Antiviral Activity of Ailanthone from Ailanthus altissima on the Rice Stripe Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sources of Virus, Vectors, and Plant Materials
2.2. Preparation of Rice Protoplasts and Virus Infection
2.3. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time RT-PCR Analysis
2.4. Sodium Dodecyl Sulfate−Polyacrylamide Gel Electrophoresis (SDS−PAGE) and Western Blot Analysis
2.5. Curative, Protective, and Inactivating Activities of Ailanthone against the RSV Pathogenic Process in N. benthamiana
2.6. Dose-Dependent Inhibitory Effects of Ailanthone on RSV Replication in N. benthamiana
2.7. Dose-Dependent Inhibitory Effects of Ailanthone on RSV Replication in Rice Protoplasts
3. Results
3.1. Effect of Ailanthone on RSV Multiplication in N. benthamiana
3.1.1. Dose-Dependent Inhibitory Activities of Ailanthone on RSV-CP Expression in N. benthamiana
3.1.2. Preventive, Inactivating, and Curative Effects against RSV Multiplication in N. benthamiana
3.1.3. Effects of Ailanthone on Coding Gene Expression of RSV in N. benthamiana
3.2. Effect of Ailanthone on RSV Replication in Rice Protoplasts
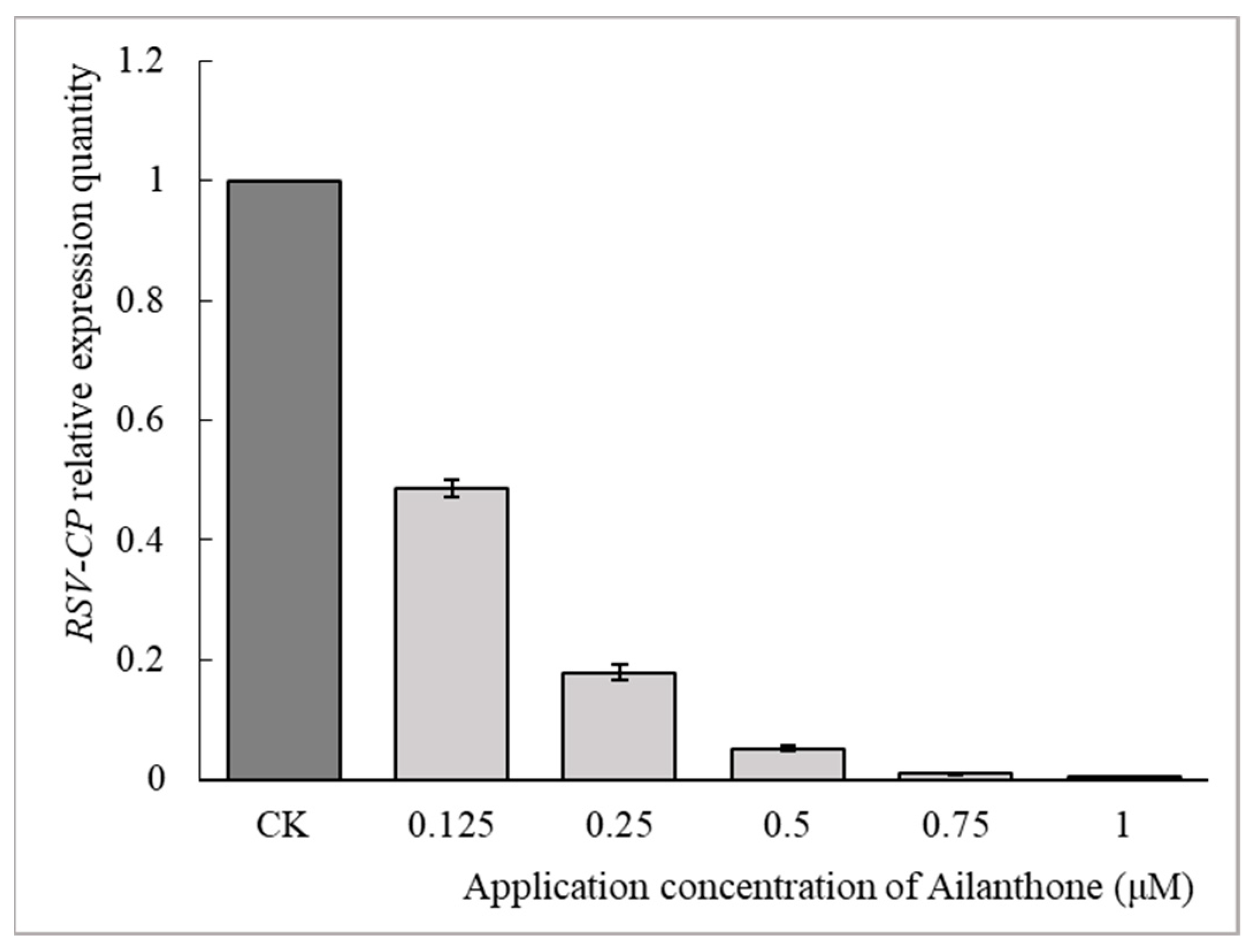
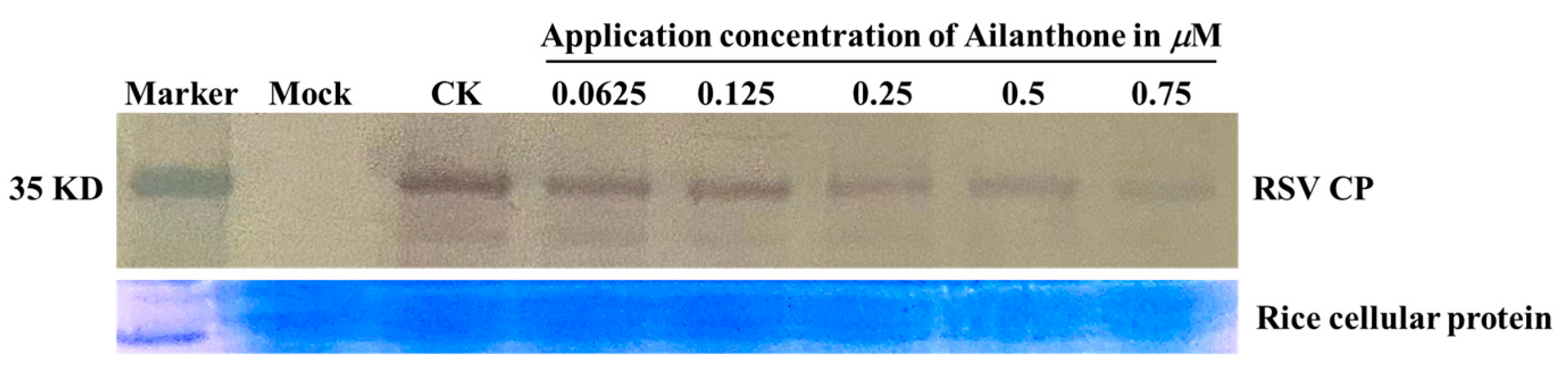
3.2.1. Dose-Dependent Inhibitory Activities of Ailanthone against RSV Multiplication in Rice Protoplasts
3.2.2. Effects of Ailanthone on Coding Gene Expression of RSV in Rice Protoplasts
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, Y.; Fu, S.; Tao, X.; Zhou, X. Rice stripe virus: Exploring molecular weapons in the arsenal of a negative-sense RNA virus. Annu. Rev. Phytopathol. 2021, 59, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Hibino, H. Biology and epidemiology of rice viruses. Annu. Rev. Phytopathol. 1996, 34, 249–274. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Yuan, X.; Zhou, T.; Fan, Y.; Zhou, Y. Arabidopsis is susceptible to Rice stripe virus infections. J. Phytopathol. 2011, 159, 767–772. [Google Scholar] [CrossRef]
- Xiong, R.; Wu, J.; Zhou, Y.; Zhou, X. Characterization and subcellular localization of an RNA silencing suppressor encoded by rice stripe tenuivirus. Virology 2009, 387, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, L.; Fu, S.; Wu, J.; Zhou, X. Population diversity of Rice stripe virus-derived siRNAs in three different hosts and RNAi-based antiviral immunity in Laodelphgax striatellus. PLoS ONE 2012, 7, e46238. [Google Scholar] [CrossRef]
- Lu, L.; Wu, S.; Jiang, J.; Liang, J.; Zhou, X.; Wu, J. Whole genome deep sequencing revealed host impact on population structure, variation and evolution of Rice stripe virus. Virology 2018, 524, 32–44. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, X. Role of Rice stripe virus NSvc4 in cell-to-cell movement and symptom development in Nicotiana benthamiana. Front. Plant Sci. 2012, 3, 269. [Google Scholar] [CrossRef]
- Zhang, J.; He, F.; Chen, J.; Wang, Y.; Yang, Y.; Hu, D.; Song, B. Purine nucleoside derivatives containing a sulfa ethylamine moiety: Design, synthesis, antiviral activity, and mechanism. J. Agri. Food Chem. 2021, 69, 5575–5582. [Google Scholar] [CrossRef]
- Sun, F.; Hu, P.; Wang, W.; Lan, Y.; Du, L.; Zhou, Y.; Zhou, T. Rice stripe virus coat protein-mediated virus resistance is associated with RNA silencing in Arabidopsis. Front. Microbiol. 2020, 11, 591619. [Google Scholar] [CrossRef]
- Otuka, A. Migration of rice planthoppers and their vectored reemerging and novel rice viruses in East Asia. Front. Microbiol. 2013, 4, 309. [Google Scholar] [CrossRef]
- Tan, Q.W.; Ni, J.C.; Zheng, L.P.; Fang, P.H.; Shi, J.T.; Chen, Q.J. Anti-Tobacco mosaic virus quassinoids from Ailanthus altissima (Mill.) Swingle. J. Agric. Food Chem. 2018, 66, 7347–7357. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Y.; He, J.; Zheng, X.; Hu, J.; Liu, Y.; Dai, H.; Zhang, Y.; Wang, B.; Wu, W.; et al. Stv11 encodes a sulphotransferase and confers durable resistance to Rice stripe virus. Nat. Commun. 2014, 5, 4768. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, X.; Wang, S.; Yie, Y.; Tien, P. Infection and replication of a planthopper transmitted virus-Rice stripe virus in rice protoplasts. J. Virol. Methods 1996, 59, 57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011, 30, 30. [Google Scholar] [CrossRef]
- Yamaji, Y.; Maejima, K.; Ozeki, J.; Komatsu, K.; Shiraishi, T.; Okano, Y.; Himeno, M.; Sugawara, K.; Neriya, Y.; Minato, N.; et al. Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell 2012, 24, 778–793. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, C.; Fu, S.; Liu, Y.; Xu, Y.; Wu, J.; Wang, Y.; Zhou, X. NSvc4 encoded by Rice stripe virus targets host chloroplasts to suppress chloroplast-mediated defense. Viruses 2022, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Wu, J.; Lu, L.; Xu, Y.; Zhou, X. Interaction between Rice stripe virus disease-specific protein and host PsbP enhance virus symptoms. Mol. Plant. 2014, 7, 691–708. [Google Scholar] [CrossRef]
- Fiaschetti, G.; Grotzer, M.A.; Shalaby, T.; Castelletti, D.; Arcaro, A. Quassinoids: From traditional drugs to new cancer therapeutics. Curr. Med. Chem. 2011, 18, 316–328. [Google Scholar] [CrossRef]
- Guo, Z.; Vangapandu, S.; Sindelar, R.W.; Walker, L.A.; Sindelar, R.D. Biologically active quassinoids and their chemistry: Potential leads for drug design. Curr. Med. Chem. 2005, 12, 173–190. [Google Scholar] [CrossRef]
- Hoüel, E.; Stien, D.; Bourdy, G.; Deharo, E. Quassinoids: Anticancer and antimalarial activities. In Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes.; Ramawat, K.G., Mérillon, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 3775–3802. [Google Scholar]
- Tan, Q.W.; Ni, J.C.; Shi, J.T.; Zhu, J.X.; Chen, Q.J. Two novel quassinoid glycosides with antiviral activity from the samara of Ailanthus altissima. Molecules 2020, 25, 5679. [Google Scholar] [CrossRef]
- Ni, J.C.; Shi, J.C.; Tan, Q.W.; Chen, Q.J. Two new compounds from the samara of Ailanthus altissima. Nat. Prod. Res. 2019, 33, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.C.; Shi, J.T.; Tan, Q.W.; Chen, Q.J. Phenylpropionamides, piperidine, and phenolic derivatives from the samara of Ailanthus altissima. Molecules 2017, 22, 2107. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.G.; Dang, Y.G.; Li, G.Y.; Guo, L.J.; Wang, W.T.; Tan, Q.W.; Lin, Q.Y.; Wu, Z.J.; Xie, L.H. Note: Anti-viral activity of Ailanthus altissima crude extract on Rice stripe virus in rice suspension cells. Phytoparasitica 2008, 36, 405–408. [Google Scholar] [CrossRef]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Fan, Y.; Cheng, Z.; Zhou, Y. Studies on the methodology for resistance identification of rice cultivar to the rice stripe disease. Plant Prot. 2008, 34, 77–80. [Google Scholar]
- Zhao, W.; Wang, Q.; Xu, Z.; Liu, R.; Cui, F. Immune responses induced by different genotypes of the disease-specific protein of Rice stripe virus in the vector insect. Virology 2019, 527, 122–131. [Google Scholar] [CrossRef]
- Zhao, W.; Xu, Z.; Zhang, X.; Yang, M.; Kang, L.; Liu, R.; Cui, F. Genomic variations in the 3′-termini of Rice stripe virus in the rotation between vector insect and host plant. New Phytol. 2018, 219, 1085–1096. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L.; He, Y.; Qin, Q.; Chen, C.; Wei, Z.; Tan, X.; Xie, K.; Zhang, R.; Hong, G. Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc. Natl. Acad. Sci. USA 2020, 117, 9112–9121. [Google Scholar] [CrossRef]
- Dai, J.M.; Mi, Q.L.; Li, X.M.; Gang, D.; Yang, G.Y.; Zhang, J.D.; Wang, J.; Li, Y.K.; Yang, H.Y.; Miao, D.; et al. The anti-TMV potency of the tobacco-derived fungus Aspergillus versicolor and its active alkaloids, as anti-TMV activity inhibitors. Phytochemistry 2023, 205, 113485. [Google Scholar] [CrossRef]
- Yan, Y.; Tang, P.; Zhang, X.; Wang, D.; Peng, M.; Yan, X.; Hu, Z.; Tang, L.; Hao, X. Anti-TMV effects of seco-pregnane C21 steroidal glycosides isolated from the roots of Cynanchum paniculatum. Fitoterapia 2022, 161, 105225. [Google Scholar] [CrossRef]
- Xu, W.T.; Tian, H.; Song, H.; Liu, Y.; Li, Y.; Wang, Q. Route development, antiviral studies, field evaluation and toxicity of an antiviral plant protectant NK0238. Front. Agric. Sci. Eng. 2022, 9, 110–119. [Google Scholar] [CrossRef]
- Li, Z.; Yang, B.; Ding, Y.; Zhou, X.; Fang, Z.; Liu, S.S.; Yang, J.; Yang, S. Discovery of phosphonate derivatives containing different substituted 1,2,3-triazole motif as promising tobacco mosaic virus (TMV) helicase inhibitors for controlling plant viral diseases. Pest Manag. Sci. 2023, 79, 3979–3992. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, R.; Hao, Y. Discovery of aldisine and its derivatives as novel antiviral, larvicidal, and antiphytopathogenic-fungus agents. J. Agric. Food Chem. 2022, 70, 12355–12363. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Wang, Z.; Liu, J.; Lv, M.; Liu, J.; Li, X.; Chen, Z.; Jin, L.; Hu, D.; Yang, S.; et al. Screening anti-southern rice black-streaked dwarf virus drugs based on S7-1 gene expression in rice suspension cells. J. Agric. Food Chem. 2013, 61, 8049–8055. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zha, Q.; Zhang, W.; Wu, G.; Yan, F.; Wu, Z.; Du, Z. Cap-snatching inhibitors of influenza virus are inhibitory to the in vitro transcription of Rice stripe virus. Phytopath. Res. 2022, 4, 36. [Google Scholar] [CrossRef]
- Yao, M.; Zhang, T.; Zhou, T.; Zhou, Y.; Zhou, X.; Tao, X. Repetitive prime-and-realign mechanism converts short capped RNA leaders into longer ones that may be more suitable for elongation during Rice stripe virus transcription initiation. J. Gen. Virol. 2012, 93, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic. Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef]
- Wu, G.; Zheng, G.; Hu, Q.; Ma, M.; Li, M.; Sun, X.; Yan, F.; Qing, L. NS3 Protein from Rice stripe virus affects the expression of endogenous genes in Nicotiana benthamiana. Virol. J. 2018, 15, 105. [Google Scholar] [CrossRef]
- Wu, W.; Zheng, L.; Chen, H.; Jia, D.; Li, F.; Wei, T. Nonstructural protein NS4 of Rice stripe virus plays a critical role in viral spread in the body of vector insects. PLoS ONE 2014, 9, e88636. [Google Scholar] [CrossRef]
- Xiong, R.Y.; Wu, J.X.; Zhou, Y.J.; Zhou, X.P. Identification of a movement protein of the Tenuivirus rice virus. J. Virol. 2008, 82, 12304–12311. [Google Scholar] [CrossRef]




| Gene | Forward Primer 5′−3′ | Reverse Primer 5′−3′ | Length (bp) |
|---|---|---|---|
| NSvc3 (CP) | GCAGGCTATGATGCTGCAAC | TTGTCAGACCACGCTCCTTC | 510 |
| Actin (N. benthamiana) | ACTGATGAAGATACTCACAGA | TGGAATTGTATGTGGTTTCAT | 236 |
| Gene | Forward Primer 5′−3′ | Reverse Primer 5′−3′ | Length (bp) |
|---|---|---|---|
| RdRp | CAGTGAAGTGGAGGCTGCT | AGCAAGTCTGTGGCTCTCAA | 166 |
| NS2 | TGGGATGCTGTGAGGAGTTCA | GGATCAGTTTCAGATGCTCAGT | 174 |
| NSvc2 | GAGAGGGATGGAGTGGACAT | TGAGGTCCCATTGAGGGATA | 180 |
| NS3 | TTCACATCGTCTGTGGGTTC | TGGAAGGGTGCCTAGATGAATG | 163 |
| NSvc3 (CP) | TGCAGAAGGCAATCAATGACAT | TGTCACCACCTTTGTCCTCAA | 150 |
| NS4 (SP) | CCTGTTAGGAGGTGAAGATGATGA | GCTCTCAGCCTTAGCCATCTTG | 180 |
| NSvc4 (MP) | TGAAGGCCCATAGGAAAGCA | TGCGGAGGGTAGTTATTCCAC | 153 |
| 18S rRNA (Nipponbare) | ATGATAACTCGACGGATCGC | CTTGGATGTGGTAGCCGTTT | 169 |
| Actin (N. benthamiana) | TGTGCTCAGTGGTGGCTCTA | GGTGCTGAGAGAAGCCAAGATA | 163 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Q.; Zhu, J.; Ju, Y.; Chi, X.; Cao, T.; Zheng, L.; Chen, Q. Antiviral Activity of Ailanthone from Ailanthus altissima on the Rice Stripe Virus. Viruses 2024, 16, 73. https://doi.org/10.3390/v16010073
Tan Q, Zhu J, Ju Y, Chi X, Cao T, Zheng L, Chen Q. Antiviral Activity of Ailanthone from Ailanthus altissima on the Rice Stripe Virus. Viruses. 2024; 16(1):73. https://doi.org/10.3390/v16010073
Chicago/Turabian StyleTan, Qingwei, Jianxuan Zhu, Yuanyuan Ju, Xinlin Chi, Tangdan Cao, Luping Zheng, and Qijian Chen. 2024. "Antiviral Activity of Ailanthone from Ailanthus altissima on the Rice Stripe Virus" Viruses 16, no. 1: 73. https://doi.org/10.3390/v16010073
APA StyleTan, Q., Zhu, J., Ju, Y., Chi, X., Cao, T., Zheng, L., & Chen, Q. (2024). Antiviral Activity of Ailanthone from Ailanthus altissima on the Rice Stripe Virus. Viruses, 16(1), 73. https://doi.org/10.3390/v16010073





