, Z.; Markov, A.; Valerievich Yumashev, A.; Ezzatifar, F.; Ahmadi, M.; Mohammad Gheibihayat, S.; Gholizadeh Navashenaq, J. Chemokine CXCL14; a Double-Edged Sword in Cancer Development. Int. Immunopharmacol. 2021, 97, 107681. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Izukuri, K.; Suzuki, K.; Yajima, N.; Ozawa, S.; Ito, S.; Kubota, E.; Hata, R.-I. Chemokine CXCL14/BRAK Transgenic Mice Suppress Growth of Carcinoma Cell Transplants. [Corrected]. Transgenic Res. 2010, 19, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Nara, N.; Nakayama, Y.; Okamoto, S.; Tamura, H.; Kiyono, M.; Muraoka, M.; Tanaka, K.; Taya, C.; Shitara, H.; Ishii, R.; et al. Disruption of CXC Motif Chemokine Ligand-14 in Mice Ameliorates Obesity-Induced Insulin Resistance. J. Biol. Chem. 2007, 282, 30794–30803. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Banisadr, G.; Bhattacharyya, B.J.; Belmadani, A.; Izen, S.C.; Ren, D.; Tran, P.B.; Miller, R.J. The Chemokine BRAK/CXCL14 Regulates Synaptic Transmission in the Adult Mouse Dentate Gyrus Stem Cell Niche. J. Neurochem. 2011, 119, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhang, W.; Wan, T.; He, L.; Chen, T.; Yuan, Z.; Ma, S.; Yu, Y.; Chen, G. Molecular Cloning and Characterization of a Novel CXC Chemokine Macrophage Inflammatory Protein-2 Gamma Chemoattractant for Human Neutrophils and Dendritic Cells. J. Immunol. 2000, 165, 2588–2595. [Google Scholar] [CrossRef]
- Chen, L.; Guo, L.; Tian, J.; He, H.; Marinova, E.; Zhang, P.; Zheng, B.; Han, S. Overexpression of CXC Chemokine Ligand 14 Exacerbates Collagen-Induced Arthritis. J. Immunol. 2010, 184, 4455–4459. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Basilico, P.; Cremona, T.P.; Collins, P.; Moser, B.; Benarafa, C.; Wolf, M. CXCL14 Displays Antimicrobial Activity against Respiratory Tract Bacteria and Contributes to Clearance of Streptococcus Pneumoniae Pulmonary Infection. J. Immunol. 2015, 194, 5980–5989. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Waldemer, R.J.; Nalluri, R.; Nuzzi, P.D.; Chen, J. RNAi Screen Reveals Potentially Novel Roles of Cytokines in Myoblast Differentiation. PLoS ONE 2013, 8, e68068. [Google Scholar] [CrossRef]
- Krohn, S.C.; Bonvin, P.; Proudfoot, A.E.I. CCL18 Exhibits a Regulatory Role through Inhibition of Receptor and Glycosaminoglycan Binding. PLoS ONE 2013, 8, e72321. [Google Scholar] [CrossRef]
- Kuang, H.; Chen, Q.; Zhang, Y.; Zhang, L.; Peng, H.; Ning, L.; Cao, Y.; Duan, E. The Cytokine Gene CXCL14 Restricts Human Trophoblast Cell Invasion by Suppressing Gelatinase Activity. Endocrinology 2009, 150, 5596–5605. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, J.; Yan, D.; Yuan, Y.; Sah, S.; Satyal, U.; Liu, M.; Han, W.; Yu, Y. Neutralization of Chemokine CXCL14 (BRAK) Expression Reduces CCl4 Induced Liver Injury and Steatosis in Mice. Eur. J. Pharmacol. 2011, 671, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Shellenberger, T.D.; Wang, M.; Gujrati, M.; Jayakumar, A.; Strieter, R.M.; Burdick, M.D.; Ioannides, C.G.; Efferson, C.L.; El-Naggar, A.K.; Roberts, D.; et al. BRAK/CXCL14 Is a Potent Inhibitor of Angiogenesis and a Chemotactic Factor for Immature Dendritic Cells. Cancer Res. 2004, 64, 8262–8270. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, M.A.; Fraser, J.K.; Murison, J.G.; Kelly, S.L.; Prestidge, R.L.; Palmer, D.J.; Watson, J.D.; Kumble, K.D. B Cell- and Monocyte-Activating Chemokine (BMAC), a Novel Non-ELR Alpha-Chemokine. Int. Immunol. 2000, 12, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chatterjee, M.; Schmid, H.; Beck, S.; Gawaz, M. CXCL14 as an Emerging Immune and Inflammatory Modulator. J. Inflamm. 2016, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chang, C.; Yang, L.-Y.; Cai, H.-Q.; Chen, D.-X.; Zhang, Y.; Cai, Y.; Wang, J.-J.; Wei, W.-Q.; Hao, J.-J.; et al. Dysregulation of CXCL14 Promotes Malignant Phenotypes of Esophageal Squamous Carcinoma Cells via Regulating SRC and EGFR Signaling. Biochem. Biophys. Res. Commun. 2022, 609, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Liu, R.; Zhang, X.; Cai, L.; Li, Y.; Dong, P.; Gao, J.; Liu, Y.; He, L. CXCL14 as a Potential Marker for Immunotherapy Response Prediction in Renal Cell Carcinoma. Ther. Adv. Med. Oncol. 2023, 15, 17588359231217966. [Google Scholar] [CrossRef]
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef]
- Wrangle, J.M.; Patterson, A.; Johnson, C.B.; Neitzke, D.J.; Mehrotra, S.; Denlinger, C.E.; Paulos, C.M.; Li, Z.; Cole, D.J.; Rubinstein, M.P. IL-2 and Beyond in Cancer Immunotherapy. J. Interferon Cytokine Res. 2018, 38, 45–68. [Google Scholar] [CrossRef]
- Tanegashima, K.; Takahashi, R.; Nuriya, H.; Iwase, R.; Naruse, N.; Tsuji, K.; Shigenaga, A.; Otaka, A.; Hara, T. CXCL14 Acts as a Specific Carrier of CpG DNA into Dendritic Cells and Activates Toll-like Receptor 9-Mediated Adaptive Immunity. EBioMedicine 2017, 24, 247–256. [Google Scholar] [CrossRef]
- Iwase, R.; Naruse, N.; Nakagawa, M.; Saito, R.; Shigenaga, A.; Otaka, A.; Hara, T.; Tanegashima, K. Identification of Functional Domains of CXCL14 Involved in High-Affinity Binding and Intracellular Transport of CpG DNA. J. Immunol. 2021, 207, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Witte, A.; Rohlfing, A.-K.; Dannenmann, B.; Dicenta, V.; Nasri, M.; Kolb, K.; Sudmann, J.; Castor, T.; Rath, D.; Borst, O.; et al. The Chemokine CXCL14 Mediates Platelet Function and Migration via Direct Interaction with CXCR4. Cardiovasc. Res. 2021, 117, 903–917. [Google Scholar] [CrossRef] [PubMed]
- Otte, M.; Kliewer, A.; Schütz, D.; Reimann, C.; Schulz, S.; Stumm, R. CXCL14 Is No Direct Modulator of CXCR4. FEBS Lett. 2014, 588, 4769–4775. [Google Scholar] [CrossRef]
- Tanegashima, K.; Suzuki, K.; Nakayama, Y.; Tsuji, K.; Shigenaga, A.; Otaka, A.; Hara, T. CXCL14 Is a Natural Inhibitor of the CXCL12-CXCR4 Signaling Axis. FEBS Lett. 2013, 587, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Tanegashima, K. CXCL14 Antagonizes the CXCL12-CXCR4 Signaling Axis. Biomol. Concepts 2014, 5, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.J.; McCully, M.L.; Martínez-Muñoz, L.; Santiago, C.; Wheeldon, J.; Caucheteux, S.; Thelen, S.; Cecchinato, V.; Laufer, J.M.; Purvanov, V.; et al. Epithelial Chemokine CXCL14 Synergizes with CXCL12 via Allosteric Modulation of CXCR4. FASEB J. 2017, 31, 3084–3097. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Humphreys, T.D.; Kremer, K.N.; Bramati, P.S.; Bradfield, L.; Edgar, C.E.; Hedin, K.E. CXCR4 Physically Associates with the T Cell Receptor to Signal in T Cells. Immunity 2006, 25, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Kouzeli, A.; Collins, P.J.; Metzemaekers, M.; Meyrath, M.; Szpakowska, M.; Artinger, M.; Struyf, S.; Proost, P.; Chevigne, A.; Legler, D.F.; et al. CXCL14 Preferentially Synergizes With Homeostatic Chemokine Receptor Systems. Front. Immunol. 2020, 11, 561404. [Google Scholar] [CrossRef]
- Song, E.Y.; Shurin, M.R.; Tourkova, I.L.; Gutkin, D.W.; Shurin, G. V Epigenetic Mechanisms of Promigratory Chemokine CXCL14 Regulation in Human Prostate Cancer Cells. Cancer Res. 2010, 70, 4394–4401. [Google Scholar] [CrossRef]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral Vector Platforms within the Gene Therapy Landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef]
- Yun, C.-O.; Hong, J.; Yoon, A.-R. Current Clinical Landscape of Oncolytic Viruses as Novel Cancer Immunotherapeutic and Recent Preclinical Advancements. Front. Immunol. 2022, 13, 953410. [Google Scholar] [CrossRef] [PubMed]
- Shalhout, S.Z.; Miller, D.M.; Emerick, K.S.; Kaufman, H.L. Therapy with Oncolytic Viruses: Progress and Challenges. Nat. Rev. Clin. Oncol. 2023, 20, 160–177. [Google Scholar] [CrossRef] [PubMed]
- Dogbey, D.M.; Torres, V.E.S.; Fajemisin, E.; Mpondo, L.; Ngwenya, T.; Akinrinmade, O.A.; Perriman, A.W.; Barth, S. Technological Advances in the Use of Viral and Non-Viral Vectors for Delivering Genetic and Non-Genetic Cargos for Cancer Therapy. Drug Deliv. Transl. Res. 2023, 13, 2719–2738. [Google Scholar] [CrossRef] [PubMed]
- Mortier, A.; Van Damme, J.; Proost, P. Regulation of Chemokine Activity by Posttranslational Modification. Pharmacol. Ther. 2008, 120, 197–217. [Google Scholar] [CrossRef] [PubMed]
- Gavas, S.; Quazi, S.; Karpiński, T.M. Nanoparticles for Cancer Therapy: Current Progress and Challenges. Nanoscale Res. Lett. 2021, 16, 173. [Google Scholar] [CrossRef]
- Zhu, J.; Martinez, J.; Huang, X.; Yang, Y. Innate Immunity against Vaccinia Virus Is Mediated by TLR2 and Requires TLR-Independent Production of IFN-Beta. Blood 2007, 109, 619–625. [Google Scholar] [CrossRef]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune Responses to Viral Gene Therapy Vectors. Mol Ther 2020, 28, 709–722. [Google Scholar] [CrossRef]
- Zaiss, A.K.; Machado, H.B.; Herschman, H.R. The Influence of Innate and Pre-existing Immunity on Adenovirus Therapy. J. Cell. Biochem. 2009, 108, 778–790. [Google Scholar] [CrossRef]
- Hillyer, P.; Male, D. Expression of Chemokines on the Surface of Different Human Endothelia. Immunol. Cell Biol. 2005, 83, 375–382. [Google Scholar] [CrossRef]
- Koren, I.; Timms, R.T.; Kula, T.; Xu, Q.; Li, M.Z.; Elledge, S.J. The Eukaryotic Proteome Is Shaped by E3 Ubiquitin Ligases Targeting C-Terminal Degrons. Cell 2018, 173, 1622–1635.e14. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-Distance Constraints Applied on Model Quality Estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-New Features and Functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated Comparative Protein Structure Modeling with SWISS-MODEL and Swiss-PdbViewer: A Historical Perspective. Electrophoresis 2009, 30 (Suppl. S1), S162–S273. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling Protein Quaternary Structure of Homo- and Hetero-Oligomers beyond Binary Interactions by Homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef]
- Peterson, F.C.; Thorpe, J.A.; Harder, A.G.; Volkman, B.F.; Schwarze, S.R. Structural Determinants Involved in the Regulation of CXCL14/BRAK Expression by the 26 S Proteasome. J. Mol. Biol. 2006, 363, 813–822. [Google Scholar] [CrossRef]
- Khalil, M.I.; Yang, C.; Vu, L.; Chadha, S.; Nabors, H.; Welbon, C.; James, C.D.; Morgan, I.M.; Spanos, W.C.; Pyeon, D. HPV Upregulates MARCHF8 Ubiquitin Ligase and Inhibits Apoptosis by Degrading the Death Receptors in Head and Neck Cancer. PLoS Pathog. 2023, 19, e1011171. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the Tumor Immune Microenvironment (TIME) for Effective Therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Melero, I.; Castanon, E.; Alvarez, M.; Champiat, S.; Marabelle, A. Intratumoural Administration and Tumour Tissue Targeting of Cancer Immunotherapies. Nat. Rev. Clin. Oncol. 2021, 18, 558–576. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Yang, W.; Su, J.; Ren, X.; Fazelzad, R.; Albert, T.; Habbous, S.; Goldstein, D.P.; de Almeida, J.R.; Hansen, A.; et al. Human Papillomavirus and P16 Immunostaining, Prevalence and Prognosis of Squamous Carcinoma of Unknown Primary in the Head and Neck Region. Int. J. Cancer. 2019, 145, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, F.; Mariz, F.; Osen, W.; Bolchi, A.; Ottonello, S.; Müller, M. Combined Prophylactic and Therapeutic Immune Responses against Human Papillomaviruses Induced by a Thioredoxin-Based L2-E7 Nanoparticle Vaccine. PLoS Pathog. 2020, 16, e1008827. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Ferrall, L.; Gaillard, S.; Wang, C.; Chi, W.-Y.; Huang, C.-H.; Roden, R.B.S.; Wu, T.-C.; Chang, Y.-N.; Hung, C.-F. Development of DNA Vaccine Targeting E6 and E7 Proteins of Human Papillomavirus 16 (HPV16) and HPV18 for Immunotherapy in Combination with Recombinant Vaccinia Boost and PD-1 Antibody. mBio 2021, 12, e03224-20. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giacobbi, N.S.; Mullapudi, S.; Nabors, H.; Pyeon, D. The Chemokine CXCL14 as a Potential Immunotherapeutic Agent for Cancer Therapy. Viruses 2024, 16, 302. https://doi.org/10.3390/v16020302
Giacobbi NS, Mullapudi S, Nabors H, Pyeon D. The Chemokine CXCL14 as a Potential Immunotherapeutic Agent for Cancer Therapy. Viruses. 2024; 16(2):302. https://doi.org/10.3390/v16020302
Chicago/Turabian StyleGiacobbi, Nicholas S., Shreya Mullapudi, Harrison Nabors, and Dohun Pyeon. 2024. "The Chemokine CXCL14 as a Potential Immunotherapeutic Agent for Cancer Therapy" Viruses 16, no. 2: 302. https://doi.org/10.3390/v16020302
APA StyleGiacobbi, N. S., Mullapudi, S., Nabors, H., & Pyeon, D. (2024). The Chemokine CXCL14 as a Potential Immunotherapeutic Agent for Cancer Therapy. Viruses, 16(2), 302. https://doi.org/10.3390/v16020302






