Abstract
In 2019–2020, dengue virus (DENV) type 4 emerged to cause the largest DENV outbreak in Paraguay’s history. This study sought to characterize dengue relative to other acute illness cases and use phylogenetic analysis to understand the outbreak’s origin. Individuals with an acute illness (≤7 days) were enrolled and tested for DENV nonstructural protein 1 (NS1) and viral RNA by real-time RT-PCR. Near-complete genome sequences were obtained from 62 DENV-4 positive samples. From January 2019 to March 2020, 799 participants were enrolled: 253 dengue (14 severe dengue, 5.5%) and 546 other acute illness cases. DENV-4 was detected in 238 dengue cases (94.1%). NS1 detection by rapid test was 52.5% sensitive (53/101) and 96.5% specific (387/401) for dengue compared to rRT-PCR. DENV-4 sequences were grouped into two clades within genotype II. No clustering was observed based on dengue severity, location, or date. Sequences obtained here were most closely related to 2018 DENV-4 sequences from Paraguay, followed by a 2013 sequence from southern Brazil. DENV-4 can result in large outbreaks, including severe cases, and is poorly detected with available rapid diagnostics. Outbreak strains seem to have been circulating in Paraguay and Brazil prior to 2018, highlighting the importance of sustained DENV genomic surveillance.
1. Introduction
Dengue virus (DENV) is the most common arbovirus worldwide, with 50–100 million symptomatic infections resulting annually from four related viruses, designated DENV types 1–4 [1]. The reported epidemiology and relative severity of DENV-4 have differed between regions and patient populations, with predominantly secondary cases and less severe disease reported in Southeast Asia compared to a mixture of primary and secondary cases with a spectrum of disease severity in the Americas [2,3,4,5,6,7,8,9,10,11,12,13,14,15]. Five distinct genotypes of DENV-4 have been identified, and genetic differences between genotypes impact both viral biology and neutralization by pre-existing antibodies [16,17]. Genotype II was introduced into the Caribbean in the early 1980s, with multiple subsequent introductions into Brazil from Colombia or Venezuela in the early 2000s and spread to neighboring countries in the Southern Cone [6,18,19,20,21]. While DENV-4 epidemics have been described in the region, with strains emerging/re-emerging from pre-existing lineages [5,6,7,14,22,23,24,25], detection and characterization of dengue cases caused by DENV-4 have been hampered by the poor performance of available rapid diagnostics for this virus type [26,27,28]. The “gold standard” for dengue diagnosis has long been considered the detection of seroconversion between acute and convalescent samples [29,30], however, paired samples are frequently unavailable in clinical practice, further limiting detection by this method.
Paraguay is hyperendemic for DENV, with sustained viral circulation since 1999 and large disease outbreaks occurring every 2–5 years [31]. Dengue occurs throughout the country, but most cases are detected in metropolitan Asunción, which is the most populated area in the country and includes the capital and surrounding Central Department. Typically, a single DENV type predominates during the high transmission season from November through April. However, other types are also detected at lower rates [19,31,32,33] or with regional transmission [34]. DENV-4 was first identified in Paraguay in 2012 and circulated at low levels from 2012 to 2018. From 2015 to 2018, DENV-1 was predominant [31,33], but in 2019–2020, DENV-4 emerged to cause the largest DENV outbreak in the country’s history [31,35].
A previous study of temporal distribution of DENV in Paraguay revealed epidemic waves yearly recurrently during the late summer months. Moreover, the mosquito-viral suitability index accurately corresponded to the seasonal timing of reported dengue cases [36]. From February 2019 to March 2020, a bimodal incidence of suspected dengue cases was observed in Paraguay. The first wave extended from March to June 2019, with a peak in April when 2164 suspected cases were reported [37], and the larger second wave began in October 2019, when an epidemiologic alert for dengue was issued and a sustained increase in suspected cases was reported, with a peak in February 2020 with more than 33,200 suspected cases registered [38].
Published genomic data indicate that 2018 DENV-4 strains were most closely related to strains circulating in southern Brazil, circa 2013 [19,31]. Furthermore, a recent study showed that DENV-4 strains that circulated in Paraguay in 2020 were also related to viruses circulating in midwestern and southwestern Brazil [36]. Despite the recent advances/studies in DENV phylogenetics, more genomic information is required to understand the epidemiologic pattern and virus population dynamics in Paraguay and the neighboring countries. Therefore, the objectives of this study were to (1) describe diagnostic test performance for and clinical manifestations of dengue cases detected in 2019–2020 in Paraguay and (2) perform phylogenetic analyses of identified DENV-4 strains.
2. Materials and Methods
2.1. Study Participants
Participants of both genders and all ages were enrolled into an ongoing study of suspected arboviral infections between January 2019 and March 2020 from the Hospital Central of the Instituto de Previsión Social or as outpatients at IICS-UNA. Hospital Central, located in Asunción, is a tertiary care hospital that provides medical attention to patients from Asunción, the surrounding metropolitan area, and transfers from throughout the country. IICS-UNA is a research institute in San Lorenzo, which is in metropolitan Asunción, Central Department. Inclusion criteria were an acute illness including two or more of the following symptoms: fever (measured or subjective), red eyes, rash, joint pain involving more than one joint, and/or diffuse muscle pain. Patients with fever and no other localizing signs or symptoms were also included. Day 1 was defined as the day on which symptoms began, and individuals were included in the current study up to 7 days post-symptom onset. Cases were classified according to the 2009 WHO criteria as dengue without warning signs (DWS-), dengue with warning signs (DWS+), and severe dengue (SD) [30].
2.2. Clinical Samples and DENV Testing
Serum was obtained at the enrollment visit, aliquoted, and stored at −80 °C. Participants were screened for DENV by testing for the non-structural protein 1 (NS1) antigen and/or DENV RNA in a multiplex rRT-PCR for Zika, chikungunya, and dengue (the ZCD assay) [33,39,40]. NS1 testing was performed at IICS-UNA using the Standard Q Dengue Duo rapid immunochromatographic test (SD Biosensor, Suwon, South Korea) according to manufacturer recommendations. Screening test results, both positive and negative, were confirmed in a DENV type-specific, quantitative rRT-PCR (the DENV multiplex test, DMPT) [41]. The Standard Q Dengue Duo rapid immunochromatographic test also detects anti-DENV IgM and IgG. Results of antibody detection were recorded but not incorporated into the diagnostic algorithm of acute dengue cases.
The ZCD assay and DMPT were performed at both IICS-UNA and Emory University, following shipment of sample aliquots on dry ice. At IICS-UNA, RNA was extracted from 140 µL of serum using the Viral RNA Mini Kit (Qiagen, Germantown, MD, USA) and eluted into 60 µL of buffer, according to manufacturer recommendations. At Emory, total nucleic acid extraction was performed using either (1) an EMAG instrument (bioMérieux, Durham, NC, USA) or (2) the MagMaxViral RNA Isolation Kit in a KingFisher Apex system (both from ThermoFisher Scientific, Waltham, MA, USA). For automated extractions, nucleic acids were extracted from 200 µL of serum and eluted in 60 μL of buffer. A total of 5 μL of eluate was then used in ZCD and DMPT reactions, and both assays were performed and interpreted as previously described [39,40,41]. Serum viral load was quantified from 4-point standard curves prepared with synthesized DENV target sequences and included on dedicated DMPT runs.
2.3. Case Definitions
Dengue case confirmation required a positive result in the DMPT. Cases that (1) tested negative for DENV in the ZCD assay or (2) had a positive screening test (NS1 or ZCD) that could not be confirmed in the DMPT were considered other acute illness (OAI). This case definition was employed to ensure rigorous confirmation of dengue cases with at least two different tests.
2.4. DENV Sequencing
Sixty-two 2019–2020 samples were selected for sequencing from individuals with confirmed DENV-4 infections and DMPT Ct values < 35. Samples were further selected to represent the distribution of all cases based on month of collection, city of residence, and severity of clinical illness. All samples from SD cases that met the Ct criterion were selected. A single DENV-4 case collected in 2018 as part of this ongoing study was also sequenced and included in phylogenetic analyses [33].
Extracted total nucleic acid underwent heat-labile dsDNase treatment (ArcticZymes, Tromso, Norway). cDNA was synthesized using random hexamer primers and SuperScript III RT (both from ThermoFisher Scientific) for first strand synthesis and New England Biolabs (New England Biolabs, Inc., Ipswich, MA, USA) reagents for second strand synthesis, without amplification. Sequencing libraries were fragmented and indexed using the Nextera XT DNA Library Prep kit (Illumina, San Diego, CA, USA) with dual indexes and 16 cycles of PCR. Libraries were quantified using the KAPA universal complete kit (Roche, Basel, Switzerland), pooled to equimolar concentration, and sequenced on a MiSeq with paired-end 150-bp reads (Illumina, San Diego, CA, USA). As a negative control, water was included with each batch of samples starting from DNase. As a positive control, in vitro transcribed ERCC spike-ins (NIST) were added to each sample prior to cDNA synthesis.
Sequencing reads underwent reference-based assembly using viral-ngs version 2.0.21.3-rc20 (github.com/broadinstitute/viral-pipelines; date accessed, 01 February 2022) and reference sequence KP188564.1. Consensus sequences from each sample were aligned and visually inspected using Geneious R8 (Biomatters, San Francisco, CA, USA). Genotyping was performed using the online Genome Detective Virus Tool (https://www.genomedetective.com; date accessed, 1 February 2022) [42]. Complete DENV-4 genomes were downloaded from the Bacterial and Viral Bioinformatics Resource Center (BV-BRC, https://www.bv-brc.org/; date accessed, 1 February 2022) as reference sequences for phylogenetic analysis. These were MAFFT aligned with our Paraguay DENV-4 sequences using Geneious Prime (Biomatters, Inc., San Diego, CA, USA), and untranslated regions in the 5′ and 3′ ends were trimmed.
Maximum-likelihood (ML) phylogenies were estimated with IQ-TREE (version 1.6.12) with ultrafast bootstrap approximation to evaluate clade probabilities. ModelFinder was used to select the GTR+F+gamma4 nucleotide substitution model [43]. Temporal signal was assessed using TempEst v1.5.1 [44], and 12 reference sequences with >0.01 distance from the best-fitting linear regression were excluded as outliers for possible low sequencing quality or misclassified dates. Downsampling was performed from this alignment to yield a set of unique sequences with high genome coverage of predominantly the same genotype identified in this study (genotype II; see Supplemental Material for complete details). Our final dataset included 61 DENV-4 sequences generated by our group from 2019 to 2020, 1 DENV-4 sequence generated by our group from 2018, 9 reference sequences from Paraguay in 2018, and 129 globally representative DENV-4 genotype II reference sequences. The final ML phylogenetic tree was rooted on the oldest DENV-4 sequence.
Time-scaled phylogenetic trees were constructed in BEAST v1.10.4 using a GTR+gamma4 substitution model with 3 codon positions, a relaxed molecular clock, and 200,000,000 Markov chain Monte Carlo steps [45]. TreeAnnotator v1.10.4 was used to summarize the maximum clade credibility (MCC) tree after 10% burn-in [45]. ML and time-scaled trees were visualized through the interactive Tree of Life v6 (iTOL, https://itol.embl.de; date accessed, 10 February 2022) and FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree; date accessed, 10 February 2022).
2.5. Statistical Analysis
Basic statistical analyses were performed using Excel software version 2312 (Microsoft, Redmond, WA, USA). Comparisons between group means and medians were made by ANOVA, Welch’s test, both pooled and non-pooled two-sample t-tests, and Kruskal–Wallis tests. Comparisons of proportions were made using chi-squared tests or Fisher exact tests. Graphs were prepared with GraphPad Prism version 9 (GraphPad, San Diego, CA, USA). Crude associations and statistical analysis were performed using SAS version 9.4. Significance was set at two-sided p-values ≤ 0.05 for all analyses.
3. Results
3.1. Geographical Distribution of Studied Cases
Participants included in the current study were enrolled between February 2019 and March 2020, and the distribution approximately mirrored country-wide numbers of suspected dengue cases, both confirmed and unconfirmed, reported to the Ministerio de Salud Pública y Bienestar Social, Paraguay (Figure 1). Patients from 14 of 17 departments and the capital district of Paraguay were included (Figure 2A). Most dengue (229/253, 90.5%) and OAI cases (501/546, 91.8%) came from the Central Department or capital district (Table S1). Dengue cases were confirmed among individuals who resided in 9 departments and the capital district (Figure 2).
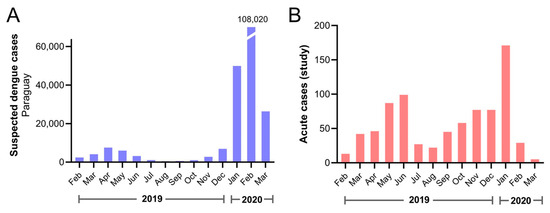
Figure 1.
Suspected dengue cases reported in the country and enrolled acute cases during the study period. (A) All suspected dengue cases reported to the Ministerio de Salud Pública y Bienestar Social, Paraguay by month. The number of cases for February 2020 is shown above the broken bar. (B) Acute cases included in the current study by date of symptom onset.
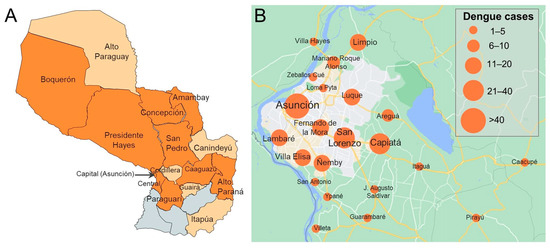
Figure 2.
(A) Map of Paraguay displaying departments from which dengue cases were enrolled (dark orange) or other acute illness cases were enrolled but dengue was not identified (light orange). Three departments from which no cases were enrolled are shown in grey (from west to east: Ñeembucú, Misiones, Caazapá). (B) Map of dengue cases detected in the study by city in the capital district (Asunción) and surrounding area. Maps were prepared using (A) Mapchart (www.mapchart.net; date accessed, 23 August 2022) and (B) Google Maps, 2022 (www.google.com/maps; date accessed, 23 August 2022).
3.2. Study Population
Seven hundred ninety-nine participants were enrolled and met inclusion criteria. This included 253 (31.7%) confirmed dengue and 546 (68.3%) OAI cases (Table 1). Dengue cases were older (mean 36.1 years, standard deviation (SD) 20.1) than OAIs (27.9, SD 19.3; p < 0.001) but were similar in gender makeup, comorbid illnesses, and days of symptoms at presentation (Table 1). DENV-4 was identified in 238 cases (94.1%), followed by DENV-2 (14, 5.5%) and DENV-1 (1, 0.4%). No mixed infections were detected, and no Zika or chikungunya cases were detected. DENV-4 serum viral load was quantifiable for 237/238 cases (99.6%, mean 6.67 log10 copies/mL, SD 1.50; Figure 3A), and viral load declined overall with days of symptoms (Figure 3B).

Table 1.
Demographic and clinical data for dengue cases versus other acute illness cases (N = 799).
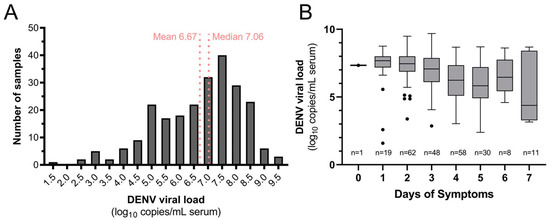
Figure 3.
Quantifiable DENV-4 serum viral load (A) distribution for dengue cases in the study population and (B) by day of symptoms at presentation.
DENV NS1 detection by rapid test demonstrated 52.5% sensitivity (53/101) and 96.5% specificity (387/401) compared to rRT-PCR (Table 2A). DENV viral load was not significantly different among samples with detectable versus undetectable NS1 or anti-DENV IgM (Figure S1). Sensitivity of NS1 detection was lowest on days 1 and 2 of symptoms (20–37%), with improved but variable detection from days 3 to 7 (44–76%; Figure S2). Anti-DENV IgM was detected in 25/96 dengue cases (26.0%) and 47/387 OAI cases (12.1%; Table 2B). Anti-DENV IgM detection did not demonstrate a consistent trend across days of symptoms (Figure S2). Of all samples analyzed for antibody detection, anti-DENV IgG was detected in 242/483 samples (50.1%) and 54/96 dengue cases (56.3%; Table 2C).

Table 2.
Comparison of (A) NS1, (B) IgM, and (C) IgG detection in acute dengue cases confirmed by rRT-PCR.
3.3. Clinical Manifestations and Severity
Symptoms reported among dengue and OAI cases are shown in Table 3. After correction for multiple comparisons, arthralgias, myalgias, and nausea remained significantly more common among dengue cases, whereas cough and sore throat were less common. Most participants reported having fever in the preceding 7 days (235/252 dengue (93.3%) and 480/531 OAI (90.4%) cases), and measured temperature did not differ between the groups (dengue, mean 38.7 °C (SD 0.7) and OAI 38.7 °C (0.8)). Of dengue cases, 136 (53.8%) were categorized as DWS-, 103 (40.7%) DWS+, and 14 (5.5%) SD.

Table 3.
Symptoms reported in the preceding 7 days among dengue and other acute illness cases.
The study population included 106 pregnant women: 22 dengue and 84 OAI cases. Two pregnant women (9.1%) had DWS+ (no SD cases). However, 19/22 (86.4%) were hospitalized, which was significantly higher than the proportion of hospitalized pregnant women with OAI cases (18/43 with disposition data (41.9%), p < 0.001).
3.4. Phylogenetic Analysis
DENV sequences in the final alignment included 138 reference and 62 newly generated sequences from Paraguay: 61 from 2019 to 2020 (Tables S2 and S3, Figure S3) and a single sequence from 2018 (NCBI-GenBank accession numbers: OP811915–OP811976). All newly generated DENV-4 sequences belonged to genotype II. Reference sequences represented samples collected from 1956 to 2018, including sequences from Asia and the Americas (Table S4).
In the ML phylogenetic analysis, all Paraguay sequences clustered together (Figure 4A, demarcated with a dashed line box), and outbreak sequences were most closely related to 2018 sequences from Paraguay, which clustered just basal to the sequences from this study (Figure S4). All ten Paraguay sequences from 2018 differed from the outbreak sequences by only two synonymous mutations, one in the NS3 gene and the other in NS5. The closest reference sequence from outside Paraguay came from a sample collected in São José do Rio Preto, Brazil in 2013 (KP188564.1). Outbreak strains comprised two clades, designated clade A (n = 24) and clade B (n = 37) (Figure 4B, tree branches shown in different shades of blue). Clades A and B differed by three synonymous single nucleotide polymorphisms, one each in the envelope, NS3, and NS5 genes. In ML analysis, Clade A appeared to be more closely related to the 2018 DENV-4 sequence generated for the current study, but in Bayesian analysis, that sequence was confirmed as ancestral to both. There was no phylogenetic clustering of cases by severity (Figure 4B), geographic location, or epidemic wave.
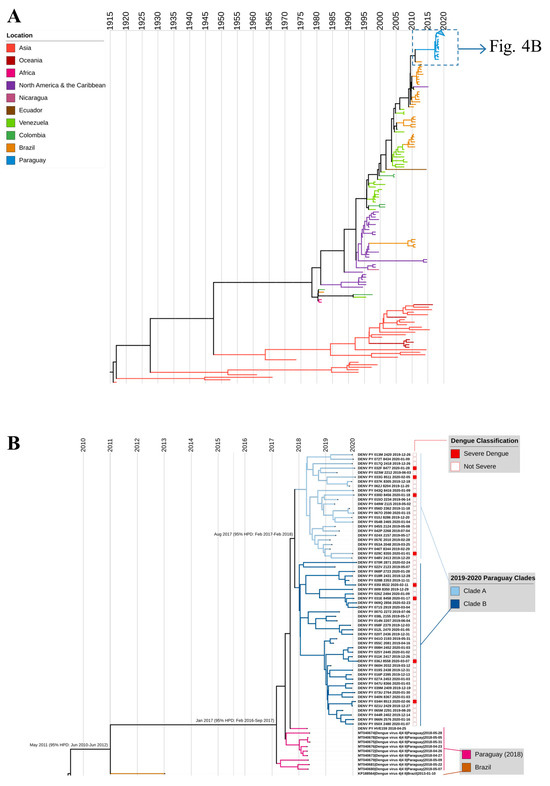
Figure 4.
MCC tree of DENV-4 genotype II. (A) Full tree with 200 sequences from the final dataset: 61 DENV-4 from Paraguay, 2019–2020 (this study); 1 DENV-4 sequence from Paraguay, 2018 (this study); 9 reference sequences from Paraguay in 2018; and 129 globally representative DENV-4 sequences. (B) Magnified and cropped MCC tree to show Paraguay 2019–2020 outbreak sequences and closest references. SD cases are indicated with a solid red box; all other dengue cases are indicated with an empty box. Years are indicated by vertical lines. Nodes for the MRCA are labeled with the month and year (95% confidence interval).
In time-scaled phylogenetic analysis, outbreak sequences again clustered together with high support and shared a most recent common ancestor in August 2017 (95% highest posterior density (HPD) February 2017–February 2018; Figure 4). Our inferred mean clock rate of 8.79 × 10−4 (95% HPD 7.85–9.80 × 10−4) is slightly higher than the median reported rate in prior studies on DENV-4, 7.91 × 10−4, but well within the range of reported rates, 6.89 × 10−4 to 20 × 10−4 [46]. All Paraguay sequences shared a common ancestor in January 2017 (95% HPD February 2016–September 2017) and diverged from their most recent ancestor, KP188564_Brazil_2013, in May 2011 (95% HPD June 2010–June 2012). These results suggest that there was unappreciated circulation of the outbreak lineage between 2011 and 2017. To assess whether the lineage was captured in prior studies of partial genome sequencing, we analyzed 743 DENV-4 reference sequences from the BV-BRC database collected between 2012 and 2018 with at least full envelope sequences (1485 bp), and we found no additional closely related reference sequences.
4. Discussion
From the end of 2019 to early 2020, DENV-4 caused the largest DENV outbreak in Paraguay’s history [31,35]. In our study population, dengue cases were poorly detected with available rapid diagnostics, associated with certain clinical manifestations, and some progressed to SD. The relative clinical severity of DENV-4 has varied in prior studies, which may reflect differences in virus strains and/or patient populations. In studies from Southeast Asia with documented transmission of all four DENV types, DENV-4 is often the least common, predominantly detected among secondary cases and associated with lower severity than other types [9,10,12]. SD risk with DENV-4 may also be lower in the Americas, particularly compared to DENV-2 [3,4,13,14,15]. However, consistent with our findings, DENV-4 still causes SD, with an overall risk similar to DENV-1 [11,13,15] and increased risk among older patients and those with secondary infections [4,5,6,7,8,9,11,13,47].
Phylogenetic analysis indicated that the DENV-4 lineage responsible for the 2019–2020 outbreak was nearly identical to viruses detected in Paraguay in 2018, consistent with a recent phylogenetic study of DENV in the country [36]. All Paraguay DENV-4 sequences shared a most recent common ancestor in 2017, and this aligns with a molecular clock analysis on 2018 DENV-4 sequences that estimated viral introduction into Paraguay in September 2017 [19]. Thus, this large outbreak was not due to introduction of a new lineage into the country in 2019 but instead resulted from local DENV-4 evolution and emergence in a susceptible population [22,23,25]. While we did not observe fixation of nonsynonymous mutations among DENV-4 sequences from Paraguay, these sequences all differed from their closest ancestor (KP188564_Brazil_2013) by seven amino acids, and the evolutionary history of this lineage over the decade preceding the outbreak is unclear due to limited DENV-4 sequences from the country and region.
DENV-4 has undergone multiple introductions into South America over the past 40 years, and genotype II, as identified in our study, has been predominant [5,6,7,16,18,20,21,24,25,47]. After being absent for three decades, DENV-4 was detected in Brazil in 2010 and resulted in explosive epidemics in the following years, probably because of the population’s susceptibility [5,48]. Similar to findings in Puerto Rico, previous research hypothesized that DENV-4 re-emergence or re-introduction in the state of Roraima in 2010 was preceded by cryptic or imperceptible circulation of the virus [20,49]. Several studies have demonstrated that densely populated states like Sao Paulo and Rio de Janeiro play a key role in the spread of DENV-4 to other Brazilian locations [48,50], and notably, the MRCA for DENV-4 strains in Paraguay was detected in Sao Paulo state. Autochthonous DENV-4 evolution that precedes viral re-emergence and the ongoing risk for introduction of new strains, such as genotype I introduction into Brazil from Asia [20], highlights the importance of sustained genomic monitoring to trace the origin of new outbreaks.
It is notable that DENV-4 emerged in Paraguay in a population where DENV-1 had been predominant for the previous four years [33], as both the change in the predominant DENV type and waning cross-protective immunity could have contributed to the high numbers of symptomatic infections seen in 2019–2020 [51,52]. The DENV-1/DENV-4 order of infections has been observed among SD cases [53], and prior DENV-1 infection has disproportionately contributed to SD elsewhere [54]. The wave dynamics observed in 2019–2020 fit with the arrival of DENV-4 outbreak strains in a susceptible population relatively late in the DENV transmission season that ended in early 2019 in Paraguay [55], and consistent with this, genetic differences were not observed between the two waves. All sequenced SD cases were detected in the second wave. However, these did not cluster in phylogenetic analyses, and this finding may be attributed to higher case numbers in the second wave increasing observed SD by chance.
Rapid NS1 testing demonstrated poor sensitivity (52.5%) for dengue cases caused by DENV-4. This was lower than the sensitivity of the same assay observed during the 2018 DENV-1 outbreak in Asunción (71.4%), though specificity was high (>96%) in both studies [33]. These data are similar to findings from Brazil, where rapid immunochromatographic tests for NS1 resulted in under-detection of DENV-4 [26,27,28,56]. Poor NS1 performance may result from lower levels of NS1 in DENV-4 cases, though data to this effect are sparse [57], or high seroprevalence of anti-DENV IgG. Sensitivity of NS1 detection may improve with heat dissociation of IgG-NS1 complexes [5,56], but this requires instrumentation and detracts from the benefits of point-of-care testing. Due to NS1 test performance, this was implemented only as a screening test to determine further work-up by rRT-PCR. NS1 rapid tests continue to be a widely used tool in clinical practice, particularly in sites with limited resources due to simplicity and relatively low cost [58,59]. However, it is important to consider the potential clinical and epidemiologic impact of their reduced sensitivity in comparison to DENV RNA detection shown in this work, particularly for DENV-4. This emphasizes the necessity of developing point-of-care diagnostic tests with improved performance features [60,61].
The clinical presentation of dengue cases differed from that of OAIs in this population, with arthralgia, myalgia, and nausea reported significantly more often among cases and cough and sore throat reported less often. DENV-4 has previously been associated with cutaneous manifestations when compared to other DENV types [34]. Although rash was also more common among dengue cases in our population, this did not remain significant after adjustment for multiple comparisons. Sore throat was also less common among DENV-1 cases from Asunción in 2018, when dengue cases more commonly experienced headache and conjunctivitis [33]. Although these remain relatively general complaints, identification of such symptom constellations will aid clinicians in the judicious use and interpretation of available diagnostics.
5. Limitations
This study focused on acute symptomatic dengue cases. Therefore, results may not be generalizable to mild or subclinical DENV-4 infections, and primary/secondary infection status could not be fully characterized. Second, although participants resided in 14/17 departments in Paraguay, over 90% of individuals lived in Asunción or the Central Department, which impacts the power of phylogenetic studies to detect regional differences in DENV-4 sequences. Third, the genomic record for DENV-4 in Paraguay dates back only to 2018 and is limited in South America as a whole. This complicated analyses of DENV-4 introduction into Paraguay and the emergence of the two clades identified in the current outbreak. Nevertheless, this study provides important information on a large dengue epidemic that occurred in an endemic country like Paraguay and could serve to improve our understanding of dengue epidemiology in the region.
6. Conclusions
Findings from the 2019–2020 DENV outbreak in Paraguay highlight the capacity of DENV-4 to cause explosive outbreaks and the need for sustained genomic monitoring of circulating DENV strains in a population. DENV-4 is poorly detected with available rapid diagnostics and, without high rates of symptomatic disease and widespread molecular testing, may remain under-reported and insufficiently characterized.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v16020181/s1, Supplemental methods. Downsampling of DENV-4 sequences. Figure S1. DENV viral load vs. (A) NS1 antigen or (B) anti-DENV IgM. Figure S2. NS1 and anti-DENV IgM rapid diagnostic test performance by day post-symptom onset. Figure S3. Epidemiologic week of symptom onset for 61 sequenced DENV-4 samples. Figure S4. ML trees of DENV-4 genotype II. Table S1. Department or district of residence of dengue and other acute illness cases. Table S2. Data for DENV-4 samples sequenced in this study. Table S3. Demographic data for participants from whom DENV-4 whole genome sequences were obtained. Table S4. Sequences retrieved from NCBI GenBank included for the phylogenetic analysis.
Author Contributions
Conceptualization, A.R. and J.J.W.; Methodology, A.R., J.S., F.C., S.P., A.P., and J.J.W.; Validation, A.R., J.S., F.C., C.B., S.P., A.K., A.H., A.P. and J.J.W.; Formal Analysis, A.R., J.S., F.C., C.B., S.P., A.P. and J.J.W.; Investigation, All authors; Resources, A.R., F.C., Y.d.G., P.L. (Patricia Langjahr), M.E.A., L.A., L.M., M.P., M.V.-H., P.L. (Patricia Luraschi), S.C., M.C.S., A.T., B.A.P., A.P. and J.J.W.; Data Curation, A.R., J.S., F.C., C.B., S.P., A.K., A.H., A.P. and J.J.W.; Writing—Original Draft Preparation, A.R., J.S., F.C., C.B., S.P., A.P. and J.J.W.; Writing—Review and Editing, All authors; Visualization, A.R., J.S., F.C., C.B., S.P., A.P. and J.J.W.; Supervision, A.R., F.C., Y.d.G., P.L. (Patricia Langjahr), L.M., B.A.P., A.P. and J.J.W.; Project Administration, A.R., F.C., B.A.P., A.P. and J.J.W.; Funding Acquisition, A.R., B.A.P. and J.J.W. All authors have read and agreed to the published version of the manuscript.
Funding
Research was supported by an award from the Doris Duke Charitable Foundation (Clinical Scientist Development Award 2019089, J.J.W.); a grant from the National Institute of Allergy and Infectious Diseases (NIAID, R21 AI146443); and the Consejo Nacional de Ciencia y Tecnología (CONACYT) with support from Fondo para la Excelencia de la Educación y la Investigación (FEEI), Paraguay (PINV18-1295, A.R.).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki. The study protocol was reviewed and approved by the Scientific and Ethics Committee of the Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción (IICS-UNA, IRB00011984) and the Emory University Institutional Review Board (IRB00000569).
Informed Consent Statement
Written informed consent was obtained from all subjects. Parents or legal guardians provided consent for children, and children older than six years of age provided assent.
Data Availability Statement
Upon publication, data supporting the presented results will be made freely available through the Emory Dataverse, which is an open data repository offered through a partnership between Emory and the Odum Institute at the University of North Carolina at Chapel Hill.
Acknowledgments
We thank the members of the study team based at the Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción; Hospital Villa Elisa; and Hospital Central del Instituto de Previsión Social, all located in Paraguay, for their dedication to this study and their excellent work. We are grateful to the study participants and their families.
Conflicts of Interest
The authors have declared that no competing interests exist.
References
- Stanaway, J.D.; Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Coffeng, L.E.; Brady, O.J.; Hay, S.I.; Bedi, N.; Bensenor, I.M.; Castaneda-Orjuela, C.A.; et al. The global burden of dengue: An analysis from the Global Burden of Disease Study 2013. Lancet Infect. Dis. 2016, 16, 712–723. [Google Scholar] [CrossRef]
- Bhoomiboonchoo, P.; Nisalak, A.; Chansatiporn, N.; Yoon, I.K.; Kalayanarooj, S.; Thipayamongkolgul, M.; Endy, T.; Rothman, A.L.; Green, S.; Srikiatkhachorn, A.; et al. Sequential dengue virus infections detected in active and passive surveillance programs in Thailand, 1994–2010. BMC Public Health 2015, 15, 250. [Google Scholar] [CrossRef]
- Heringer, M.; Nogueira, R.M.; de Filippis, A.M.; Lima, M.R.; Faria, N.R.; Nunes, P.C.; Nogueira, F.B.; dos Santos, F.B. Impact of the emergence and re-emergence of different dengue viruses’ serotypes in Rio de Janeiro, Brazil, 2010 to 2012. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 268–274. [Google Scholar] [CrossRef]
- Soo, K.M.; Khalid, B.; Ching, S.M.; Chee, H.Y. Meta-Analysis of Dengue Severity during Infection by Different Dengue Virus Serotypes in Primary and Secondary Infections. PLoS ONE 2016, 11, e0154760. [Google Scholar] [CrossRef]
- Heringer, M.; Souza, T.M.A.; Lima, M.; Nunes, P.C.G.; Faria, N.; de Bruycker-Nogueira, F.; Chouin-Carneiro, T.; Nogueira, R.M.R.; Dos Santos, F.B. Dengue type 4 in Rio de Janeiro, Brazil: Case characterization following its introduction in an endemic region. BMC Infect. Dis. 2017, 17, 410. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.B.; Gutierrez, G.F.; Bruno, A.; Cordoba, M.T.; Bono, M.M.; Polack, F.P.; Talarico, L.B.; Quipildor, M.O. Age-associated differences in clinical manifestations and laboratory parameters during a dengue virus type 4 outbreak in Argentina. J. Med. Virol. 2018, 90, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, I.L.C.; Araujo, F.M.C.; Cavalcanti, L.P.G.; Braga, D.N.M.; Perdigao, A.C.B.; Santos, F.B.D.; Nogueira, F.B.; Escossia, K.; Guedes, M.I.F. Dengue 4 in Ceara, Brazil: Characterisation of epidemiological and laboratorial aspects and causes of death during the first epidemic in the state. Mem. Inst. Oswaldo Cruz 2018, 113, e180320. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.; Verlaeten, O.; Cabie, A.; Kaidomar, S.; Moravie, V.; Martial, J.; Najioullah, F.; Plumelle, Y.; Fonteau, C.; Dussart, P.; et al. Influence of the dengue serotype, previous dengue infection, and plasma viral load on clinical presentation and outcome during a dengue-2 and dengue-4 co-epidemic. Am. J. Trop. Med. Hyg. 2008, 78, 990–998. [Google Scholar] [CrossRef]
- Fried, J.R.; Gibbons, R.V.; Kalayanarooj, S.; Thomas, S.J.; Srikiatkhachorn, A.; Yoon, I.K.; Jarman, R.G.; Green, S.; Rothman, A.L.; Cummings, D.A. Serotype-specific differences in the risk of dengue hemorrhagic fever: An analysis of data collected in Bangkok, Thailand from 1994 to 2006. PLoS Neglected Trop. Dis. 2010, 4, e617. [Google Scholar] [CrossRef]
- Sabchareon, A.; Sirivichayakul, C.; Limkittikul, K.; Chanthavanich, P.; Suvannadabba, S.; Jiwariyavej, V.; Dulyachai, W.; Pengsaa, K.; Margolis, H.S.; Letson, G.W. Dengue infection in children in Ratchaburi, Thailand: A cohort study. I. Epidemiology of symptomatic acute dengue infection in children, 2006–2009. PLoS Neglected Trop. Dis. 2012, 6, e1732. [Google Scholar] [CrossRef]
- Sharp, T.M.; Hunsperger, E.; Santiago, G.A.; Munoz-Jordan, J.L.; Santiago, L.M.; Rivera, A.; Rodriguez-Acosta, R.L.; Gonzalez Feliciano, L.; Margolis, H.S.; Tomashek, K.M. Virus-specific differences in rates of disease during the 2010 Dengue epidemic in Puerto Rico. PLoS Neglected Trop. Dis. 2013, 7, e2159. [Google Scholar] [CrossRef]
- Yung, C.F.; Lee, K.S.; Thein, T.L.; Tan, L.K.; Gan, V.C.; Wong, J.G.X.; Lye, D.C.; Ng, L.C.; Leo, Y.S. Dengue serotype-specific differences in clinical manifestation, laboratory parameters and risk of severe disease in adults, singapore. Am. J. Trop. Med. Hyg. 2015, 92, 999–1005. [Google Scholar] [CrossRef]
- Vicente, C.R.; Herbinger, K.H.; Froschl, G.; Malta Romano, C.; de Souza Areias Cabidelle, A.; Cerutti Junior, C. Serotype influences on dengue severity: A cross-sectional study on 485 confirmed dengue cases in Vitoria, Brazil. BMC Infect. Dis. 2016, 16, 320. [Google Scholar] [CrossRef] [PubMed]
- Rocha, B.A.M.; Guilarde, A.O.; Argolo, A.; Tassara, M.P.; da Silveira, L.A.; Junqueira, I.C.; Turchi, M.D.; Feres, V.C.R.; Martelli, C.M.T. Dengue-specific serotype related to clinical severity during the 2012/2013 epidemic in centre of Brazil. Infect. Dis. Poverty 2017, 6, 116. [Google Scholar] [CrossRef]
- Nunes, P.C.G.; de Filippis, A.M.B.; Lima, M.; Faria, N.; de Bruycker-Nogueira, F.; Santos, J.B.; Heringer, M.; Chouin-Carneiro, T.; Couto-Lima, D.; de Santis Goncalves, B.; et al. 30 years of dengue fatal cases in Brazil: A laboratorial-based investigation of 1047 cases. BMC Infect. Dis. 2018, 18, 346. [Google Scholar] [CrossRef] [PubMed]
- Waman, V.P.; Kasibhatla, S.M.; Kale, M.M.; Kulkarni-Kale, U. Population genomics of dengue virus serotype 4: Insights into genetic structure and evolution. Arch. Virol. 2016, 161, 2133–2148. [Google Scholar] [CrossRef]
- Gallichotte, E.N.; Baric, T.J.; Nivarthi, U.; Delacruz, M.J.; Graham, R.; Widman, D.G.; Yount, B.L.; Durbin, A.P.; Whitehead, S.S.; de Silva, A.M.; et al. Genetic Variation between Dengue Virus Type 4 Strains Impacts Human Antibody Binding and Neutralization. Cell Rep. 2018, 25, 1214–1224. [Google Scholar] [CrossRef]
- Foster, J.E.; Bennett, S.N.; Vaughan, H.; Vorndam, V.; McMillan, W.O.; Carrington, C.V. Molecular evolution and phylogeny of dengue type 4 virus in the Caribbean. Virology 2003, 306, 126–134. [Google Scholar] [CrossRef]
- Graf, T.; Vazquez, C.; Giovanetti, M.; de Bruycker-Nogueira, F.; Fonseca, V.; Claro, I.M.; de Jesus, J.G.; Gomez, A.; Xavier, J.; de Mendonca, M.C.L.; et al. Epidemiologic History and Genetic Diversity Origins of Chikungunya and Dengue Viruses, Paraguay. Emerg. Infect. Dis. 2021, 27, 1393–1404. [Google Scholar] [CrossRef]
- Nunes, M.R.; Faria, N.R.; Vasconcelos, H.B.; Medeiros, D.B.; Silva de Lima, C.P.; Carvalho, V.L.; Pinto da Silva, E.V.; Cardoso, J.F.; Sousa, E.C., Jr.; Nunes, K.N.; et al. Phylogeography of dengue virus serotype 4, Brazil, 2010–2011. Emerg. Infect. Dis. 2012, 18, 1858–1864. [Google Scholar] [CrossRef]
- Carrington, C.V.; Foster, J.E.; Pybus, O.G.; Bennett, S.N.; Holmes, E.C. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J. Virol. 2005, 79, 14680–14687. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S.N.; Drummond, A.J.; Kapan, D.D.; Suchard, M.A.; Munoz-Jordan, J.L.; Pybus, O.G.; Holmes, E.C.; Gubler, D.J. Epidemic dynamics revealed in dengue evolution. Mol. Biol. Evol. 2010, 27, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S.N.; Holmes, E.C.; Chirivella, M.; Rodriguez, D.M.; Beltran, M.; Vorndam, V.; Gubler, D.J.; McMillan, W.O. Selection-driven evolution of emergent dengue virus. Mol. Biol. Evol. 2003, 20, 1650–1658. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Silva, C.L.; Carreno, M.F.; Ortiz-Baez, A.S.; Rey, L.A.; Villabona-Arenas, C.J.; Ocazionez, R.E. Evolutionary history and spatio-temporal dynamics of dengue virus serotypes in an endemic region of Colombia. PLoS ONE 2018, 13, e0203090. [Google Scholar] [CrossRef]
- Martin, E.; Chirivella, M.; Co, J.K.G.; Santiago, G.A.; Gubler, D.J.; Munoz-Jordan, J.L.; Bennett, S.N. Insights into the molecular evolution of Dengue virus type 4 in Puerto Rico over two decades of emergence. Virus Res. 2016, 213, 23–31. [Google Scholar] [CrossRef]
- Acosta, P.O.; Granja, F.; Meneses, C.A.; Nascimento, I.A.; Sousa, D.D.; Lima Junior, W.P.; Naveca, F.G. False-negative dengue cases in Roraima, Brazil: An approach regarding the high number of negative results by NS1 ag kits. Rev. Inst. Med. Trop. Sao Paulo 2014, 56, 447–450. [Google Scholar] [CrossRef]
- Colombo, T.E.; Vedovello, D.; Araki, C.S.; Cogo-Moreira, H.; dos Santos, I.N.; Reis, A.F.; Costa, F.R.; Cruz, L.E.; Casagrande, L.; Regatieri, L.J.; et al. Dengue-4 false negative results by Panbio(R) Dengue Early ELISA assay in Brazil. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2013, 58, 710–712. [Google Scholar] [CrossRef]
- Sea, V.R.; Cruz, A.C.; Gurgel, R.Q.; Nunes, B.T.; Silva, E.V.; Dolabella, S.S.; dos Santos, R.L. Underreporting of Dengue-4 in Brazil due to low sensitivity of the NS1 Ag test in routine control programs. PLoS ONE 2013, 8, e64056. [Google Scholar] [CrossRef]
- Hunsperger, E.A.; Sharp, T.M.; Lalita, P.; Tikomaidraubuta, K.; Cardoso, Y.R.; Naivalu, T.; Khan, A.S.; Marfel, M.; Hancock, W.T.; Tomashek, K.M.; et al. Use of a Rapid Test for Diagnosis of Dengue during Suspected Dengue Outbreaks in Resource-Limited Regions. J. Clin. Microbiol. 2016, 54, 2090–2095. [Google Scholar] [CrossRef][Green Version]
- World Health Organization. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control; WHO Press: Paris, France, 2009. [Google Scholar]
- Dirección de Vigilancia de la Salud, Ministerio de Salud Pública y Bienestar Social. Arbovirosis. Available online: https://dgvs.mspbs.gov.py/page/#arbovirosis.html (accessed on 20 September 2022).
- Rojas, A.; Moreira Soares, A.; Mendoza, L.P.; Acosta, M.E.; Aria, L.; Paez, M.; Herebia, L.; Vallejos, M.A.; de Guillen, Y.; Aquino, V.H. Revisiting the dengue epidemic of 2011 in Paraguay: Molecular epidemiology of dengue virus in the Asuncion metropolitan area. BMC Infect. Dis. 2021, 21, 769. [Google Scholar] [CrossRef]
- Rojas, A.; Cardozo, F.; Cantero, C.; Stittleburg, V.; Lopez, S.; Bernal, C.; Gimenez Acosta, F.E.; Mendoza, L.; Pinsky, B.A.; Arevalo de Guillen, I.; et al. Characterization of dengue cases among patients with an acute illness, Central Department, Paraguay. PeerJ 2019, 7, e7852. [Google Scholar] [CrossRef]
- Halsey, E.S.; Marks, M.A.; Gotuzzo, E.; Fiestas, V.; Suarez, L.; Vargas, J.; Aguayo, N.; Madrid, C.; Vimos, C.; Kochel, T.J.; et al. Correlation of serotype-specific dengue virus infection with clinical manifestations. PLoS Neglected Trop. Dis. 2012, 6, e1638. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization. Dengue. Available online: https://www3.paho.org/data/index.php/es/temas/indicadores-dengue.html (accessed on 1 July 2022).
- Vazquez, C.; Alcantara, L.C.J.; Fonseca, V.; Lima, M.; Xavier, J.; Adelino, T.; Fritsch, H.; Castro, E.; de Oliveira, C.; Schuab, G.; et al. Retrospective Spatio-Temporal Dynamics of Dengue Virus 1, 2 and 4 in Paraguay. Viruses 2023, 15, 1275. [Google Scholar] [CrossRef] [PubMed]
- Dirección de Vigilancia de la Salud, Ministerio de Salud Pública y Bienestar Social. Boletín Epidemiológico SE 1a la SE 21. 2019, (21). Available online: https://dgvs.mspbs.gov.py/boletin-epidemiologico-semanal (accessed on 20 September 2022).
- Dirección de Vigilancia de la Salud, Ministerio de Salud Pública y Bienestar Social. Boletín Epidemiológico, SE 1 a la SE 52 (29-12-2019 al 26-12-2020). 2020, (52). Available online: https://dgvs.mspbs.gov.py/boletin-epidemiologico-semanal (accessed on 20 September 2022).
- Cantero, C.; Cardozo, F.; Waggoner, J.J.; Pinsky, B.A.; Espinola, A.; Infanzon, B.; Acosta, M.E.; Aria, L.; Arevalo de Guillen, Y.; Cuevas, T.; et al. Implementation of a Multiplex rRT-PCR for Zika, Chikungunya, and Dengue Viruses: Improving Arboviral Detection in an Endemic Region. Am. J. Trop. Med. Hyg. 2020, 102, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, J.J.; Gresh, L.; Mohamed-Hadley, A.; Ballesteros, G.; Davila, M.J.; Tellez, Y.; Sahoo, M.K.; Balmaseda, A.; Harris, E.; Pinsky, B.A. Single-Reaction Multiplex Reverse Transcription PCR for Detection of Zika, Chikungunya, and Dengue Viruses. Emerg. Infect. Dis. 2016, 22, 1295–1297. [Google Scholar] [CrossRef]
- Waggoner, J.J.; Abeynayake, J.; Sahoo, M.K.; Gresh, L.; Tellez, Y.; Gonzalez, K.; Ballesteros, G.; Pierro, A.M.; Gaibani, P.; Guo, F.P.; et al. Single-reaction, multiplex, real-time rt-PCR for the detection, quantitation, and serotyping of dengue viruses. PLoS Neglected Trop. Dis. 2013, 7, e2116. [Google Scholar] [CrossRef] [PubMed]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.M.; et al. Genome Detective: An automated system for virus identification from high-throughput sequencing data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kuhnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Pollett, S.; Melendrez, M.C.; Maljkovic Berry, I.; Duchêne, S.; Salje, H.; Cummings, D.A.T.; Jarman, R.G. Understanding dengue virus evolution to support epidemic surveillance and counter-measure development. Infect. Genet. Evol. 2018, 62, 279–295. [Google Scholar] [CrossRef]
- Campos, G.S.; Pinho, A.C.; Brandao, C.J.; Bandeira, A.C.; Sardi, S.I. Dengue virus 4 (DENV-4) re-emerges after 30 years in Brazil: Cocirculation of DENV-2, DENV-3, and DENV-4 in Bahia. Jpn. J. Infect. Dis. 2015, 68, 45–49. [Google Scholar] [CrossRef]
- Ortiz-Baez, A.S.; Cunha, M.D.P.; Vedovello, D.; Colombo, T.E.; Nogueira, M.L.; Villabona-Arenas, C.J.; Zanotto, P.M.A. Origin, tempo, and mode of the spread of DENV-4 Genotype IIB across the state of São Paulo, Brazil during the 2012–2013 outbreak. Mem. Inst. Oswaldo Cruz 2019, 114, e180251. [Google Scholar] [CrossRef] [PubMed]
- de Souza, R.P.; Rocco, I.M.; Maeda, A.Y.; Spenassatto, C.; Bisordi, I.; Suzuki, A.; Silveira, V.R.; Silva, S.J.; Azevedo, R.M.; Tolentino, F.M.; et al. Dengue virus type 4 phylogenetics in Brazil 2011: Looking beyond the veil. PLoS Neglected Trop. Dis. 2011, 5, e1439. [Google Scholar] [CrossRef] [PubMed]
- Macedo, G.A.; de Araújo, J.M.; Schatzmayr, H.G.; Costa, F.A.; de Filippis, A.M.; Santos, F.B.; Nogueira, R.M. Virological surveillance for early warning of dengue epidemics in the State of Rio de Janeiro, Brazil. Trans. R. Soc. Trop. Med. Hyg. 2013, 107, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, J.J.; Gresh, L.; Mohamed-Hadley, A.; Balmaseda, A.; Soda, K.J.; Abeynayake, J.; Sahoo, M.K.; Liu, Y.; Kuan, G.; Harris, E.; et al. Characterization of Dengue Virus Infections Among Febrile Children Clinically Diagnosed With a Non-Dengue Illness, Managua, Nicaragua. J. Infect. Dis. 2017, 215, 1816–1823. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Montoya, M.; Gresh, L.; Balmaseda, A.; Harris, E. Neutralizing antibody titers against dengue virus correlate with protection from symptomatic infection in a longitudinal cohort. Proc. Natl. Acad. Sci. USA 2016, 113, 728–733. [Google Scholar] [CrossRef]
- Gibbons, R.V.; Kalanarooj, S.; Jarman, R.G.; Nisalak, A.; Vaughn, D.W.; Endy, T.P.; Mammen, M.P., Jr.; Srikiatkhachorn, A. Analysis of repeat hospital admissions for dengue to estimate the frequency of third or fourth dengue infections resulting in admissions and dengue hemorrhagic fever, and serotype sequences. Am. J. Trop. Med. Hyg. 2007, 77, 910–913. [Google Scholar] [CrossRef]
- Sangkawibha, N.; Rojanasuphot, S.; Ahandrik, S.; Viriyapongse, S.; Jatanasen, S.; Salitul, V.; Phanthumachinda, B.; Halstead, S.B. Risk factors in dengue shock syndrome: A prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am. J. Epidemiol. 1984, 120, 653–669. [Google Scholar] [CrossRef]
- Romeo-Aznar, V.; Picinini Freitas, L.; Goncalves Cruz, O.; King, A.A.; Pascual, M. Fine-scale heterogeneity in population density predicts wave dynamics in dengue epidemics. Nat. Commun. 2022, 13, 996. [Google Scholar] [CrossRef]
- Lima Mda, R.; Nogueira, R.M.; Filippis, A.M.; Nunes, P.C.; Sousa, C.S.; Silva, M.H.; Santos, F.B. A simple heat dissociation method increases significantly the ELISA detection sensitivity of the nonstructural-1 glycoprotein in patients infected with DENV type-4. J. Virol. Methods 2014, 204, 105–108. [Google Scholar] [CrossRef]
- Nunes, P.C.G.; Nogueira, R.M.R.; Heringer, M.; Chouin-Carneiro, T.; Damasceno Dos Santos Rodrigues, C.; de Filippis, A.M.B.; Lima, M.; Dos Santos, F.B. NS1 Antigenemia and Viraemia Load: Potential Markers of Progression to Dengue Fatal Outcome? Viruses 2018, 10, 326. [Google Scholar] [CrossRef]
- Shukla, M.K.; Singh, N.; Sharma, R.K.; Barde, P.V. Utility of dengue NS1 antigen rapid diagnostic test for use in difficult to reach areas and its comparison with dengue NS1 ELISA and qRT-PCR. J. Med. Virol. 2017, 89, 1146–1150. [Google Scholar] [CrossRef]
- Teoh, B.T.; Sam, S.S.; Tan, K.K.; Johari, J.; Abd-Jamil, J.; Hooi, P.S.; AbuBakar, S. The Use of NS1 Rapid Diagnostic Test and qRT-PCR to Complement IgM ELISA for Improved Dengue Diagnosis from Single Specimen. Sci. Rep. 2016, 6, 27663. [Google Scholar] [CrossRef] [PubMed]
- Hunsperger, E.A.; Munoz-Jordan, J.; Beltran, M.; Colon, C.; Carrion, J.; Vazquez, J.; Acosta, L.N.; Medina-Izquierdo, J.F.; Horiuchi, K.; Biggerstaff, B.J.; et al. Performance of Dengue Diagnostic Tests in a Single-Specimen Diagnostic Algorithm. J. Infect. Dis. 2016, 214, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Hunsperger, E.A.; Yoksan, S.; Buchy, P.; Nguyen, V.C.; Sekaran, S.D.; Enria, D.A.; Vazquez, S.; Cartozian, E.; Pelegrino, J.L.; Artsob, H.; et al. Evaluation of commercially available diagnostic tests for the detection of dengue virus NS1 antigen and anti-dengue virus IgM antibody. PLoS Neglected Trop. Dis. 2014, 8, e3171. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).