Phage Display’s Prospects for Early Diagnosis of Prostate Cancer
Abstract
1. Introduction
2. Advanced Phage-Driven Analytical Tools for the Diagnosis of PC
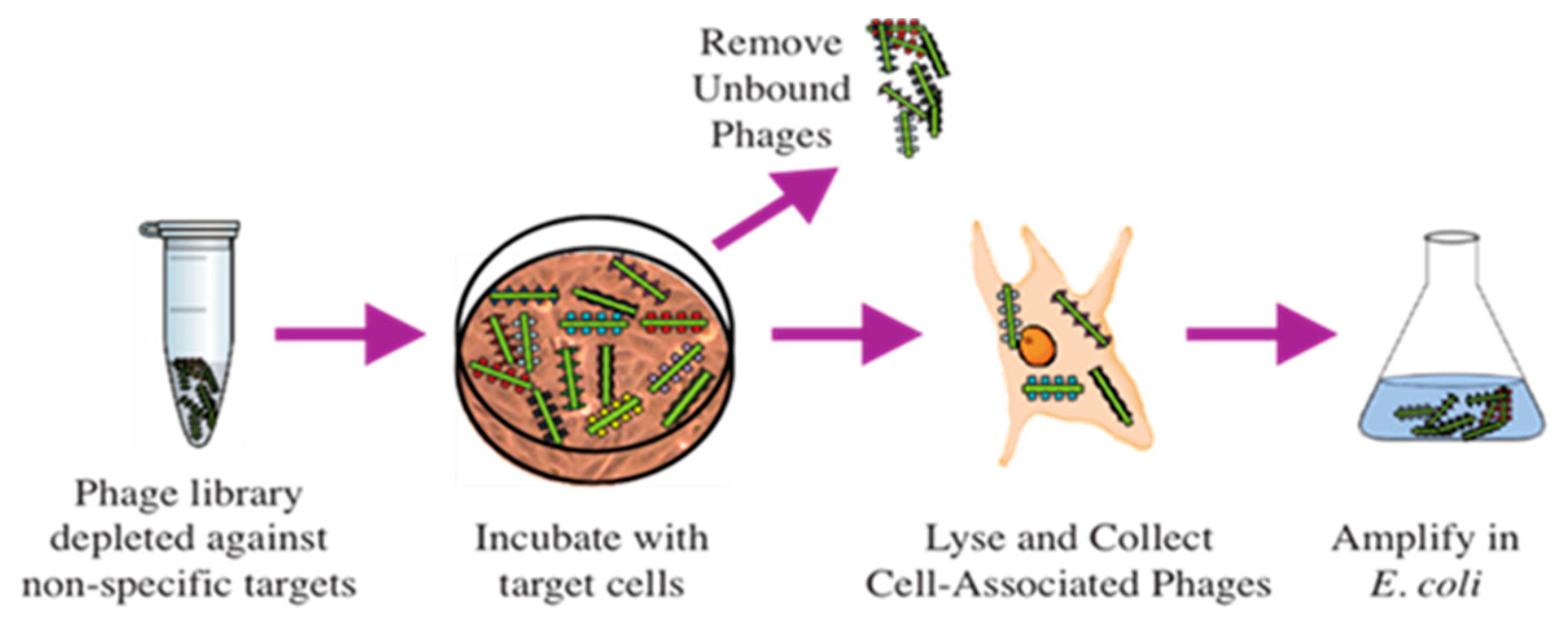
2.1. Selection of Phage Probes against PC-Cell-Associated Antigens
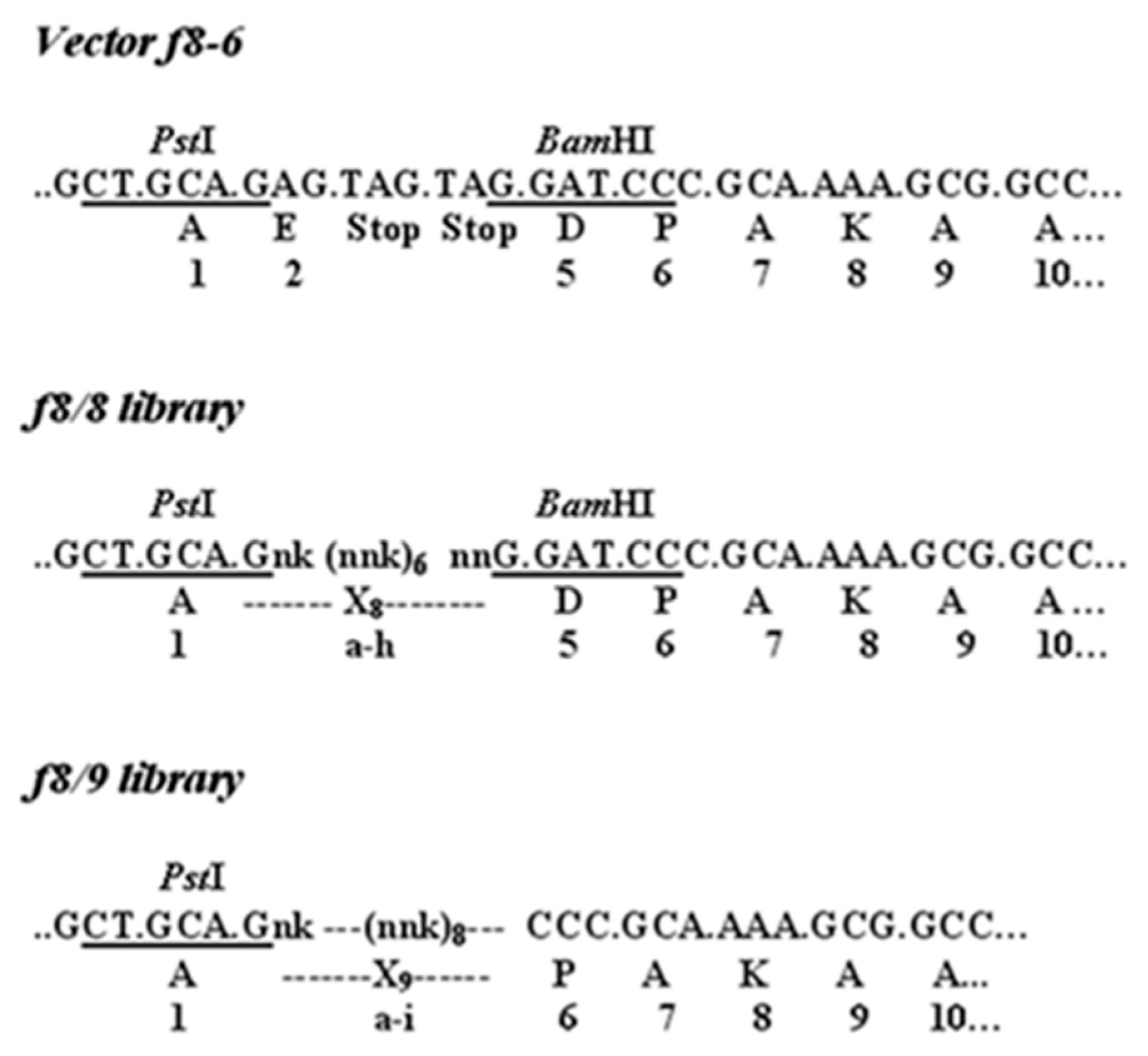
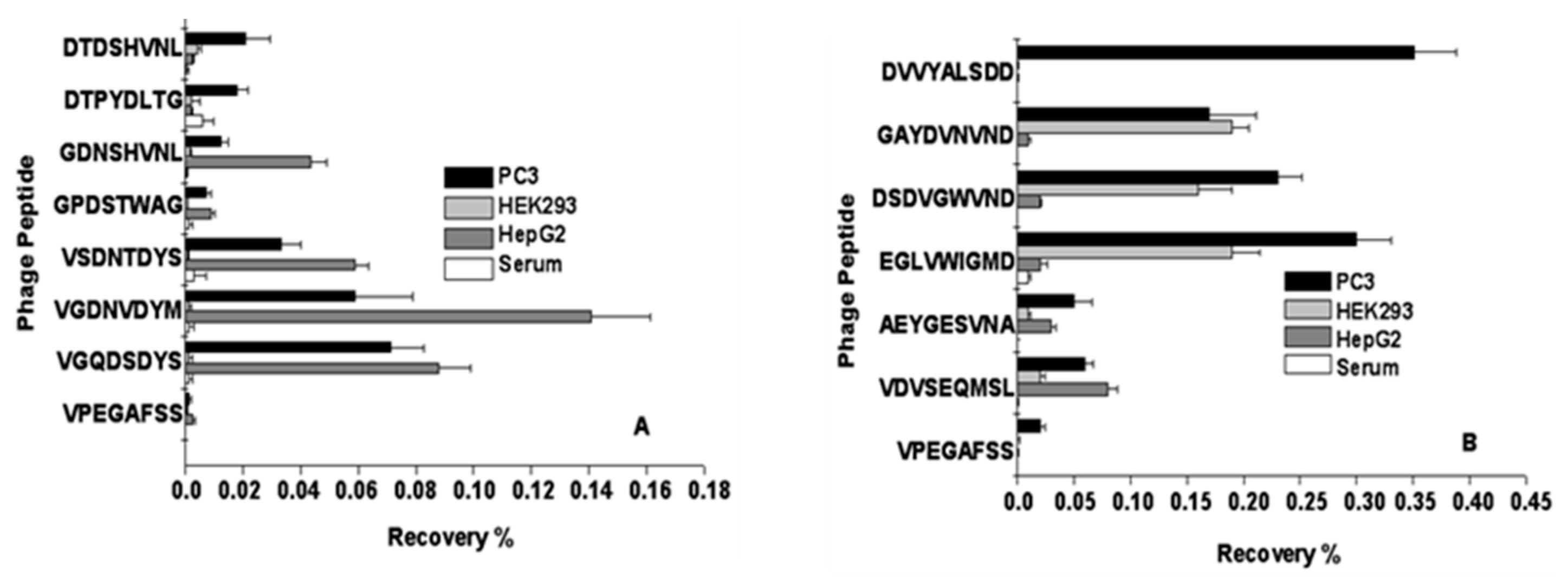
2.1.1. Selection of PC-Cell-Binding Phages from f8-Type (Landscape) Libraries
2.1.2. Selection of PC Cell Binders from p3-Type Phage-Displayed Antibody Libraries
2.2. Selection of Phage Probes against Prostate-Specific Antigen (PSA)
2.2.1. PSA as a PC Biomarker
2.2.2. Selection of p3-Type Phage-Displayed Peptides against PSA
2.2.3. Development and Affinity Maturation of p3-Type Phage-Displayed Antibodies against PSA
2.2.4. Selection of PSA-Binding p8-Type Multivalent Landscape Phage Probes
3. Development of Phage-Driven Biosensors for the Detection of Different forms of PSA
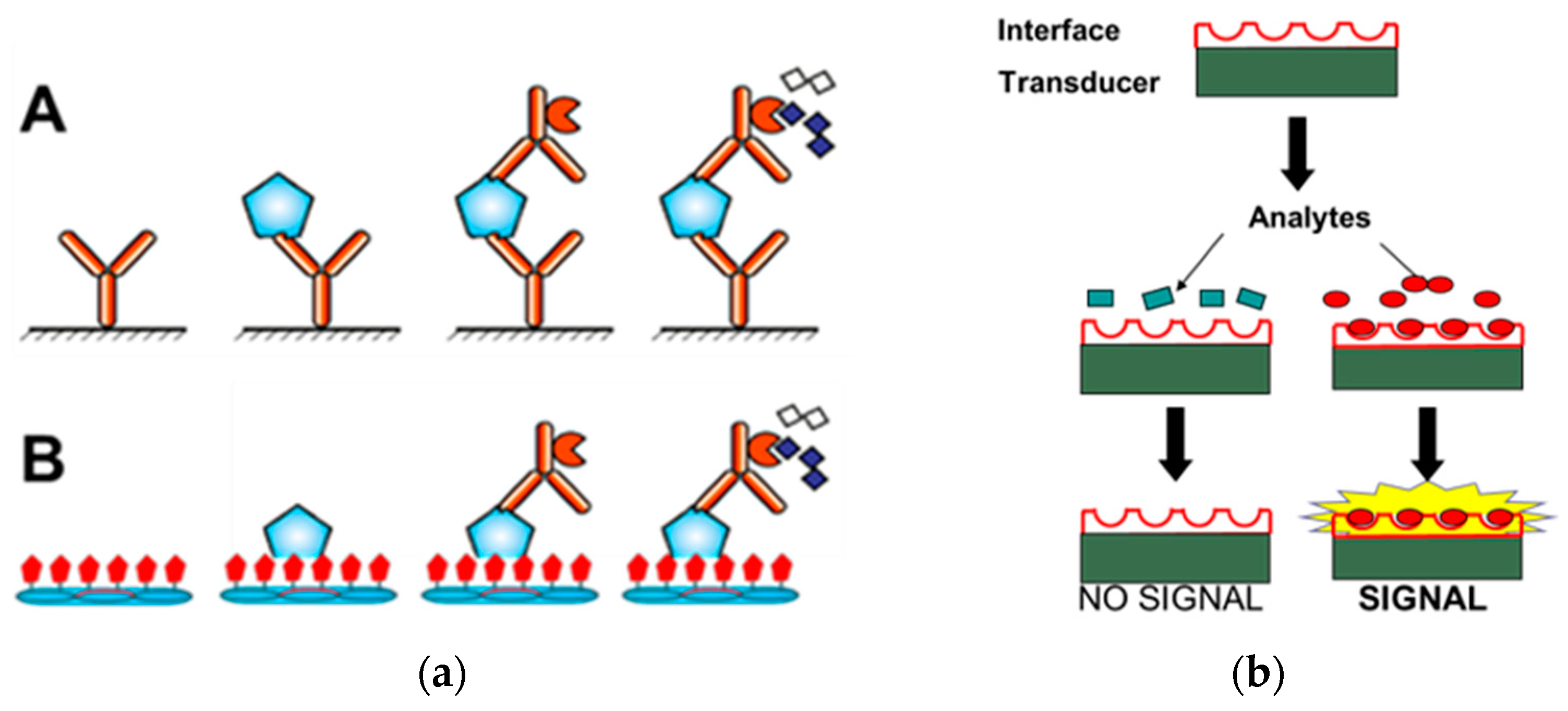
3.1. Landscape-Phage-Driven Enzyme-Linked Immunosorbent Assay (Phage ELISA)
3.2. Phage-Driven Electrochemical Immunosensors for Detection of PSA
4. Conclusions
Funding
Conflicts of Interest
References
- Chhikara, B.S.; Parang, K. Global Cancer Statistics 2022: The trends projection analysis. Chem. Biol. Lett. 2023, 10, 451. [Google Scholar]
- Hugosson, J.; Roobol, M.J.; Mansson, M.; Tammela, T.L.J.; Zappa, M.; Nelen, V.; Kwiatkowski, M.; Lujan, M.; Carlsson, S.V.; Talala, K.M.; et al. A 16-yr Follow-up of the European Randomized study of Screening for Prostate Cancer. Eur. Urol. 2019, 76, 43–51. [Google Scholar] [CrossRef]
- Hawkes, N. Cancer survival data emphasize importance of early diagnosis. BMJ 2019, 364, l408. [Google Scholar] [CrossRef]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early detection of cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Ioffe, V. Early Detection of Prostate Cancer: AUA/SUO Guideline Part I: Prostate Cancer Screening. Letter. J. Urol. 2023, 210, 731. [Google Scholar] [CrossRef] [PubMed]
- Moghul, M.; Cazzaniga, W.; Croft, F.; Kinsella, N.; Cahill, D.; James, N.D. Mobile Health Solutions for Prostate Cancer Diagnostics—A Systematic Review. Clin. Pr. 2023, 13, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J. Tumor Markers in Clinical Practice: A Review Focusing on Common Solid Cancers. Med. Princ. Pract. 2013, 22, 4–11. [Google Scholar] [CrossRef]
- Jatho, A.; Mugisha, N.M.; Kafeero, J.; Holoya, G.; Okuku, F.; Niyonzima, N. Mobile cancer prevention and early detection outreach in Uganda: Partnering with communities toward bridging the cancer health disparities through “asset-based community development model”. Cancer Med. 2020, 9, 7317–7329. [Google Scholar] [CrossRef]
- Zhu, M.; Liang, Z.; Feng, T.; Mai, Z.; Jin, S.; Wu, L.; Zhou, H.; Chen, Y.; Yan, W. Up-to-Date Imaging and Diagnostic Techniques for Prostate Cancer: A Literature Review. Diagnostics 2023, 13, 2283. [Google Scholar] [CrossRef]
- Nasimi, H.; Madsen, J.S.; Zedan, A.H.; Malmendal, A.; Osther, P.J.S.; Alatraktchi, F.A. Protein biomarker detection in prostate cancer: A comprehensive review of electrochemical biosensors. Sens. Actuators Rep. 2023, 6, 100168. [Google Scholar] [CrossRef]
- Guliy, O.I.; Staroverov, S.A.; Dykman, L.A. Heat Shock Proteins in Cancer Diagnostics. Appl. Biochem. Microbiol. 2023, 59, 395–407. [Google Scholar] [CrossRef]
- Fenton, J.J.; Weyrich, M.S.; Durbin, S.; Liu, Y.; Bang, H.; Melnikow, J. Prostate-Specific Antigen-Based Screening for Prostate Cancer: Evidence Report and Systematic Review for the US Preventive Services Task Force. J. Am. Med. Assoc. 2018, 319, 1914–1931. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Ma, S.L.; Hara, T.O.; Singh, S. Nanomaterials-Based Biosensors for the Detection of Prostate Cancer Biomarkers: Recent Trends and Future Perspective. Adv. Mater. Technol. 2023, 8, 2201860. [Google Scholar] [CrossRef]
- Liu, X.; Wang, D.; Chu, J.S.; Xu, Y.; Wang, W.J. Sandwich pair nanobodies, a pOKor electrochemical immunosensing serum prostate-specific antigen with preferable specificity. J. Pharm. Biomed. Anal. 2018, 158, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kukkar, D.; Hashemi, B.; Kim, K.H.; Deep, A. Advanced Functional Structure-Based Sensing and Imaging Strategies for Cancer Detection: Possibilities, Opportunities, Challenges, and Prospects. Adv. Funct. Mater. 2019, 29, 1807859. [Google Scholar] [CrossRef]
- Garg, S.; Sachdeva, A.; Peeters, M.; McClements, J. Point-of-Care Prostate Specific Antigen Testing: Examining Translational Progress toward Clinical Implementation. Acs Sens. 2023, 8, 3643–3658. [Google Scholar] [CrossRef]
- Panteghini, M. Implementation of standardization in clinical practice: Not always an easy task. Clin. Chem. Lab. Med. 2012, 50, 1237–1241. [Google Scholar] [CrossRef]
- Ferraro, S.; Bussetti, M.; Rizzardi, S.; Braga, F.; Panteghini, M. Verification of Harmonization of Serum Total and Free Prostate-Specific Antigen (PSA) Measurements and Implications for Medical Decisions. Clin. Chem. 2021, 67, 543–553. [Google Scholar] [CrossRef]
- Stenman, U.H.; Paus, E.; Allard, W.J.; Andersson, I.; Andrès, C.; Barnett, T.R.; Becker, C.; Belenky, A.; Bellanger, L.; Pellegrino, C.M.; et al. Summary report of the TD-3 workshop: Characterization of 83 antibodies against prostate-specific antigen. Tumor Biol. 1999, 20, 1–12. [Google Scholar] [CrossRef]
- Srinivasan, B.; Nanus, D.M.; Erickson, D.; Mehta, S. Highly portable quantitative screening test for prostate-specific antigen at point of care. Curr. Res. Biotechnol. 2021, 3, 288–299. [Google Scholar] [CrossRef]
- Stephan, C.; Kramer, J.; Meyer, H.A.; Kristiansen, G.; Ziemer, S.; Deger, S.; Lein, M.; Loening, S.A.; Jung, K. Different prostate-specific antigen assays give different results on the same blood sample: An obstacle to recommending uniform limits for prostate biopsies. BJU Int. 2007, 99, 1427–1431. [Google Scholar] [CrossRef]
- Dukle, A.; Nathanael, A.J.; Panchapakesan, B.; Oh, T.H. Role of Paper-Based Sensors in Fight against Cancer for the Developing World. Biosensors 2022, 12, 737. [Google Scholar] [CrossRef]
- Graves, H.C.; Wehner, N.; Stamey, T.A. Ultrasensitive Radioimmunoassay of Prostate-Specific Antigen. Clin. Chem. 1992, 38, 735–742. [Google Scholar] [CrossRef]
- Myrtle, J.F.; Shackelford, W.; Bartholomew, R.M.; Wampler, J. Prostate-Specific Antigen-Quantitation in Serum by IMMUNORADIOMETRIC ASSAY. Clin. Chem. 1983, 29, 1216. [Google Scholar]
- Lang, Q.; Wang, F.; Yin, L.; Liu, M.; Petrenko, V.A.; Liu, A. Specific Probe Selection from Landscape Phage Display Library and Its Application in Enzyme-Linked Immunosorbent Assay of Free Prostate-Specific Antigen. Anal. Chem. 2014, 86, 2767–2774. [Google Scholar] [CrossRef]
- Eriksson, S.; Vehniäinen, M.; Jansén, T.; Meretoja, V.; Saviranta, P.; Pettersson, K.; Lövgren, T. Dual-label time-resolved immunofluorometric assay of free and total prostate-specific antigen based on recombinant Fab fragments. Clin. Chem. 2000, 46, 658–666. [Google Scholar] [CrossRef]
- Peltomaa, R.; Benito-Peña, E.; Barderas, R.; Moreno-Bondi, M.C. Phage Display in the Quest for New Selective Recognition Elements for Biosensors. Acs Omega 2019, 4, 11569–11580. [Google Scholar] [CrossRef] [PubMed]
- Kierny, M.R.; Cunningham, T.D.; Kay, B.K. Detection of biomarkers using recombinant antibodies coupled to nanostructured platforms. Nano Rev. Exp. 2012, 3, 17240. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Guliy, O.I.; Evstigneeva, S.S.; Dykman, L.A. Recombinant antibodies by phage display for bioanalytical applications. Biosens. Bioelectron. 2023, 222, 114909. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Phage Display: Simple Evolution in a Petri Dish (Nobel Lecture). Angew. Chem. Int. Ed. 2019, 58, 14428–14437. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, V.A.; Smith, G.P.; Gong, X.; Quinn, T. A library of organic landscapes on filamentous phage. Protein Eng. 1996, 9, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, V.A.; Smith, G.P. Phages from landscape libraries as substitute antibodies. Protein Eng. 2000, 13, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, V.A. Landscape phage as a molecular recognition interface for detection devices. Microelectron. J. 2008, 39, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Petrenko, V.A.; Vodyanoy, V.J. Phage display for detection of biological threat agents. J. Microbiol. Methods 2003, 53, 253–262. [Google Scholar] [CrossRef]
- Nanduri, V.; Sorokulova, I.B.; Samoylov, A.M.; Simonian, A.L.; Petrenko, V.A.; Vodyanoy, V. Phage as a molecular recognition element in biosensors immobilized by physical adsorption. Biosens. Bioelectron. 2007, 22, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Brigati, J.R.; Samoylova, T.I.; Jayanna, P.K.; Petrenko, V.A. Phage display for generating peptide reagents. Curr. Protoc. Protein Sci. 2008, 51, 18.9.1–18.9.27. [Google Scholar] [CrossRef] [PubMed]
- Kuzmicheva, G.A.; Jayanna, P.K.; Sorokulova, I.B.; Petrenko, V.A. Diversity and censoring of landscape phage libraries. Protein Eng. Des. Sel. 2009, 22, 9–18. [Google Scholar] [CrossRef]
- Horikawa, S.; Bedi, D.; Li, S.; Shen, W.; Huang, S.; Chen, I.H.; Chai, Y.; Auad, M.L.; Bozack, M.J.; Barbaree, J.M.; et al. Effects of surface functionalization on the surface phage coverage and the subsequent performance of phage-immobilized magnetoelastic biosensors. Biosens. Bioelectron. 2011, 26, 2361–2367. [Google Scholar] [CrossRef]
- Scott, A.M.; Welt, S. Antibody-based immunological therapies. Curr. Opin. Immunol. 1997, 9, 717–722. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Smith, G.P.; Mazooji, M.M.; Quinn, T. Alpha-helically constrained phage display library. Protein Eng. 2002, 15, 943–950. [Google Scholar] [CrossRef]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef]
- Kuzmicheva, G.A.; Jayanna, P.K.; Eroshkin, A.M.; Grishina, M.A.; Pereyaslavskaya, E.S.; Potemkin, V.A.; Petrenko, V.A. Mutations in fd phage major coat protein modulate affinity of the displayed peptide. Protein Eng. Des. Sel. 2009, 22, 631–639. [Google Scholar] [CrossRef]
- Knez, K.; Noppe, W.; Geukens, N.; Janssen, K.P.F.; Spasic, D.; Heyligen, J.; Vriens, K.; Thevissen, K.; Cammue, B.P.A.; Petrenko, V.; et al. Affinity Comparison of p3 and p8 Peptide Displaying Bacteriophages Using Surface Plasmon Resonance. Anal. Chem. 2013, 85, 10075–10082. [Google Scholar] [CrossRef]
- Gomes, M.; Fleck, A.; Degaugue, A.; Gourmelon, F.; Léger, C.; Aumont-Nicaise, M.; Mesneau, A.; Jean-Jacques, H.; Hassaine, G.; Urvoas, A.; et al. Design of an artificial phage-display library based on a new scaffold improved for average stability of the randomized proteins. Sci. Rep. 2023, 13, 1339. [Google Scholar] [CrossRef]
- Petrenko, V.A. Landscape Phage: Evolution from Phage Display to Nanobiotechnology. Viruses 2018, 10, 311. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Gillespie, J.W.; De Plano, L.M.; Shokhen, M.A. Phage-Displayed Mimotopes of SARS-CoV-2 Spike Protein Targeted to Authentic and Alternative Cellular Receptors. Viruses 2022, 14, 384. [Google Scholar] [CrossRef]
- Han, L.; Xia, H.; Yin, L.; Petrenko, V.A.; Liu, A. Selected landscape phage probe as selective recognition interface for sensitive total prostate-specific antigen immunosensor. Biosens. Bioelectron. 2018, 106, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Wang, D.; Yan, L.; Petrenko, V.A.; Liu, A.H. Specific phages-based electrochemical impedimetric immunosensors for label-free and ultrasensitive detection of dual prostate-specific antigens. Sens. Actuators B-Chem. 2019, 297, 126727. [Google Scholar] [CrossRef]
- Qi, H.; Wang, F.; Petrenko, V.A.; Liu, A. Peptide Microarray with Ligands at High Density Based on Symmetrical Carrier Landscape Phage for Detection of Cellulase. Anal. Chem. 2014, 86, 5844–5850. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.R.; Kelly, K.A.; Mahmood, U.; Weissleder, R.; Deutscher, S.L. In vivo selection of phage for the optical imaging of PC-3 human prostate carcinoma in mice. Neoplasia 2006, 8, 772–780. [Google Scholar] [CrossRef]
- Romanov, V.I.; Durand, D.B.; Petrenko, V.A. Phage display selection of peptides that affect prostate carcinoma cells attachment and invasion. Prostate 2001, 47, 239–251. [Google Scholar] [CrossRef]
- Fagbohun, O.A.; Kazmierczak, R.A.; Petrenko, V.A.; Eisenstark, A. Metastatic prostate cancer cell-specific phage-like particles as a targeted gene-delivery system. J. Nanobiotechnology 2013, 11, 31. [Google Scholar] [CrossRef]
- Bhasin, A.; Drago, N.P.; Majumdar, S.; Sanders, E.C.; Weiss, G.A.; Penner, R.M. Viruses Masquerading as Antibodies in Biosensors: The Development of the Virus BioResistor. Acc. Chem. Res. 2020, 53, 2384–2394. [Google Scholar] [CrossRef]
- Jayanna, P.K.; Bedi, D.; Deinnocentes, P.; Bird, R.C.; Petrenko, V.A. Landscape phage ligands for PC3 prostate carcinoma cells. Protein Eng. Des. Sel. 2010, 23, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Jayanna, P.K.; Bedi, D.; Gillespie, J.W.; DeInnocentes, P.; Wang, T.; Torchilin, V.P.; Bird, R.C.; Petrenko, V.A. Landscape phage fusion protein-mediated targeting of nanomedicines enhances their prostate tumor cell association and cytotoxic efficiency. Nanomedicine 2010, 6, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Liu, P.; Petrenko, V.A.; Liu, A. A Label-Free Electrochemical Impedance Cytosensor Based on Specific Peptide-Fused Phage Selected from Landscape Phage Library. Sci. Rep. 2016, 6, 22199. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Han, L.; Wang, F.; Petrenko, V.A.; Liu, A. Gold nanoprobe functionalized with specific fusion protein selection from phage display and its application in rapid, selective and sensitive colorimetric biosensing of Staphylococcus aureus. Biosens. Bioelectron. 2016, 82, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, Y.B.; Han, L.; Cai, Y.Y.; Ren, H.; Ma, T.X.; Li, X.Q.; Petrenko, V.A.; Liu, A.H. Colorimetric Assay of Bacterial Pathogens Based on Co3O4 Magnetic Nanozymes Conjugated with Specific Fusion Phage Proteins and Magnetophoretic Chromatography. ACS Appl. Mater. Interfaces 2020, 12, 9090–9097. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Lu, H.; Qiu, H.J.; Petrenko, V.; Liu, A. Phagemid vectors for phage display: Properties, characteristics and construction. J. Mol. Biol. 2012, 417, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, P.; Sun, L.; Li, C.; Petrenko, V.A.; Liu, A. Bio-mimetic nanostructure self-assembled from Au@Ag heterogeneous nanorods and phage fusion proteins for targeted tumor optical detection and photothermal therapy. Sci. Rep. 2014, 4, 6808. [Google Scholar] [CrossRef]
- Shen, W.; Li, S.; Park, M.-K.; Zhang, Z.; Cheng, Z.; Petrenko, V.A.; Chin, B.A. Blocking Agent Optimization for Nonspecific Binding on Phage Based Magnetoelastic Biosensors. J. Electrochem. Soc. 2012, 159, B818. [Google Scholar] [CrossRef]
- Li, S.; Lakshmanan, R.S.; Petrenko, V.A.; Chin, B.A.; Blois, H.; Cao, B.; Chin, B.; Deutscher, S.L.; Gea, M.; Iris, F.; et al. Phage-based Pathogen Biosensors. In Phage Nanobiotechnology; Petrenko, V., Smith, G.P., O’Brien, P., Craighead, H., Kroto, H., Eds.; The Royal Society of Chemistry: London, UK, 2011. [Google Scholar]
- Lakshmanan, R.S.; Guntupalli, R.; Hu, J.; Kim, D.J.; Petrenko, V.A.; Barbaree, J.M.; Chin, B.A. Phage immobilized magnetoelastic sensor for the detection of Salmonella typhimurium. J. Microbiol. Methods 2007, 71, 55–60. [Google Scholar] [CrossRef]
- Fu, L.; Li, S.; Zhang, K.; Chen, I.-H.; Petrenko, V.A.; Cheng, Z. Magnetostrictive Microcantilever as an Advanced Transducer for Biosensors. Sensors 2007, 7, 2929–2941. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Lee, M.; Kim, D. Detection of early-stage prostate cancer by using a simple carbon nanotube@paper biosensor. Biosens. Bioelectron. 2018, 102, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Singh, J.; Goyat, R.; Saharan, Y.; Chaudhry, V.; Umar, A.; Ibrahim, A.A.; Akbar, S.; Ameen, S.; Baskoutas, S. Nanomaterials-based biosensor and their applications: A review. Heliyon 2023, 9, e19929. [Google Scholar] [CrossRef]
- Pillay, T.S.; Muyldermans, S. Application of Single-Domain Antibodies (“Nanobodies”) to Laboratory Diagnosis. Ann. Lab. Med. 2021, 41, 549–558. [Google Scholar] [CrossRef]
- Fernández-Sánchez, C.; McNeil, C.J.; Rawson, K.; Nilsson, O. Disposable Noncompetitive Immunosensor for Free and Total Prostate-Specific Antigen Based on Capacitance Measurement. Anal. Chem. 2004, 76, 5649–5656. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, Y.; Sadeghi, M.; Ehzari, H.; Derakhshankhah, H. Label-free electrochemical immunosensor based on antibody-immobilized Fe-Cu layered double hydroxide nanosheetas an electrochemical probe for the detection of ultra trace amount of prostate cancer biomarker (PSA). Microchem. J. 2023, 195, 109460. [Google Scholar] [CrossRef]
- Nakhjavani, S.A.; Tokyay, B.K.; Soylemez, C.; Sarabi, M.R.; Yetisen, A.K.; Tasoglu, S. Biosensors for prostate cancer detection. Trends Biotechnol. 2023, 41, 1248–1267. [Google Scholar] [CrossRef]
- Saerens, D.; Frederix, F.; Reekmans, G.; Conrath, K.; Jans, K.; Brys, L.; Huang, L.; Bosmans, E.; Maes, G.; Borghs, G.; et al. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal. Chem. 2005, 77, 7547–7555. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Boon, T. Tumor antigens recognised by T lymphocytes. Eur. J. Cancer 1999, 35, S216. [Google Scholar]
- Jayanna, P.K.; Deinnocentes, P.; Bird, R.C.; Petrenko, V.A. Landscape Phage Probes for PC3 Prostate Carcinoma cells. In Proceedings of the Nanotechnology Conference and Trade Show (Nanotech 2008), Boston, MA, USA, 1–5 June 2008; pp. 457–460. [Google Scholar]
- Winter, G. Harnessing Evolution to Make Medicines (Nobel Lecture). Angew. Chem.-Int. Edit. 2019, 58, 14438–14445. [Google Scholar] [CrossRef] [PubMed]
- Popkov, M.; Rader, C.; Barbas, C.F. Isolation of human prostate cancer cell reactive antibodies using phage display technology. J. Immunol. Methods 2004, 291, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Rader, C.; Ritter, G.; Nathan, S.; Elia, M.; Gout, I.; Jungbluth, A.A.; Cohen, L.S.; Welt, S.; Old, L.J.; Barbas, C.F. The rabbit antibody repertoire as a novel source for the generation of therapeutic human antibodies. J. Biol. Chem. 2000, 275, 13668–13676. [Google Scholar] [CrossRef] [PubMed]
- Stenman, U.H.; Leinonen, J.; Alfthan, H.; Rannikko, S.; Tuhkanen, K.; Alfthan, O. A Complex between Prostate-Specific Antigen and Alpha-1-Antichymotrypsin Is the Major Form of Prostate-Specific Antigen in Serum of Patients with Prostatic-Cancer-assay of the Complex Improves Clinical Sensitivity for Cancer. Cancer Res. 1991, 51, 222–226. [Google Scholar] [PubMed]
- Stamey, T.A.; Yang, N.; Hay, A.R.; McNeal, J.E.; Freiha, F.S.; Redwine, E. Prostate-Specific Antigen as A Serum Marker for Adenocarcinoma of the Prostate. N. Engl. J. Med. 1987, 317, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Rowe, E.W.J.; Laniado, M.E.; Walker, M.M.; Patel, A. Prostate cancer detection in men with a ‘normal’ total prostate-specific antigen (PSA) level using percentage free PSA: A prospective screening study. BJU Int. 2005, 95, 1249–1252. [Google Scholar] [CrossRef]
- Jung, K.; Brux, B.; Lein, M.; Rudolph, B.; Kristiansen, G.; Hauptmann, S.; Schnorr, D.; Loening, S.A.; Sinha, P. Molecular forms of prostate-specific antigen in malignant and benign prostatic tissue: Biochemical and diagnostic implications. Clin. Chem. 2000, 46, 47–54. [Google Scholar] [CrossRef]
- Huang, H.Q.; Zhang, Y.; Xu, H.G. Different free prostate-specific antigen to total prostate-specific antigen ratios using three detecting systems. J. Clin. Lab. Anal. 2018, 32, e22231. [Google Scholar] [CrossRef] [PubMed]
- Lilja, H.; Oldbring, J.; Rannevik, G.; Laurell, C.B. Seminal Vesicle-Secreted Proteins and Their Reactions during Gelation and Liquefaction of Human-Semen. J. Clin. Investig. 1987, 80, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Loeb, S.; Lilja, H.; Vickers, A. Beyond prostate-specific antigen: Utilizing novel strategies to screen men for prostate cancer. Curr. Opin. Urol. 2016, 26, 459–465. [Google Scholar] [CrossRef]
- McJimpsey, E.L. Molecular Form Differences Between Prostate-Specific Antigen (PSA) Standards Create Quantitative Discordances in PSA ELISA Measurements. Sci. Rep. 2016, 6, 22050. [Google Scholar] [CrossRef] [PubMed]
- Stephan, C.; Jung, K.; Lein, M.; Sinha, P.; Schnorr, D.; Loening, S.A. Molecular forms of prostate-specific antigen and human kallikrein 2 as promising tools for early diagnosis of prostate cancer. Cancer Epidemiol. Biomark. Prev. 2000, 9, 1133–1147. [Google Scholar]
- Filella, X.; Truan, D.; Alcover, J.; Quintó, L.; Molina, R.; Luque, P.; Coca, F.; Ballesta, A.M. Comparison of several combinations of free, complexed, and total PSA in the diagnosis of prostate cancer in patients with urologic symptoms. Urology 2004, 63, 1100–1103. [Google Scholar] [CrossRef]
- Grossklaus, D.J.; Shappell, S.B.; Gautam, S.; Smith, J.A.; Cookson, M.S. Ratio of Free-to-Total Prostate Specific Antigen Correlates with Tumor Volume in Patients with Increased Prostate Specific Antigen. J. Urol. 2001, 165, 455–458. [Google Scholar] [CrossRef]
- Jung, K.; Elgeti, U.; Lein, M.; Brux, B.; Sinha, P.; Rudolph, B.; Hauptmann, S.; Schnorr, D.; Loening, S.A. Ratio of Free or Complexed Prostate-specific Antigen (PSA) to Total PSA: Which Ratio Improves Differentiation between Benign Prostatic Hyperplasia and Prostate Cancer? Clin. Chem. 2000, 46, 55–62. [Google Scholar] [CrossRef]
- Djavan, B.; Zlotta, A.; Kratzik, C.; Remzi, M.; Seitz, C.; Schulman, C.C.; Marberger, M. PSA, PSA density, PSA density of transition zone, free/total PSA ratio, and PSA velocity for early detection of prostate cancer in men with serum PSA 2.5 to 4.0 ng/ml. Urology 1999, 54, 517–522. [Google Scholar] [CrossRef]
- Dorizzi, R.M.; Maltoni, P.; Sgarzani, C.; Torello, M.; Montanari, F. Spurious results for total and free prostatespecific antigen (PSA); sometimes really “a riddle wrapped in a mystery inside an enigma”. Clin. Chem. Lab. Med. 2022, 60, E91–E94. [Google Scholar] [CrossRef]
- Ferraro, S.; Biganzoli, E.M.; Plebani, M. Reply to: Spurious results for total and free prostate-specific antigen (PSA); sometimes really “a riddle wrapped in a mystery inside an enigma”. Clin. Chem. Lab. Med. 2022, 60, E95–E96. [Google Scholar] [CrossRef]
- Ferrieu-Weisbuch, C.; Michel, S.; Collomb-Clerc, E.; Pothion, C.; Deléage, G.; Jolivet-Reynaud, C. Characterization of prostate-specific antigen binding peptides selected by phage display technology. J. Mol. Recognit. 2006, 19, 10–20. [Google Scholar] [CrossRef]
- Wang, Y.B.; Wang, M.Y.; Yu, H.P.; Wang, G.; Ma, P.X.; Pang, S.; Jiao, Y.M.; Liu, A.H. Screening of peptide selectively recognizing prostate-specific antigen and its application in detecting total prostate-specific antigen. Sens. Actuators B Chem. 2022, 367, 132009. [Google Scholar] [CrossRef]
- Wu, P.; Leinonen, J.; Koivunen, E.; Lankinen, H.; Stenman, U.H. Identification of novel prostate-specific antigen-binding peptides modulating its enzyme activity. Eur. J. Biochem. 2000, 267, 6212–6220. [Google Scholar] [CrossRef]
- Wu, P.; Zhu, L.; Stenman, U.H.; Leinonen, J. Immunopeptidometric assay for enzymatically active prostate-specific antigen. Clin. Chem. 2004, 50, 125–129. [Google Scholar] [CrossRef][Green Version]
- Koivunen, E.; Wang, B.C.; Dickinson, C.D.; Ruoslahti, E. Peptides in Cell-Adhesion Research. Extracell. Matrix Compon. 1994, 245, 346–369. [Google Scholar]
- Muller, B.H.; Savatier, A.; L’Hostis, G.; Costa, N.; Bossus, M.; Michel, S.; Ott, C.; Becquart, L.; Ruffion, A.; Stura, E.A.; et al. In Vitro Affinity Maturation of an Anti-PSA Antibody for Prostate Cancer Diagnostic Assay. J. Mol. Biol. 2011, 414, 545–562. [Google Scholar] [CrossRef]
- Petrenko, V.A.; Brigati, J.R. Phage as Bispecific Probes. In Immunoassay and Other Bioanalytical Techniques; Van Emon, J.M., Ed.; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Boca Raton, FL, USA, 2007. [Google Scholar]
- Ferraro, S.; Bussetti, M.; Panteghini, M. Serum Prostate-Specific Antigen Testing for Early Detection of Prostate Cancer: Managing the Gap between Clinical and Laboratory Practice. Clin. Chem. 2021, 67, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Sandúa, A.; Sanmamed, M.F.; Rodríguez, M.; Ancizu-Marckert, J.; Gúrpide, A.; Perez-Gracia, J.L.; Alegre, E.; González, A. PSA reactivity in extracellular microvesicles to commercial immunoassays. Clin. Chim. Acta 2023, 543. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Zhao, F.; Zhao, Y.; Shangguan, L.; Liu, S. A portable chemiluminescence imaging immunoassay for simultaneous detection of different isoforms of prostate specific antigen in serum. Biosens. Bioelectron. 2016, 81, 97–102. [Google Scholar] [CrossRef]
- Andreeva, I.P.; Grigorenko, V.G.; Egorov, A.M.; Osipov, A.P. Quantitative Lateral Flow Immunoassay for Total Prostate Specific Antigen in Serum. Anal. Lett. 2016, 49, 579–588. [Google Scholar] [CrossRef]
- Barbosa, A.I.; Castanheira, A.P.; Edwards, A.D.; Reis, N.M. A lab-in-a-briefcase for rapid prostate specific antigen (PSA) screening from whole blood. Lab. Chip 2014, 14, 2918–2928. [Google Scholar] [CrossRef] [PubMed]
- Woodrum, D.L.; French, C.M.; Hill, T.M.; Roman, S.J.; Slatore, H.L.; Shaffer, J.L.; York, L.G.; Eure, K.L.; Loveland, K.G.; Gasior, G.H.; et al. Analytical performance of the Tandem®-R free PSA immunoassay measuring free prostate-specific antigen. Clin. Chem. 1997, 43, 1203–1208. [Google Scholar] [CrossRef] [PubMed]
- Galkin, A.; Komar, A.; Gorshunov, Y.; Besarab, A.; Soloviova, V. New Monoclonal Antibodies to the Prostate-Specific Antigen: Obtaining and Studying Biological Properties. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 573–577. [Google Scholar] [CrossRef]
- Zhang, X.M.; Soori, G.; Dobleman, T.J.; Xiao, G.G. The application of monoclonal antibodies in cancer diagnosis. Expert. Rev. Mol. Diagn. 2014, 14, 97–106. [Google Scholar] [CrossRef]
- Lee, S.; Xie, J.; Chen, X.Y. Peptide-Based Probes for Targeted Molecular Imaging. Biochemistry 2010, 49, 1364–1376. [Google Scholar] [CrossRef]
- Stura, E.A.; Muller, B.H.; Bossus, M.; Michel, S.; Jolivet-Reynaud, C.; Ducancel, F. Crystal Structure of Human Prostate-Specific Antigen in a Sandwich Antibody Complex. J. Mol. Biol. 2011, 414, 530–544. [Google Scholar] [CrossRef]
- Brigati, J.; Williams, D.D.; Sorokulova, I.B.; Nanduri, V.; Chen, I.H.; Turnbough, C.L., Jr.; Petrenko, V.A. Diagnostic probes for Bacillus anthracis spores selected from a landscape phage library. Clin. Chem. 2004, 50, 1899–1906. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P.; Petrenko, V.A.; Matthews, L.J. Cross-linked filamentous phage as an affinity matrix. J. Immunol. Methods 1998, 215, 151–161. [Google Scholar] [CrossRef]
- Goldberg, M.E.; Djavadi-Ohaniance, L. Methods for measurement of antibody/antigen affinity based on ELISA and RIA. Curr. Opin. Immunol. 1993, 5, 278–281. [Google Scholar] [CrossRef]
- Kubota, S.; Kawaki, H.; Takigawa, M. ELISA of CCN Family Proteins in Body Fluids Including Serum and Plasma. In CCN Proteins: Methods and Protocols; Takigawa, M., Ed.; Springer: New York, NY, USA, 2017; pp. 127–138. [Google Scholar]
- Sanders, E.C.; Santos, A.M.; Nguyen, E.K.; Gelston, A.A.; Majumdar, S.; Weiss, G.A. Phage vs. Phage: Direct Selections of Sandwich Binding Pairs. Viruses 2023, 15, 807. [Google Scholar] [CrossRef]
- Arévalo, F.J.; González-Techera, A.; Zon, M.A.; González-Sapienza, G.; Fernández, H. Ultra-sensitive electrochemical immunosensor using analyte peptidomimetics selected from phage display peptide libraries. Biosens. Bioelectron. 2012, 32, 231–237. [Google Scholar] [CrossRef]
- Luo, Z.B.; Qi, Q.G.; Zhang, L.J.; Zeng, R.J.; Su, L.S.; Tang, D.P. Branched Polyethylenimine-Modified up conversion Nanohybrid-Mediated Photoelectrochemical Immunoassay with Synergistic Effect of Dual-Purpose Copper Ions. Anal. Chem. 2019, 91, 4149–4156. [Google Scholar] [CrossRef]
- Yu, Z.Z.; Tang, Y.; Cai, G.N.; Ren, R.R.; Tang, D.P. Paper Electrode-Based Flexible Pressure Sensor for Point-of-Care Immunoassay with Digital Multimeter. Anal. Chem. 2019, 91, 1222–1226. [Google Scholar] [CrossRef] [PubMed]
- Achi, F.; Attar, A.M.; Lahcen, A.A. Electrochemical nanobiosensors for the detection of cancer biomarkers in real samples: Trends and challenges. TrAC Trends Anal. Chem. 2024, 170, 117423. [Google Scholar] [CrossRef]
- Guo, X.F.; Kulkarni, A.; Doepke, A.; Halsall, H.B.; Iyer, S.; Heineman, W.R. Carbohydrate-Based Label-Free Detection of Escherichia coli ORN 178 Using Electrochemical Impedance Spectroscopy. Anal. Chem. 2012, 84, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Daniels, J.S.; Pourmand, N. Label-free impedance biosensors: Opportunities and challenges. Electroanalysis 2007, 19, 1239–1257. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, S.P.; Yu, L.Z.; Zhang, W.Z.; Wei, Y.; Feng, D.X. Electrochemical Immunosensor for Detection of Prostate Specific Antigen Based on CNSs/Thi@AuNPs Nanocomposites as Sensing Platform. Int. J. Electrochem. Sci. 2022, 17, 22086. [Google Scholar] [CrossRef]
- Hou, L.; Tang, Y.; Xu, M.; Gao, Z.; Tang, D. Tyramine-Based Enzymatic Conjugate Repeats for Ultrasensitive Immunoassay Accompanying Tyramine Signal Amplification with Enzymatic Biocatalytic Precipitation. Anal. Chem. 2014, 86, 8352–8358. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, K.; Shen, Y.Z.; Yuan, J.S.; Waheed, I.; Mao, C.B.; Zhou, X. Advances in the Development of Phage-Based Probes for Detection of Bio-Species. Biosensors 2022, 12, 30. [Google Scholar] [CrossRef]
- Huang, S.; Yang, H.; Lakshmanan, R.S.; Johnson, M.L.; Chen, I.; Wan, J.; Wikle, H.C.; Petrenko, V.A.; Barbaree, J.M.; Cheng, Z.Y.; et al. The effect of salt and phage concentrations on the binding sensitivity of magnetoelastic biosensors for Bacillus anthracis detection. Biotechnol. Bioeng. 2008, 101, 1014–1021. [Google Scholar] [CrossRef]
- Sorokulova, I.B.; Olsen, E.V.; Chen, I.H.; Fiebor, B.; Barbaree, J.M.; Vodyanoy, V.J.; Chin, B.A.; Petrenko, V.A. Landscape phage probes for Salmonella typhimurium. J. Microbiol. Methods 2005, 63, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Towards Lateral Flow Quantitative Assays: Detection Approaches. Biosensors 2019, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Gogoi, M.; Mahato, M.; Joshi, A.B.; Baruah, A.J.; Kodgire, P.; Boruah, P. Biosensors for detection of prostate cancer: A review. Biomed. Microdevices 2022, 24, 32. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.F.; Wang, Y.L.; Huang, Z.Q.; Liu, Q. Recent advances in analysis technology for detection of prostate cancer biomarkers. Microchem. J. 2023, 190, 108740. [Google Scholar] [CrossRef]


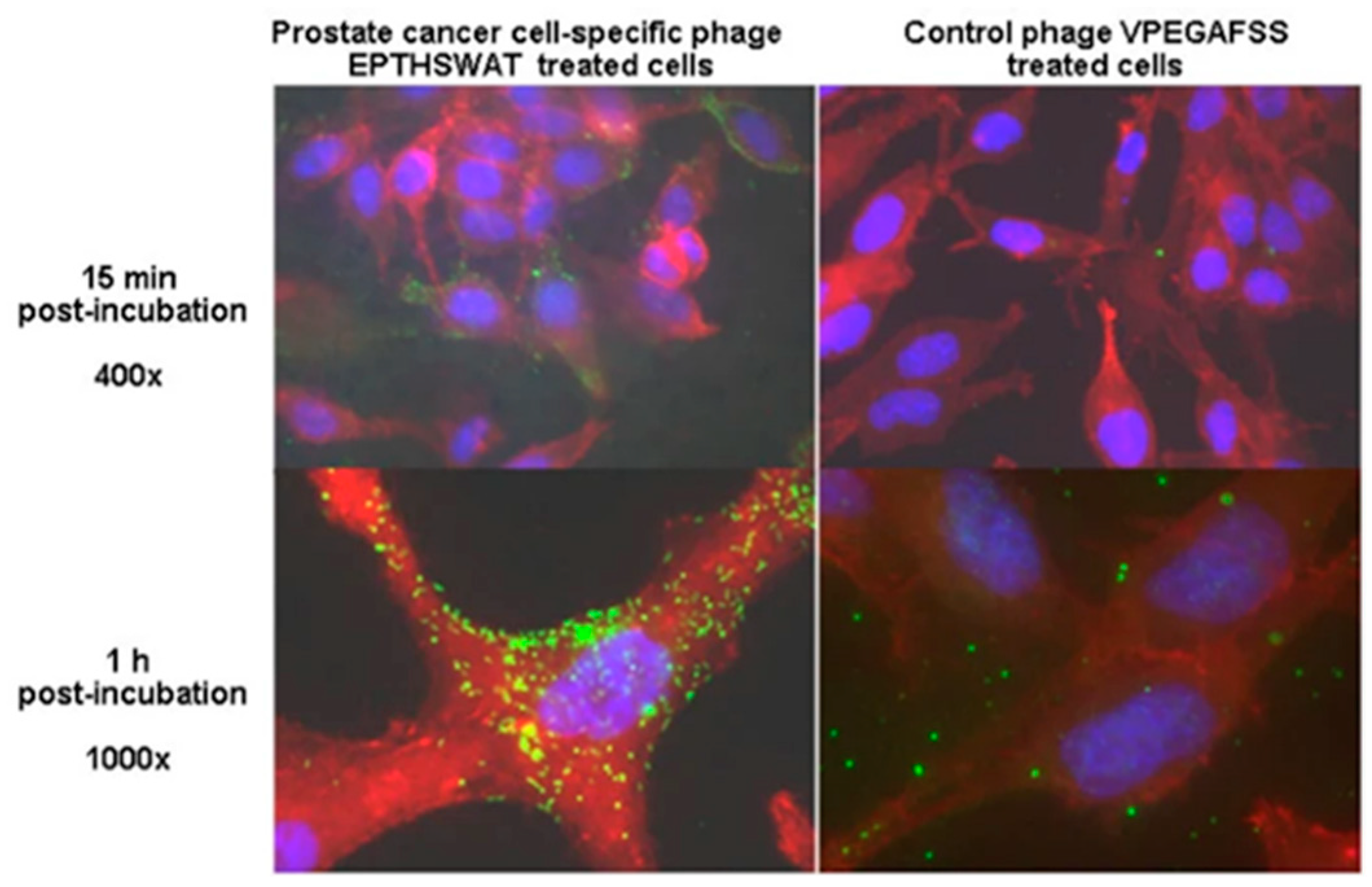


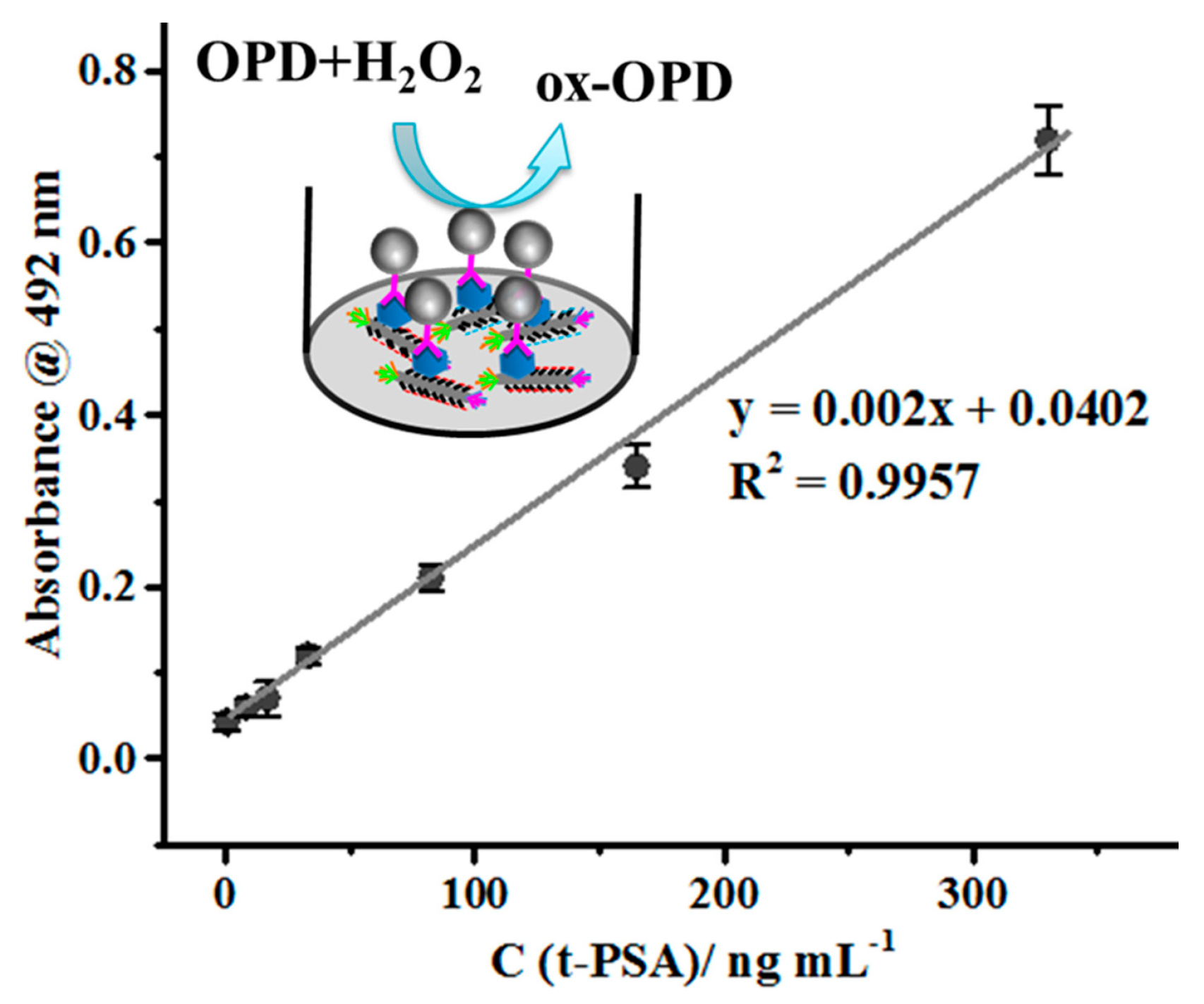
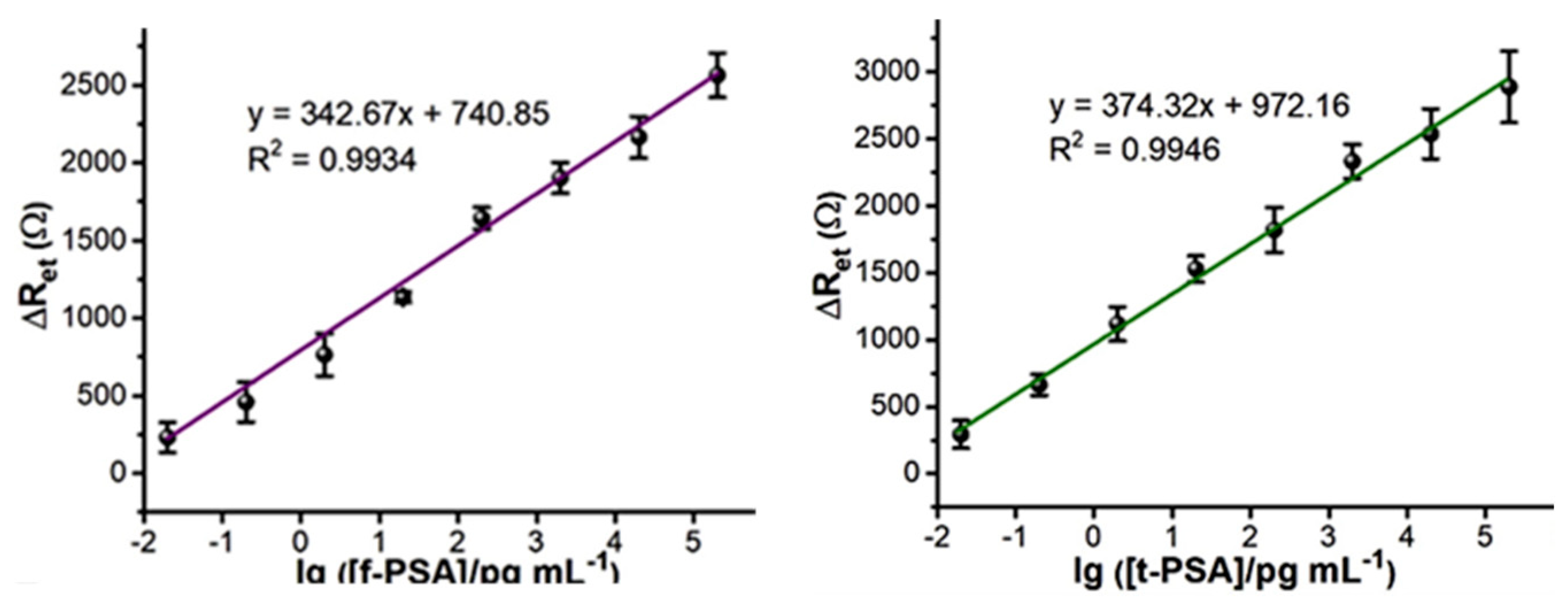
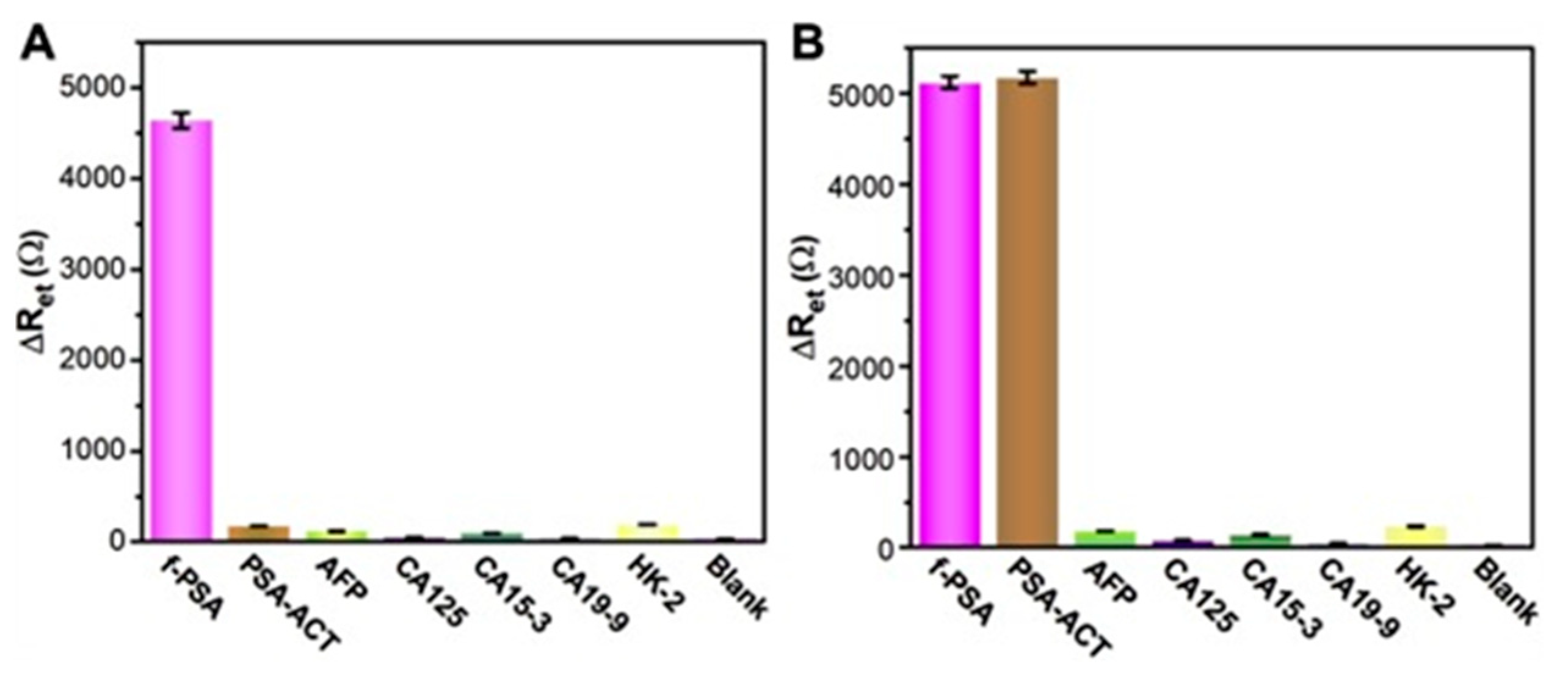
| f-PSA (ng mL−1) | t-PSA (ng mL−1) | f-PSA/ t-PSA Ratio | |||||
|---|---|---|---|---|---|---|---|
| Known | Detected | Relative Error (%) | Known | Detected | Relative Error (%) | ||
| 1# | 1.26 | 1.24 ± 0.04 | −1.58 | 4.92 | 4.81 ± 0.13 | 2.23 | 0.258 |
| 2# | 1.71 | 1.76 ± 0.05 | 2.92 | 6.83 | 6.98 ± 0.21 | 2.19 | 0.252 |
| 3# | 3.58 | 3.67 ± 0.10 | 2.51 | 10.21 | 10.47 ± 0.35 | 2.54 | 0.351 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrenko, V.A. Phage Display’s Prospects for Early Diagnosis of Prostate Cancer. Viruses 2024, 16, 277. https://doi.org/10.3390/v16020277
Petrenko VA. Phage Display’s Prospects for Early Diagnosis of Prostate Cancer. Viruses. 2024; 16(2):277. https://doi.org/10.3390/v16020277
Chicago/Turabian StylePetrenko, Valery A. 2024. "Phage Display’s Prospects for Early Diagnosis of Prostate Cancer" Viruses 16, no. 2: 277. https://doi.org/10.3390/v16020277
APA StylePetrenko, V. A. (2024). Phage Display’s Prospects for Early Diagnosis of Prostate Cancer. Viruses, 16(2), 277. https://doi.org/10.3390/v16020277






