Prior Influenza Infection Mitigates SARS-CoV-2 Disease in Syrian Hamsters
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Propagation
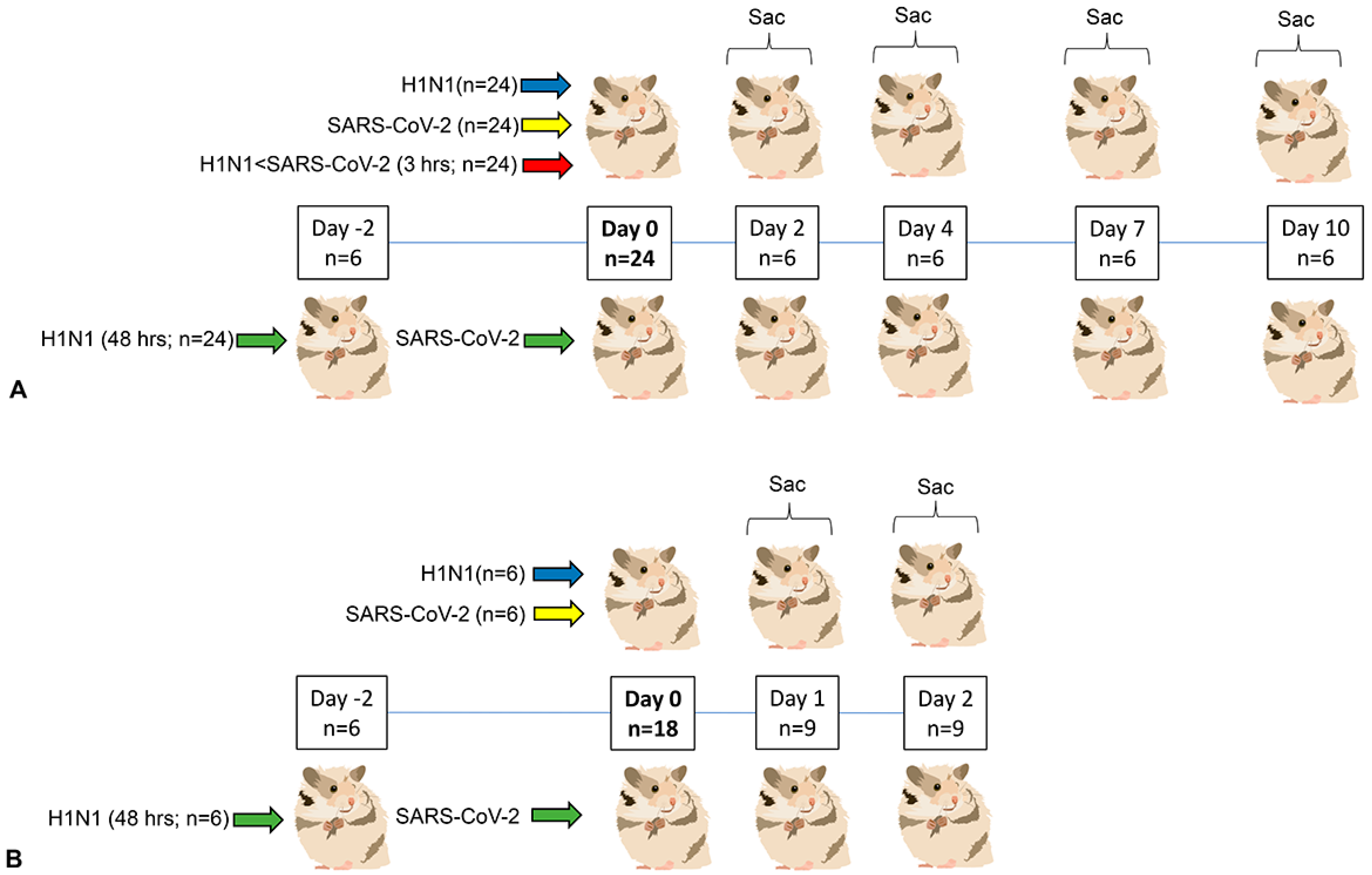
2.2. Animals and Housing
2.3. Anesthesia, Viral Inoculation, Clinical Evaluation and Euthanasia
2.4. Pathology, Immunohistochemistry and Microscopy
2.5. Flow Cytometry (Blood and Bronchoalveolar Lavage Fluid)
2.6. Nasal Transcriptomics
2.7. Lung Cytokine Expression
2.8. Statistics
3. Results
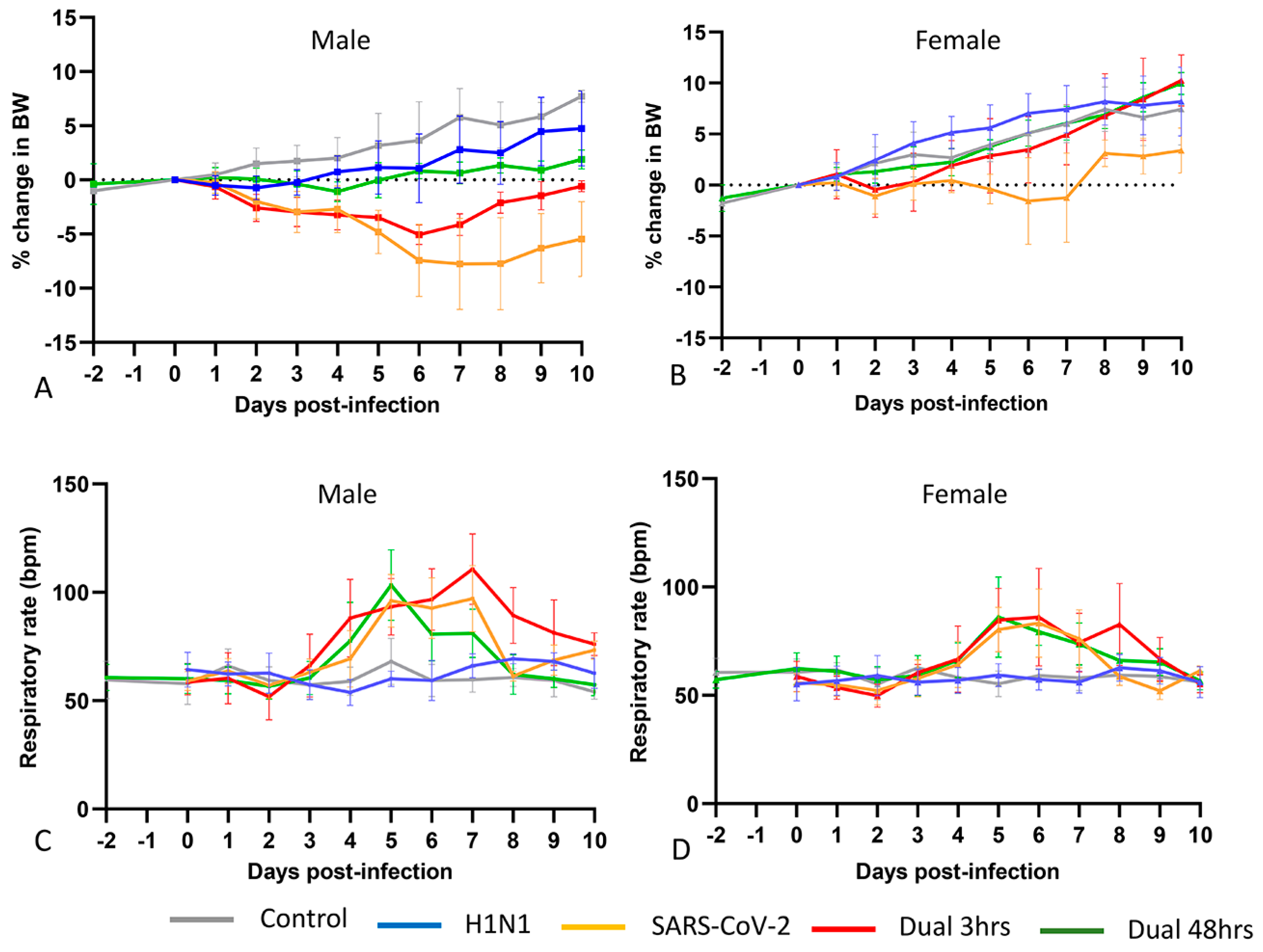
3.1. Clinical Phenotype (Experiment 1)
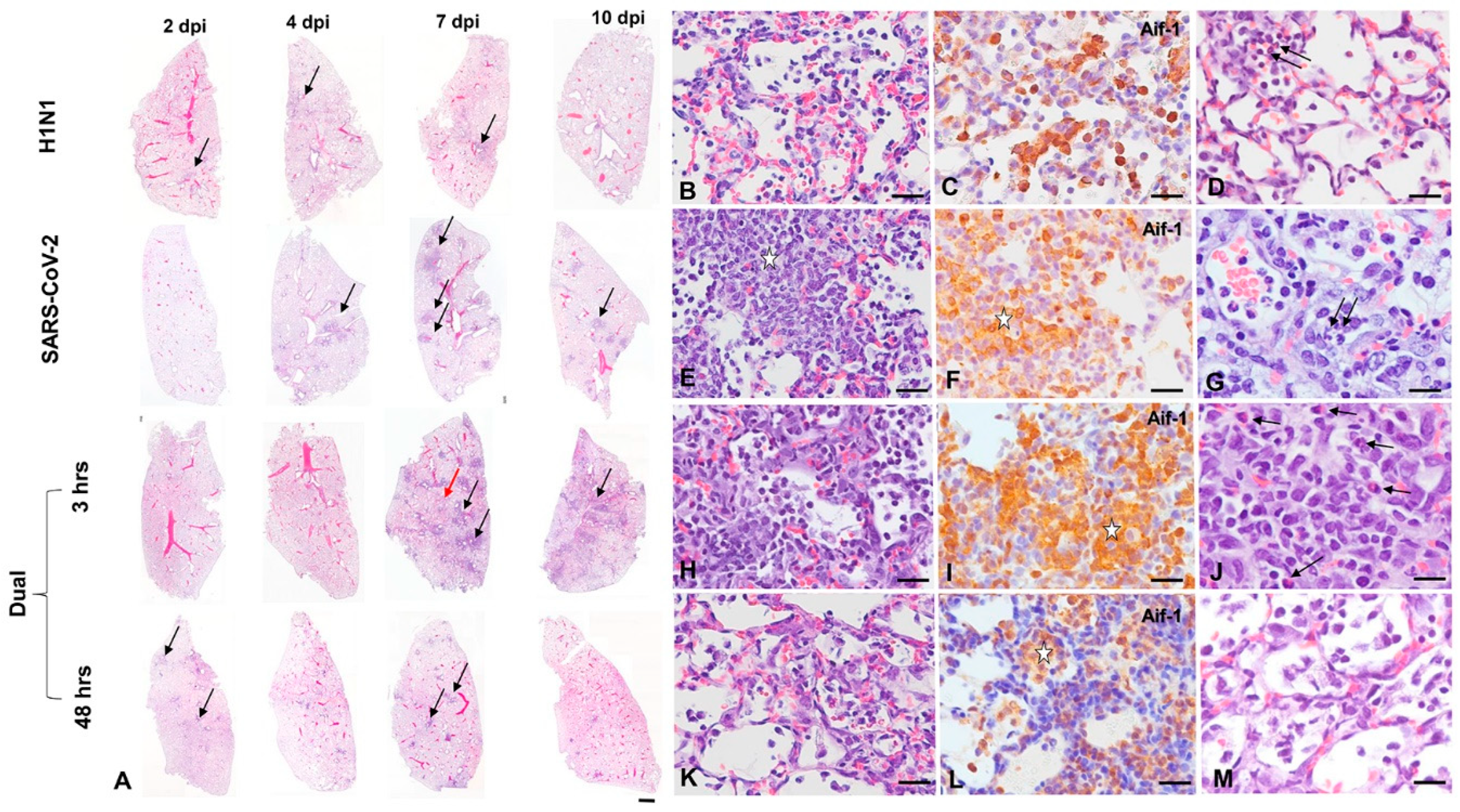
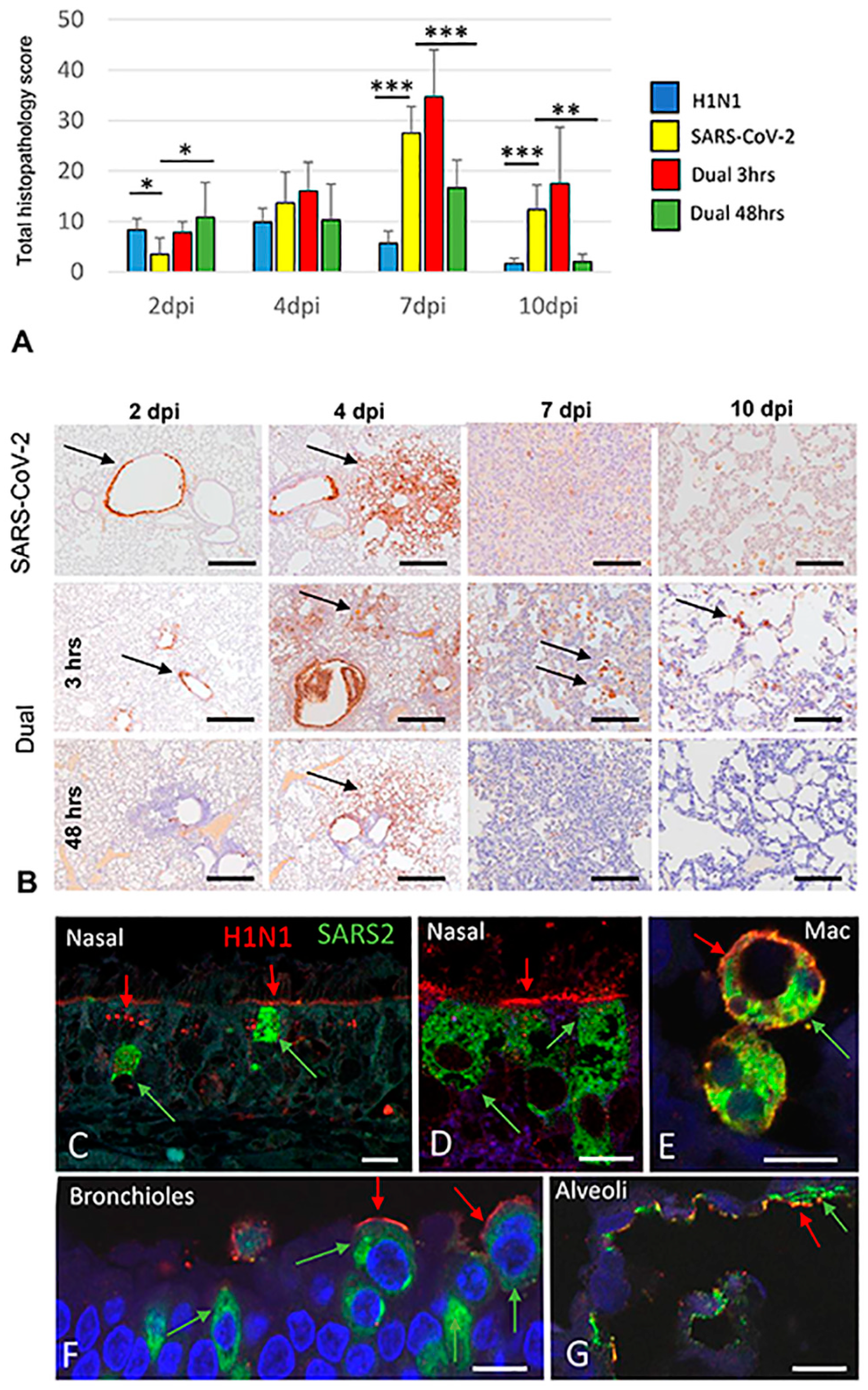
3.2. Pathology and Immunohistochemistry (Experiment 1)
3.2.1. H1N1pdm09 Single Infection
3.2.2. SARS-CoV-2-only infection
3.2.3. H1N1 and SARS-CoV-2 Coinfections
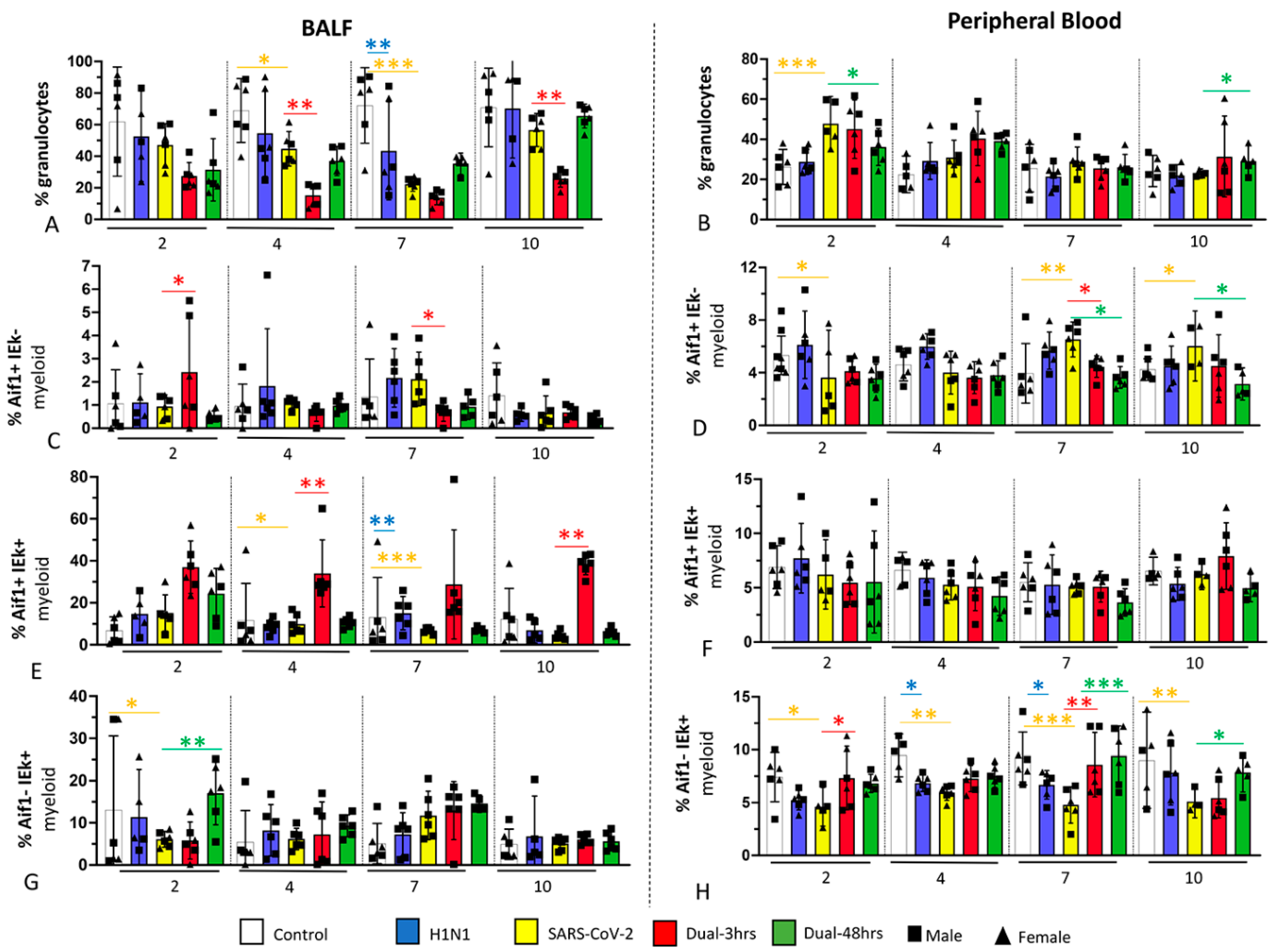
3.3. Immune Cell Populations in Bronchoalveolar Lavage Fluid (BALF) and Blood (Experiment 1)
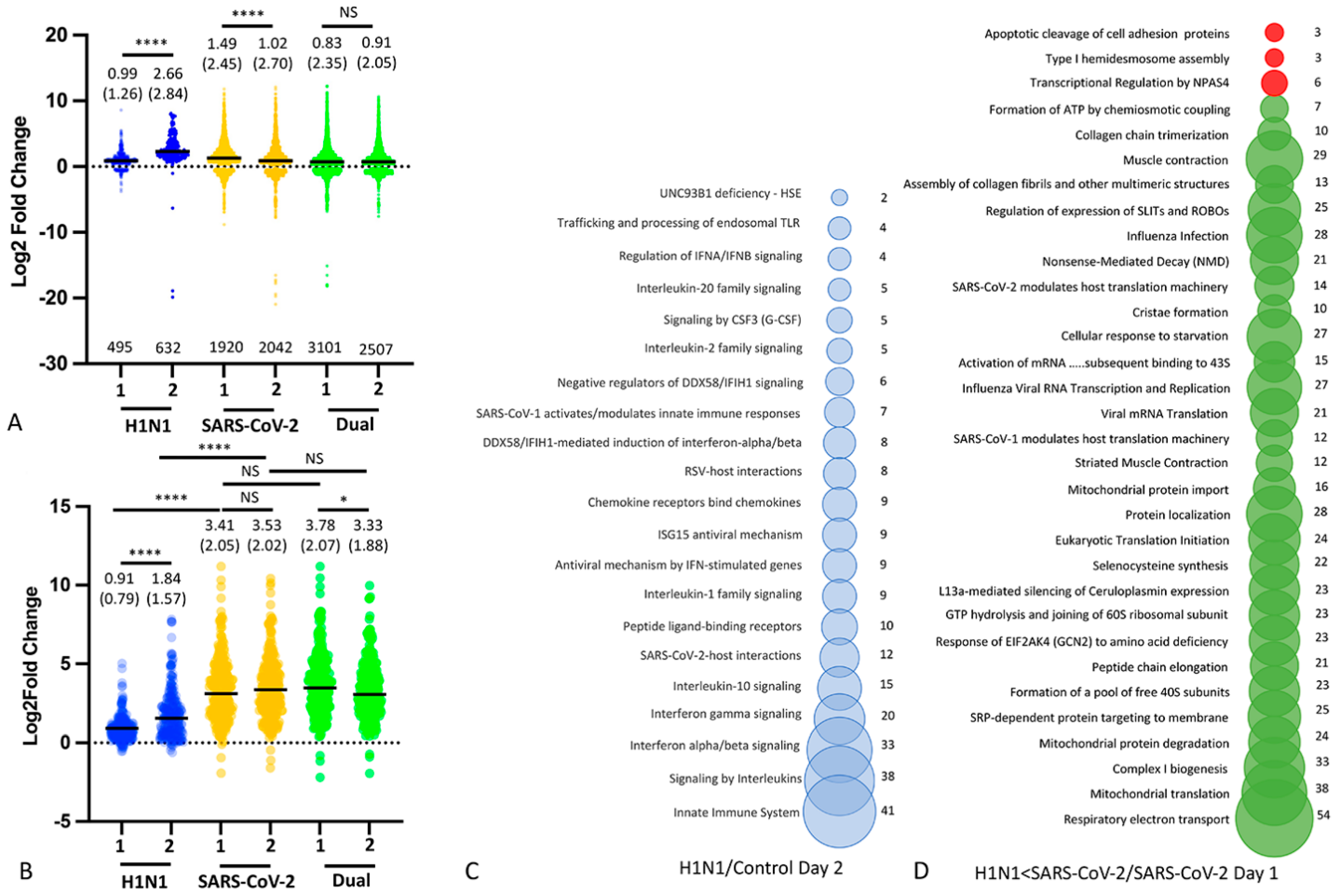
3.4. Nasal Transcriptomics (Experiment 2)
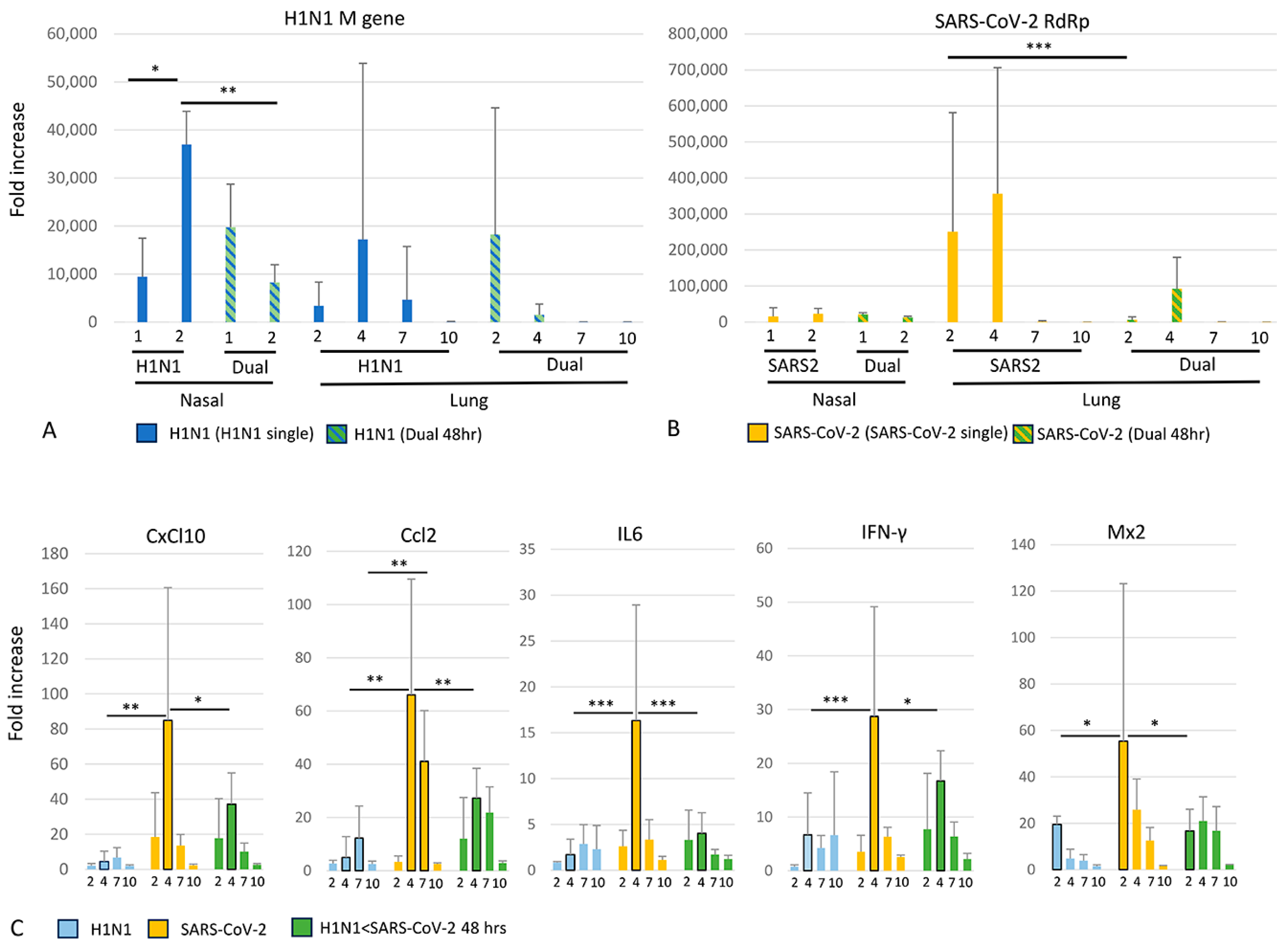
3.5. Viral and Cytokine Gene Expression (qRT-PCR)
4. Discussion
5. Conclusions and Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tokars, J.I.; Olsen, S.J.; Reed, C. Seasonal Incidence of Symptomatic Influenza in the United States. Clin. Infect. Dis. 2018, 66, 1511–1518. [Google Scholar] [CrossRef] [PubMed]
- Nichols, G.L.; Gillingham, E.L.; Macintyre, H.L.; Vardoulakis, S.; Hajat, S.; Sarran, C.E.; Amankwaah, D.; Phalkey, R. Coronavirus seasonality, respiratory infections and weather. BMC Infect. Dis. 2021, 21, 1101. [Google Scholar] [CrossRef]
- Phillips, N. The coronavirus is here to stay—Here’s what that means. Nature 2021, 590, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Musuuza, J.S.; Watson, L.; Parmasad, V.; Putman-Buehler, N.; Christensen, L.; Safdar, N. Prevalence and outcomes of coinfection and superinfection with SARS-CoV-2 and other pathogens: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0251170. [Google Scholar] [CrossRef]
- Stowe, J.; Tessier, E.; Zhao, H.; Guy, R.; Muller-Pebody, B.; Zambon, M.; Andrews, N.; Ramsay, M.; Lopez Bernal, J. Interactions between SARS-CoV-2 and influenza, and the impact of coinfection on disease severity: A test-negative design. Int. J. Epidemiol. 2021, 50, 1124–1133. [Google Scholar] [CrossRef]
- Akhtar, Z.; Islam, M.A.; Aleem, M.A.; Mah, E.M.S.; Ahmmed, M.K.; Ghosh, P.K.; Rahman, M.; Rahman, M.Z.; Sumiya, M.K.; Rahman, M.M.; et al. SARS-CoV-2 and influenza virus coinfection among patients with severe acute respiratory infection during the first wave of COVID-19 pandemic in Bangladesh: A hospital-based descriptive study. BMJ Open 2021, 11, e053768. [Google Scholar] [CrossRef]
- Khorramdelazad, H.; Kazemi, M.H.; Najafi, A.; Keykhaee, M.; Zolfaghari Emameh, R.; Falak, R. Immunopathological similarities between COVID-19 and influenza: Investigating the consequences of Coinfection. Microb. Pathog. 2021, 152, 104554. [Google Scholar] [CrossRef] [PubMed]
- Flerlage, T.; Boyd, D.F.; Meliopoulos, V.; Thomas, P.G.; Schultz-Cherry, S. Influenza virus and SARS-CoV-2: Pathogenesis and host responses in the respiratory tract. Nat. Rev. Microbiol. 2021, 19, 425–441. [Google Scholar] [CrossRef]
- Ma, S.; Lai, X.; Chen, Z.; Tu, S.; Qin, K. Clinical characteristics of critically ill patients coinfected with SARS-CoV-2 and the influenza virus in Wuhan, China. Int. J. Infect. Dis. 2020, 96, 683–687. [Google Scholar] [CrossRef]
- Yue, H.; Zhang, M.; Xing, L.; Wang, K.; Rao, X.; Liu, H.; Tian, J.; Zhou, P.; Deng, Y.; Shang, J. The epidemiology and clinical characteristics of coinfection of SARS-CoV-2 and influenza viruses in patients during COVID-19 outbreak. J. Med. Virol. 2020, 92, 2870–2873. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, F.; Wu, K.; Wang, J.; Li, F.; Huang, S.; Lu, J.; Huang, J.; Liu, H.; Zhou, R.; et al. Clinical and virological impact of single and dual infections with influenza A (H1N1) and SARS-CoV-2 in adult inpatients. PLoS Neglected Trop. Dis. 2021, 15, e0009997. [Google Scholar] [CrossRef]
- Huang, Y.; Skarlupka, A.L.; Jang, H.; Blas-Machado, U.; Holladay, N.; Hogan, R.J.; Ross, T.M. SARS-CoV-2 and Influenza A virus Coinfections in Ferrets. J. Virol. 2021, 96, JVI0179121. [Google Scholar] [CrossRef]
- Kinoshita, T.; Watanabe, K.; Sakurai, Y.; Nishi, K.; Yoshikawa, R.; Yasuda, J. Coinfection of SARS-CoV-2 and influenza virus causes more severe and prolonged pneumonia in hamsters. Sci. Rep. 2021, 11, 21259. [Google Scholar] [CrossRef]
- Zhang, A.J.; Lee, A.C.; Chan, J.F.; Liu, F.; Li, C.; Chen, Y.; Chu, H.; Lau, S.Y.; Wang, P.; Chan, C.C.; et al. Coinfection by Severe Acute Respiratory Syndrome Coronavirus 2 and Influenza A(H1N1)pdm09 Virus Enhances the Severity of Pneumonia in Golden Syrian Hamsters. Clin. Infect. Dis. 2021, 72, e978–e992. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhao, Y.; Dong, J.; Liang, S.; Guo, M.; Liu, X.; Wang, X.; Huang, Z.; Sun, X.; Zhang, Z.; et al. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity. Cell Res. 2021, 31, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado-Payán, E.; Montagud-Marrahi, E.; Torres-Elorza, M.; Bodro, M.; Blasco, M.; Poch, E.; Soriano, A.; Piñeiro, G.J. SARS-CoV-2 and influenza virus coinfection. Lancet 2020, 395, e84. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ma, J.; Wang, H.; Wang, X.; Hu, Z.; Li, H.; Zhang, H.; Liu, X. Coinfection of influenza A virus and SARS-CoV-2: A retrospective cohort study. J. Med. Virol. 2021, 93, 2947–2954. [Google Scholar] [CrossRef] [PubMed]
- Nickbakhsh, S.; Mair, C.; Matthews, L.; Reeve, R.; Johnson, P.C.D.; Thorburn, F.; von Wissmann, B.; Reynolds, A.; McMenamin, J.; Gunson, R.N.; et al. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. USA 2019, 116, 27142–27150. [Google Scholar] [CrossRef] [PubMed]
- Kloepfer, K.M.; Gern, J.E. Ecological and individual data both indicate that influenza inhibits rhinovirus infection. Proc. Natl. Acad. Sci. USA 2020, 117, 6987. [Google Scholar] [CrossRef] [PubMed]
- Waner, J.L. Mixed viral infections: Detection and management. Clin. Microbiol. Rev. 1994, 7, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Asner, S.A.; Science, M.E.; Tran, D.; Smieja, M.; Merglen, A.; Mertz, D. Clinical disease severity of respiratory viral coinfection versus single viral infection: A systematic review and meta-analysis. PLoS ONE 2014, 9, e99392. [Google Scholar] [CrossRef] [PubMed]
- Walzl, G.; Tafuro, S.; Moss, P.; Openshaw, P.J.; Hussell, T. Influenza virus lung infection protects from respiratory syncytial virus-induced immunopathology. J. Exp. Med. 2000, 192, 1317–1326. [Google Scholar] [CrossRef]
- Wissinger, E.; Goulding, J.; Hussell, T. Immune homeostasis in the respiratory tract and its impact on heterologous infection. Semin. Immunol. 2009, 21, 147–155. [Google Scholar] [CrossRef]
- Hua, X.; Vijay, R.; Channappanavar, R.; Athmer, J.; Meyerholz, D.K.; Pagedar, N.; Tilley, S.; Perlman, S. Nasal priming by a murine coronavirus provides protective immunity against lethal heterologous virus pneumonia. JCI Insight 2018, 3, e99025. [Google Scholar] [CrossRef] [PubMed]
- Cheemarla, N.R.; Watkins, T.A.; Mihaylova, V.T.; Wang, B.; Zhao, D.; Wang, G.; Landry, M.L.; Foxman, E.F. Dynamic innate immune response determines susceptibility to SARS-CoV-2 infection and early replication kinetics. J. Exp. Med. 2021, 218, e20210583. [Google Scholar] [CrossRef]
- Dee, K.; Goldfarb, D.M.; Haney, J.; Amat, J.A.R.; Herder, V.; Stewart, M.; Szemiel, A.M.; Baguelin, M.; Murcia, P.R. Human Rhinovirus Infection Blocks Severe Acute Respiratory Syndrome Coronavirus 2 Replication Within the Respiratory Epithelium: Implications for COVID-19 Epidemiology. J. Infect. Dis. 2021, 224, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Essaidi-Laziosi, M.; Alvarez, C.; Puhach, O.; Sattonnet-Roche, P.; Torriani, G.; Tapparel, C.; Kaiser, L.; Eckerle, I. Sequential infections with rhinovirus and influenza modulate the replicative capacity of SARS-CoV-2 in the upper respiratory tract. Emerg. Microbes Infect. 2022, 11, 412–423. [Google Scholar] [CrossRef]
- Ryan, K.A.; Schewe, K.E.; Crowe, J.; Fotheringham, S.A.; Hall, Y.; Humphreys, R.; Marriott, A.C.; Paterson, J.; Rayner, E.; Salguero, F.J.; et al. Sequential Delivery of Live Attenuated Influenza Vaccine and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in the Ferret Model Can Reduce SARS-CoV-2 Shedding and Does Not Result in Enhanced Lung Pathology. J. Infect. Dis. 2022, 225, 404–412. [Google Scholar] [CrossRef]
- Thi Nhu Thao, T.; Labroussaa, F.; Ebert, N.; V’Kovski, P.; Stalder, H.; Portmann, J.; Kelly, J.; Steiner, S.; Holwerda, M.; Kratzel, A.; et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 2020, 582, 561–565. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 1997, 71, 9608–9617. [Google Scholar] [CrossRef]
- Davidson, A.D.; Williamson, M.K.; Lewis, S.; Shoemark, D.; Carroll, M.W.; Heesom, K.J.; Zambon, M.; Ellis, J.; Lewis, P.A.; Hiscox, J.A.; et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020, 12, 68. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.D.; Osterrieder, N.; Bertzbach, L.D.; Vladimirova, D.; Greuel, S.; Ihlow, J.; Horst, D.; Trimpert, J.; Dietert, K. Standardization of Reporting Criteria for Lung Pathology in SARS-CoV-2-infected Hamsters: What Matters? Am. J. Respir. Cell Mol. Biol. 2020, 63, 856–859. [Google Scholar] [CrossRef]
- Adams, D.R.; McFarland, L.Z. Morphology of the nasal fossae and associated structures of the hamster (Mesocricetus auratus). J. Morphol. 1972, 137, 161–179. [Google Scholar] [CrossRef]
- Zeiss, C.J.; Allore, H.G.; Towle, V.; Tao, W. CNTF induces dose-dependent alterations in retinal morphology in normal and rcd-1 canine retina. Exp. Eye Res. 2006, 82, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Dunston, D.; Ashby, S.; Krosnowski, K.; Ogura, T.; Lin, W. An effective manual deboning method to prepare intact mouse nasal tissue with preserved anatomical organization. J. Vis. Exp. 2013, 78, e50538. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Bluyssen, H.A.; Nakamura, N.; Vlietstra, R.J.; Smit, E.M.; Hagemeijer, A.; Trapman, J. Isolation, properties and chromosomal localization of four closely linked hamster interferon-alpha-encoding genes. Gene 1995, 158, 295–300. [Google Scholar] [CrossRef]
- Zivcec, M.; Safronetz, D.; Haddock, E.; Feldmann, H.; Ebihara, H. Validation of assays to monitor immune responses in the Syrian golden hamster (Mesocricetus auratus). J. Immunol. Methods 2011, 368, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Neter, J.; Kutner, M.; Wasserman, W.; Nachtsheim, C. Applied Linear Statistical Models; Irwin: Chicago, IL, USA, 1996. [Google Scholar]
- Bednash, J.S.; Kagan, V.E.; Englert, J.A.; Farkas, D.; Tyurina, Y.Y.; Tyurin, V.A.; Samovich, S.N.; Farkas, L.; Elhance, A.; Johns, F.; et al. Syrian hamsters as a model of lung injury with SARS-CoV-2 infection: Pathologic, physiologic, and detailed molecular profiling. Transl. Res. 2022, 240, 1–16. [Google Scholar] [CrossRef]
- Rosenke, K.; Meade-White, K.; Letko, M.; Clancy, C.; Hansen, F.; Liu, Y.; Okumura, A.; Tang-Huau, T.L.; Li, R.; Saturday, G.; et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020, 9, 2673–2684. [Google Scholar] [CrossRef]
- Boudewijns, R.; Thibaut, H.J.; Kaptein, S.J.F.; Li, R.; Vergote, V.; Seldeslachts, L.; Van Weyenbergh, J.; De Keyzer, C.; Bervoets, L.; Sharma, S.; et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020, 11, 5838. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Zhang, A.J.; Yuan, S.; Poon, V.K.; Chan, C.C.; Lee, A.C.; Chan, W.M.; Fan, Z.; Tsoi, H.W.; Wen, L.; et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020, 71, 2428–2446. [Google Scholar] [CrossRef]
- Dhakal, S.; Ruiz-Bedoya, C.A.; Zhou, R.; Creisher, P.S.; Villano, J.S.; Littlefield, K.; Ruelas Castillo, J.; Marinho, P.; Jedlicka, A.E.; Ordonez, A.A.; et al. Sex Differences in Lung Imaging and SARS-CoV-2 Antibody Responses in a COVID-19 Golden Syrian Hamster Model. mBio 2021, 12, e0097421. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. The pathology of influenza virus infections. Annu. Rev. Pathol. 2008, 3, 499–522. [Google Scholar] [CrossRef]
- Nin, N.; Sánchez-Rodríguez, C.; Ver, L.S.; Cardinal, P.; Ferruelo, A.; Soto, L.; Deicas, A.; Campos, N.; Rocha, O.; Ceraso, D.H.; et al. Lung histopathological findings in fatal pandemic influenza A (H1N1). Med. Intensiv. 2012, 36, 24–31. [Google Scholar] [CrossRef]
- Nouailles, G.; Wyler, E.; Pennitz, P.; Postmus, D.; Vladimirova, D.; Kazmierski, J.; Pott, F.; Dietert, K.; Muelleder, M.; Farztdinov, V.; et al. Temporal omics analysis in Syrian hamsters unravel cellular effector responses to moderate COVID-19. Nat. Commun. 2021, 12, 4869. [Google Scholar] [CrossRef]
- Iwatsuki-Horimoto, K.; Nakajima, N.; Ichiko, Y.; Sakai-Tagawa, Y.; Noda, T.; Hasegawa, H.; Kawaoka, Y. Syrian Hamster as an Animal Model for the Study of Human Influenza Virus Infection. J. Virol. 2018, 92, 10–1128. [Google Scholar] [CrossRef]
- Ju, X.; Yan, Y.; Liu, Q.; Li, N.; Sheng, M.; Zhang, L.; Li, X.; Liang, Z.; Huang, F.; Liu, K.; et al. Neuraminidase of Influenza A Virus Binds Lysosome-Associated Membrane Proteins Directly and Induces Lysosome Rupture. J. Virol. 2015, 89, 10347–10358. [Google Scholar] [CrossRef]
- Bryche, B.; St Albin, A.; Murri, S.; Lacôte, S.; Pulido, C.; Ar Gouilh, M.; Lesellier, S.; Servat, A.; Wasniewski, M.; Picard-Meyer, E.; et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav. Immun. 2020, 89, 579–586. [Google Scholar] [CrossRef]
- Saris, A.; Reijnders, T.D.Y.; Nossent, E.J.; Schuurman, A.R.; Verhoeff, J.; Asten, S.V.; Bontkes, H.; Blok, S.; Duitman, J.; Bogaard, H.J.; et al. Distinct cellular immune profiles in the airways and blood of critically ill patients with COVID-19. Thorax 2021, 76, 1010–1019. [Google Scholar] [CrossRef]
- Olbei, M.; Hautefort, I.; Modos, D.; Treveil, A.; Poletti, M.; Gul, L.; Shannon-Lowe, C.D.; Korcsmaros, T. SARS-CoV-2 Causes a Different Cytokine Response Compared to Other Cytokine Storm-Causing Respiratory Viruses in Severely Ill Patients. Front. Immunol. 2021, 12, 629193. [Google Scholar] [CrossRef]
- Hedberg, P.; Karlsson Valik, J.; van der Werff, S.; Tanushi, H.; Requena Mendez, A.; Granath, F.; Bell, M.; Mårtensson, J.; Dyrdak, R.; Hertting, O.; et al. Clinical phenotypes and outcomes of SARS-CoV-2, influenza, RSV and seven other respiratory viruses: A retrospective study using complete hospital data. Thorax 2022, 77, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Orellana, P.; Martorell, J.; Vidaña, B.; Majó, N.; Martínez, J.; Falcón, A.; Rodríguez-Frandsen, A.; Casas, I.; Pozo, F.; García-Migura, L.; et al. Clinical response to pandemic H1N1 influenza virus from a fatal and mild case in ferrets. Virol. J. 2015, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L. Sex influences immune responses to viruses, and efficacy of prophylaxis and treatments for viral diseases. BioEssays News Rev. Mol. Cell. Dev. Biol. 2012, 34, 1050–1059. [Google Scholar] [CrossRef]
- Jacobsen, H.; Klein, S.L. Sex Differences in Immunity to Viral Infections. Front. Immunol. 2021, 12, 720952. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Nguyen, T.Q.; Casel, M.A.B.; Rollon, R.; Kim, S.M.; Kim, Y.I.; Yu, K.M.; Jang, S.G.; Yang, J.; Poo, H.; et al. Coinfection of SARS-CoV-2 and Influenza A virus increased disease severity, impaired neutralizing antibody, and CD4+ T cell responses. J. Virol. 2022, 96, jvi0187321. [Google Scholar] [CrossRef]
- Mudd, P.A.; Crawford, J.C.; Turner, J.S.; Souquette, A.; Reynolds, D.; Bender, D.; Bosanquet, J.P.; Anand, N.J.; Striker, D.A.; Martin, R.S.; et al. Distinct inflammatory profiles distinguish COVID-19 from influenza with limited contributions from cytokine storm. Sci. Adv. 2020, 6, eabe3024. [Google Scholar] [CrossRef]
- Mao, T.; Israelow, B.; Lucas, C.; Vogels, C.B.F.; Gomez-Calvo, M.L.; Fedorova, O.; Breban, M.I.; Menasche, B.L.; Dong, H.; Linehan, M.; et al. A stem-loop RNA RIG-I agonist protects against acute and chronic SARS-CoV-2 infection in mice. J. Exp. Med. 2022, 219, e20211818. [Google Scholar] [CrossRef] [PubMed]
- Bessière, P.; Wasniewski, M.; Picard-Meyer, E.; Servat, A.; Figueroa, T.; Foret-Lucas, C.; Coggon, A.; Lesellier, S.; Boué, F.; Cebron, N.; et al. Intranasal type I interferon treatment is beneficial only when administered before clinical signs onset in the SARS-CoV-2 hamster model. PLoS Pathog. 2021, 17, e1009427. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wohlford-Lenane, C.; Zhao, J.; Fleming, E.; Lane, T.E.; McCray, P.B., Jr.; Perlman, S. Intranasal treatment with poly(I•C) protects aged mice from lethal respiratory virus infections. J. Virol. 2012, 86, 11416–11424. [Google Scholar] [CrossRef]
- Zhang, A.J.; Lee, A.C.; Chu, H.; Chan, J.F.; Fan, Z.; Li, C.; Liu, F.; Chen, Y.; Yuan, S.; Poon, V.K.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 Infects and Damages the Mature and Immature Olfactory Sensory Neurons of Hamsters. Clin. Infect. Dis. 2021, 73, e503–e512. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Nakajima, N.; Sato, Y.; Takahashi, K.; Accola, M.; Chiba, S.; Fan, S.; Neumann, G.; Rehrauer, W.; Suzuki, T.; et al. SARS-CoV-2 Interference of Influenza Virus Replication in Syrian Hamsters. J. Infect. Dis. 2022, 225, 282–286. [Google Scholar] [CrossRef]
- Oishi, K.; Horiuchi, S.; Minkoff, J.M.; tenOever, B.R. The Host Response to Influenza A Virus Interferes with SARS-CoV-2 Replication during Coinfection. J. Virol. 2022, 96, e0076522. [Google Scholar] [CrossRef]
- Bruns, A.M.; Leser, G.P.; Lamb, R.A.; Horvath, C.M. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol. Cell 2014, 55, 771–781. [Google Scholar] [CrossRef]
- Kulkarni, R.R.; Rasheed, M.A.; Bhaumik, S.K.; Ranjan, P.; Cao, W.; Davis, C.; Marisetti, K.; Thomas, S.; Gangappa, S.; Sambhara, S.; et al. Activation of the RIG-I pathway during influenza vaccination enhances the germinal center reaction, promotes T follicular helper cell induction, and provides a dose-sparing effect and protective immunity. J. Virol. 2014, 88, 13990–14001. [Google Scholar] [CrossRef]
- Loske, J.; Röhmel, J.; Lukassen, S.; Stricker, S.; Magalhães, V.G.; Liebig, J.; Chua, R.L.; Thürmann, L.; Messingschlager, M.; Seegebarth, A.; et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat. Biotechnol. 2022, 40, 319–324. [Google Scholar] [CrossRef]
- Pekayvaz, K.; Leunig, A.; Kaiser, R.; Joppich, M.; Brambs, S.; Janjic, A.; Popp, O.; Nixdorf, D.; Fumagalli, V.; Schmidt, N.; et al. Protective immune trajectories in early viral containment of non-pneumonic SARS-CoV-2 infection. Nat. Commun. 2022, 13, 1018. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, C.; Horner, S.M. MAVS Coordination of Antiviral Innate Immunity. J. Virol. 2015, 89, 6974–6977. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, A.; Tiwari-Pandey, R.; Pandey, N.R. Mitochondria: The indispensable players in innate immunity and guardians of the inflammatory response. J. Cell Commun. Signal. 2019, 13, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Foo, J.; Bellot, G.; Pervaiz, S.; Alonso, S. Mitochondria-mediated oxidative stress during viral infection. Trends Microbiol. 2022, 30, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Prasada Kabekkodu, S.; Chakrabarty, S.; Jayaram, P.; Mallya, S.; Thangaraj, K.; Singh, K.K.; Satyamoorthy, K. Severe acute respiratory syndrome coronaviruses contributing to mitochondrial dysfunction: Implications for post-COVID complications. Mitochondrion 2023, 69, 43–56. [Google Scholar] [CrossRef]
- Guarnieri, J.W.; Dybas, J.M.; Fazelinia, H.; Kim, M.S.; Frere, J.; Zhang, Y.; Soto Albrecht, Y.; Murdock, D.G.; Angelin, A.; Singh, L.N.; et al. Core mitochondrial genes are down-regulated during SARS-CoV-2 infection of rodent and human hosts. Sci. Transl. Med. 2023, 15, eabq1533. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, J.W.; Angelin, A.; Murdock, D.G.; Schaefer, P.; Portluri, P.; Lie, T.; Huang, J.; Wallace, D.C. SARS-COV-2 viroporins activate the NLRP3-inflammasome by the mitochondrial permeability transition pore. Front. Immunol. 2023, 14, 1064293. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Daveluy, S. Pemphigus vulgaris after COVID-19 infection and vaccination. J. Am. Acad. Dermatol. 2022, 87, 709–710. [Google Scholar] [CrossRef]
- Ghalamkarpour, F.; Pourani, M.R. Aggressive course of pemphigus vulgaris following COVID-19 infection. Dermatol. Ther. 2020, 33, e14398. [Google Scholar] [CrossRef]
- De Simone, C.; Caldarola, G.; D’Agostino, M.; Zampetti, A.; Amerio, P.; Feliciani, C. Exacerbation of pemphigus after influenza vaccination. Clin. Exp. Dermatol. 2008, 33, 718–720. [Google Scholar] [CrossRef]
- Mignogna, M.D.; Lo Muzio, L.; Ruocco, E. Pemphigus induction by influenza vaccination. Int. J. Dermatol. 2000, 39, 800. [Google Scholar] [CrossRef]
- Kim, H.K.; Kang, J.A.; Lyoo, K.S.; Le, T.B.; Yeo, Y.H.; Wong, S.S.; Na, W.; Song, D.; Webby, R.J.; Zanin, M.; et al. Severe acute respiratory syndrome coronavirus 2 and influenza A virus coinfection alters viral tropism and haematological composition in Syrian hamsters. Transbound. Emerg. Dis. 2022, 69, e3297–e3304. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- Meningher, T.; Hindiyeh, M.; Regev, L.; Sherbany, H.; Mendelson, E.; Mandelboim, M. Relationships between A(H1N1)pdm09 influenza infection and infections with other respiratory viruses. Influenza Other Respir. Viruses 2014, 8, 422–430. [Google Scholar] [CrossRef] [PubMed]
- van Asten, L.; Bijkerk, P.; Fanoy, E.; van Ginkel, A.; Suijkerbuijk, A.; van der Hoek, W.; Meijer, A.; Vennema, H. Early occurrence of influenza A epidemics coincided with changes in occurrence of other respiratory virus infections. Influenza Other Respir. Viruses 2016, 10, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Paget, J.; Spreeuwenberg, P.; Charu, V.; Taylor, R.J.; Iuliano, A.D.; Bresee, J.; Simonsen, L.; Viboud, C. Global mortality associated with seasonal influenza epidemics: New burden estimates and predictors from the GLaMOR Project. J. Glob. Health 2019, 9, 020421. [Google Scholar] [CrossRef] [PubMed]
- Piroth, L.; Cottenet, J.; Mariet, A.S.; Bonniaud, P.; Blot, M.; Tubert-Bitter, P.; Quantin, C. Comparison of the characteristics, morbidity, and mortality of COVID-19 and seasonal influenza: A nationwide, population-based retrospective cohort study. Lancet Respir. Med. 2021, 9, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Bautista, E.; Chotpitayasunondh, T.; Gao, Z.; Harper, S.A.; Shaw, M.; Uyeki, T.M.; Zaki, S.R.; Hayden, F.G.; Hui, D.S.; Kettner, J.D.; et al. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 2010, 362, 1708–1719. [Google Scholar] [PubMed]
- Borczuk, A.C.; Salvatore, S.P.; Seshan, S.V.; Patel, S.S.; Bussel, J.B.; Mostyka, M.; Elsoukkary, S.; He, B.; Del Vecchio, C.; Fortarezza, F.; et al. COVID-19 pulmonary pathology: A multi-institutional autopsy cohort from Italy and New York City. Mod. Pathol. 2020, 33, 2156–2168. [Google Scholar] [CrossRef] [PubMed]
- Angeles Montero-Fernandez, M.; Pardo-Garcia, R. Histopathology features of the lung in COVID-19 patients. Diagn. Histopathol. 2021, 27, 123–127. [Google Scholar] [CrossRef]
- Vincent, J.L.; Levi, M.; Hunt, B.J. Prevention and management of thrombosis in hospitalised patients with COVID-19 pneumonia. Lancet Respir. Med. 2022, 10, 214–220. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pietro, C.; Haberman, A.M.; Lindenbach, B.D.; Smith, P.C.; Bruscia, E.M.; Allore, H.G.; Vander Wyk, B.; Tyagi, A.; Zeiss, C.J. Prior Influenza Infection Mitigates SARS-CoV-2 Disease in Syrian Hamsters. Viruses 2024, 16, 246. https://doi.org/10.3390/v16020246
Di Pietro C, Haberman AM, Lindenbach BD, Smith PC, Bruscia EM, Allore HG, Vander Wyk B, Tyagi A, Zeiss CJ. Prior Influenza Infection Mitigates SARS-CoV-2 Disease in Syrian Hamsters. Viruses. 2024; 16(2):246. https://doi.org/10.3390/v16020246
Chicago/Turabian StyleDi Pietro, Caterina, Ann M. Haberman, Brett D. Lindenbach, Peter C. Smith, Emanuela M. Bruscia, Heather G. Allore, Brent Vander Wyk, Antariksh Tyagi, and Caroline J. Zeiss. 2024. "Prior Influenza Infection Mitigates SARS-CoV-2 Disease in Syrian Hamsters" Viruses 16, no. 2: 246. https://doi.org/10.3390/v16020246
APA StyleDi Pietro, C., Haberman, A. M., Lindenbach, B. D., Smith, P. C., Bruscia, E. M., Allore, H. G., Vander Wyk, B., Tyagi, A., & Zeiss, C. J. (2024). Prior Influenza Infection Mitigates SARS-CoV-2 Disease in Syrian Hamsters. Viruses, 16(2), 246. https://doi.org/10.3390/v16020246





