Development of Stable Infectious cDNA Clones of Tomato Black Ring Virus Tagged with Green Fluorescent Protein
Abstract
1. Introduction
2. Materials and Methods
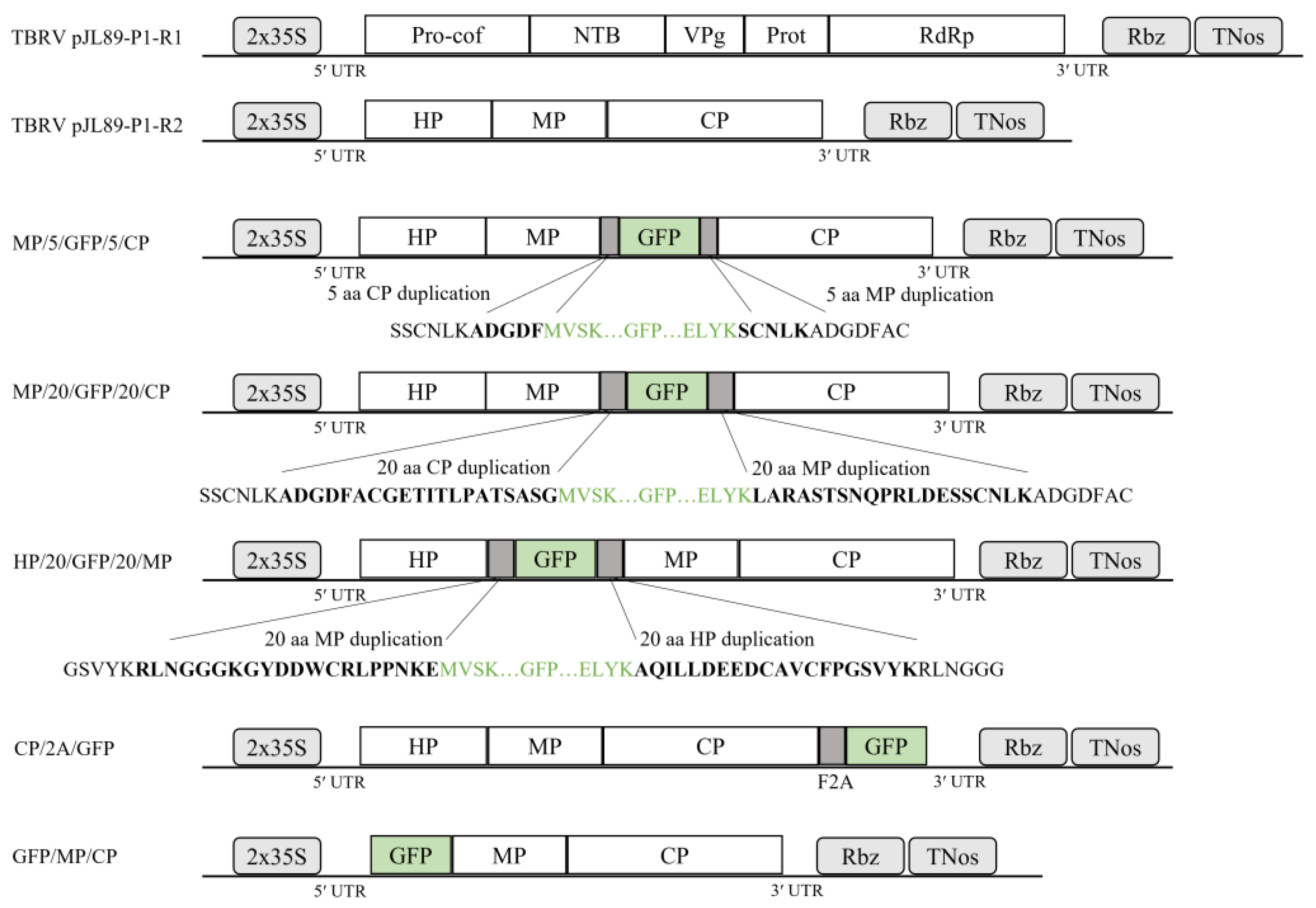
2.1. Generation of GFP-Tagged Infectious TBRV Clones Utilizing the In-Fusion Cloning Method
2.2. Agrobacterium Infiltration
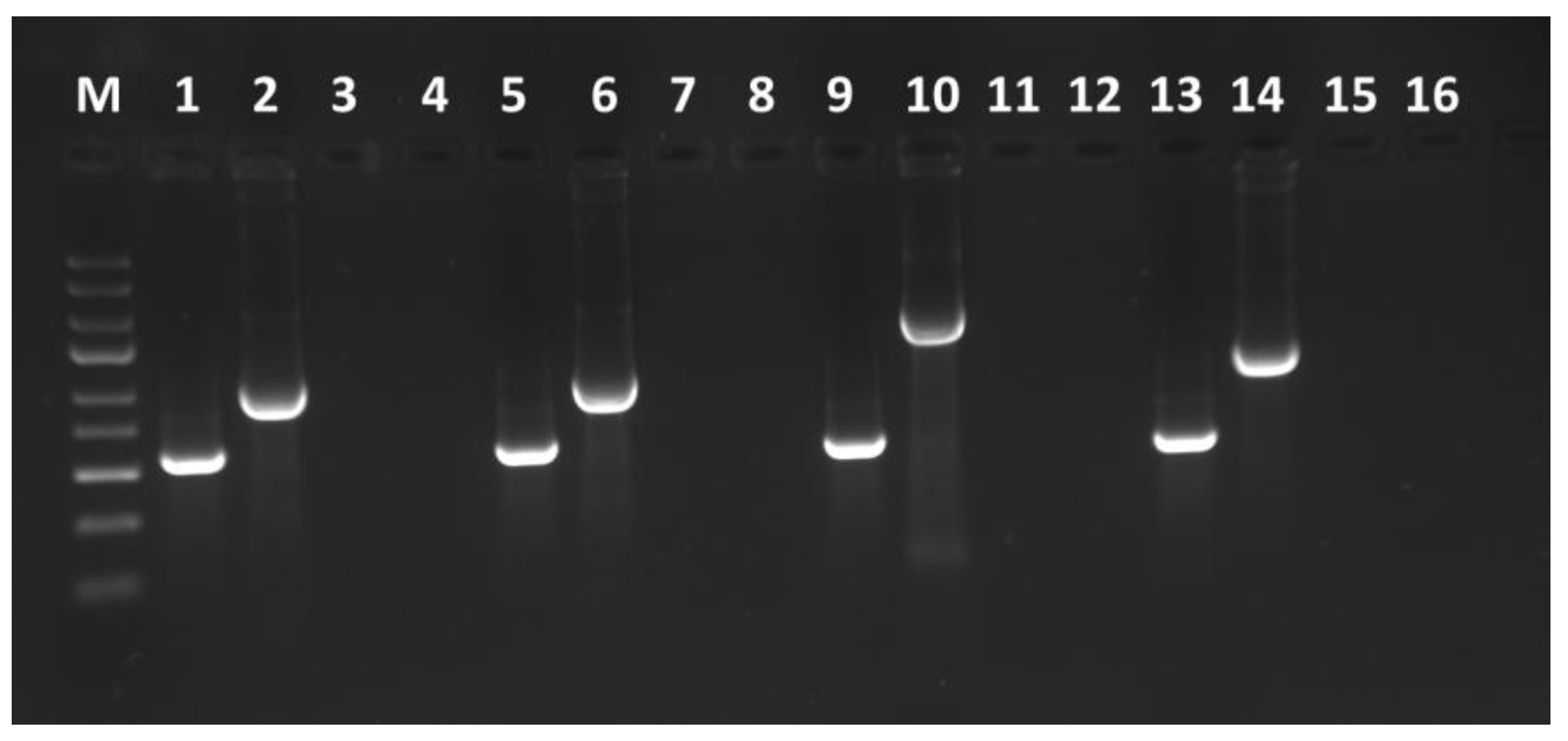
2.3. TBRV and GFP Coding Sequence Detection
2.4. Confirmation of Biological Activity, Stability, and Infectibility of Constructed Clones
2.5. GFP Expression Observation
3. Results
3.1. Construction of TBRV pJL89-P1-R2-GFP Infectious Clones
3.2. Biological Activity of TBRV pJL89-P1-R2-GFP Clones in Various Hosts
3.3. Systemic Expression of the Gfp Gene in Tested Plant Hosts
3.4. Stability Confirmation of Constructed pJL89-TBRV-P1-R2-GFP Clones
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahlquist, P.; Dasgupta, R.; Kaesberg, P. Nucleotide Sequence of the Brome Mosaic Virus Genome and Its Implications for Viral Replication. J. Mol. Biol. 1984, 172, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Chen, S.; Wang, J. Construction of Stable Infectious Full-Length and eGFP-Tagged cDNA Clones of Mirabilis Crinkle Mosaic Virus via In-Fusion Cloning. Virus Res. 2020, 286, 198039. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.-J.; Tian, Y.-P.; Geng, C.; Guo, Y.; Jia, M.-A.; Li, X.-D. Development and Application of a Full-Length Infectious Clone of Potato Virus Y Isolate Belonging to SYR-I Strain. Virus Res. 2020, 276, 197827. [Google Scholar] [CrossRef] [PubMed]
- Masuta, C.; Yamana, T.; Tacahashi, Y.; Uyeda, I.; Sato, M.; Ueda, S.; Matsumura, T. Development of Clover Yellow Vein Virus as an Efficient, Stable Gene-Expression System for Legume Species. Plant J. 2000, 23, 539–546. [Google Scholar] [CrossRef]
- Yin, J.; Hong, X.; Luo, S.; Tan, J.; Zhang, Y.; Qiu, Y.; Latif, M.F.; Gao, T.; Yu, H.; Bai, J.; et al. The Characterization of the Tobacco-Derived Wild Tomato Mosaic Virus by Employing Its Infectious DNA Clone. Biology 2022, 11, 1467. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Xiao, C.; Han, K.; Peng, J.; Lin, L.; Lu, Y.; Xie, L.; Wu, X.; Xu, P.; Li, G.; et al. Development of an Agroinoculation System for Full-Length and GFP-Tagged cDNA Clones of Cucumber Green Mottle Mosaic Virus. Arch. Virol. 2015, 160, 2867–2872. [Google Scholar] [CrossRef] [PubMed]
- Casper, S.J.; Holt, C.A. Expression of the Green Fluorescent Protein-Encoding Gene from a Tobacco Mosaic Virus-Based Vector. Gene 1996, 173, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Song, Y.S.; Ryu, K.H. Development of Infectious Transcripts from Full-Length and GFP-Tagged cDNA Clones of Pepper Mottle Virus and Stable Systemic Expression of GFP in Tobacco and Pepper. Virus Res. 2011, 155, 487–494. [Google Scholar] [CrossRef]
- MacFarlane, S.A.; Popovich, A.H. Efficient Expression of Foreign Proteins in Roots from Tobravirus Vectors. Virology 2000, 267, 29–35. [Google Scholar] [CrossRef]
- Sadowy, E.; Pluta, K.; Gronenborn, B.; Hulanicka, D. Infectious Transcripts from Cloned cDNA of Potato Leafroll Luteovirus. Acta Biochim. Pol. 1998, 45, 611–619. [Google Scholar] [CrossRef]
- Franco-Lara, L.F.; McGeachy, K.D.; Commandeur, U.; Martin, R.R.; Mayo, M.A.; Barker, H. Transformation of Tobacco and Potato with cDNA Encoding the Full-Length Genome of Potato Leafroll Virus: Evidence for a Novel Virus Distribution and Host Effects on Virus Multiplication. J. Gen. Virol. 1999, 80, 2813–2822. [Google Scholar] [CrossRef] [PubMed]
- Leiser, R.M.; Ziegler-Graff, V.; Reutenauer, A.; Herrbach, E.; Lemaire, O.; Guilley, H.; Richards, K.; Jonard, G. Agroinfection as an Alternative to Insects for Infecting Plants with Beet Western Yellows Luteovirus. Proc. Natl. Acad. Sci. USA 1992, 89, 9136–9140. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, P.; Budziszewska, M.; Frąckowiak, P.; Obrępalska-Stęplowska, A. Development of a New Tomato Torrado Virus-Based Vector Tagged with GFP for Monitoring Virus Movement in Plants. Viruses 2020, 12, 1195. [Google Scholar] [CrossRef] [PubMed]
- Ferriol, I.; Turina, M.; Zamora-Macorra, E.J.; Falk, B.W. RNA1-Independent Replication and GFP Expression from Tomato Marchitez Virus Isolate M Cloned cDNA. Phytopathology 2016, 106, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Zarzyńska-Nowak, A.; Ferriol, I.; Falk, B.W.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B. Construction of Agrobacterium Tumefaciens-Mediated Tomato Black Ring Virus Infectious cDNA Clones. Virus Res. 2017, 230, 59–62. [Google Scholar] [CrossRef]
- Kumar, M.; Zarreen, F.; Chakraborty, S. Roles of Two Distinct Alphasatellites Modulating Geminivirus Pathogenesis. Virol. J. 2021, 18, 249. [Google Scholar] [CrossRef]
- Stevens, M.; Viganó, F. Production of a Full-Length Infectious GFP-Tagged cDNA Clone of Beet Mild Yellowing Virus for the Study of Plant–Polerovirus Interactions. Virus Genes 2007, 34, 215–221. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Zarzyńska-Nowak, A. Tomato Black Ring Virus (Ring Spot of Beet). CABI Compend. 2022, 54060. [Google Scholar] [CrossRef]
- Pospieszny, H.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B.; Czerwonka, B.; Elena, S.F. An Assessment of the Transmission Rate of Tomato Black Ring Virus through Tomato Seeds. Plant Prot. Sci. 2019, 56, 9–12. [Google Scholar] [CrossRef]
- Tomato Black Ring Virus (TBRV00) [World Distribution]|EPPO Global Database. Available online: https://gd.eppo.int/taxon/TBRV00/distribution (accessed on 1 March 2023).
- Dewasme, C.; Mary, S.; Darrieutort, G.; Audeguin, L.; van Helden, M.; van Leeuwen, C. Impact of Tomato Black Ring Virus (TBRV) on Quantitative and Qualitative Features of Vitis vinifera L. Cv. Merlot and Cabernet Franc. OENO One 2019, 53, 161–169. [Google Scholar] [CrossRef]
- Gallitelli, D.; Rana, G.L.; Vovlas, C.; Martelli, G.P. Viruses of Globe Artichoke: An Overview. J. Plant Pathol. 2004, 86, 267–281. [Google Scholar]
- Dąbrowska, E.; Paduch-Cichal, E.; Piasna, P.; Malewski, T.; Mirzwa-Mróz, E. First Report of Tomato Black Ring Virus Infecting Raspberry and Blackberry in Poland. Plant Dis. 2021, 105, 3310. [Google Scholar] [CrossRef] [PubMed]
- Al-Shahwan, I.; Al-Shudifat, A.; Al-Saleh, M.; Abdalla, O.; Amer, M. First Report of Tomato Black Ring Virus on Tomato (Solanum lycopersicum) in Saudi Arabia. Plant Dis. 2021, 105, 1231. [Google Scholar] [CrossRef]
- Oncino, C.; Hemmer, O.; Fritsch, C. Specificity in the Association of Tomato Black Ring Virus Satellite RNA with Helper Virus. Virology 1995, 213, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Jończyk, M.; Le Gall, O.; Pałucha, A.; Borodynko, N.; Pospieszny, H. Cloning and Sequencing of Full-Length cDNAs of RNA1 and RNA2 of a Tomato Black Ring Virus Isolate from Poland. Arch. Virol. 2004, 149, 799–807. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Minicka, J.; Zarzyńska-Nowak, A.; Budzyńska, D.; Elena, S.F. Defective RNA Particles Derived from Tomato Black Ring Virus Genome Interfere with the Replication of Parental Virus. Virus Res. 2018, 250, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Gnanasekaran, P.; Chakraborty, S. Biology of Viral Satellites and Their Role in Pathogenesis. Curr. Opin. Virol. 2018, 33, 96–105. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N.; Figlerowicz, M.; Pospieszny, H. Two Types of Defective RNAs Arising from the Tomato Black Ring Virus Genome. Arch. Virol. 2012, 157, 569–572. [Google Scholar] [CrossRef]
- Budzyńska, D.; Minicka, J.; Hasiów-Jaroszewska, B.; Elena, S.F. Molecular Evolution of Tomato Black Ring Virus and de Novo Generation of a New Type of Defective RNAs during Long-Term Passaging in Different Hosts. Plant Pathol. 2020, 69, 1767–1776. [Google Scholar] [CrossRef]
- Minicka, J.; Taberska, A.; Zarzyńska-Nowak, A.; Kubska, K.; Budzyńska, D.; Elena, S.F.; Hasiów-Jaroszewska, B. Genetic Diversity of Tomato Black Ring Virus Satellite RNAs and Their Impact on Virus Replication. Int. J. Mol. Sci. 2022, 23, 9393. [Google Scholar] [CrossRef]
- Pelczyk, M.; Obrępalska-Stęplowska, A.; Pospieszny, H. Subviral Molecules of RNA Associated with Plant Ss(+)RNA Viruses. Postep. Biochem. 2006, 52, 212–221. [Google Scholar]
- Hemmer, O.; Oncino, C.; Fritsch, C. Efficient Replication of the in Vitro Transcripts from Cloned cDNA of Tomato Black Ring Virus Satellite RNA Requires the 48K Satellite RNA-Encoded Protein. Virology 1993, 194, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Zarzyńska-Nowak, A.; Hasiów-Jaroszewska, B.; Budzyńska, D.; Trzmiel, K. Genetic Variability of Polish Tomato Black Ring Virus Isolates and Their Satellite RNAs. Plant Pathol. 2020, 69, 1034–1041. [Google Scholar] [CrossRef]
- Zarzyńska-Nowak, A.; Jeżewska, M.; Hasiów-Jaroszewska, B.; Zielińska, L. A Comparison of Ultrastructural Changes of Barley Cells Infected with Mild and Aggressive Isolates of Barley Stripe Mosaic Virus. J. Plant Dis. Prot. 2015, 122, 153–160. [Google Scholar] [CrossRef]
- Brewer, H.C.; Hird, D.L.; Bailey, A.M.; Seal, S.E.; Foster, G.D. A Guide to the Contained Use of Plant Virus Infectious Clones. Plant Biotechnol. J. 2018, 16, 832–843. [Google Scholar] [CrossRef]
- Oparka, K.J.; Boevink, P.; Santa Cruz, S. Studying the Movement of Plant Viruses Using Green Fluorescent Protein. Trends Plant Sci. 1996, 1, 412–418. [Google Scholar] [CrossRef]
- Zhou, X.; Lin, W.; Sun, K.; Wang, S.; Zhou, X.; Jackson, A.O.; Li, Z. Specificity of Plant Rhabdovirus Cell-to-Cell Movement. J. Virol. 2019, 93, e00296-19. [Google Scholar] [CrossRef]
- Liu, D.; Pan, L.; Zhai, H.; Qiu, H.-J.; Sun, Y. Virus Tracking Technologies and Their Applications in Viral Life Cycle: Research Advances and Future Perspectives. Front. Immunol. 2023, 14, 1204730. [Google Scholar] [CrossRef]
- Digiaro, M.; Yahyaoui, E.; Martelli, G.P.; Elbeaino, T. The Sequencing of the Complete Genome of a Tomato Black Ring Virus (TBRV) and of the RNA2 of Three Grapevine Chrome Mosaic Virus (GCMV) Isolates from Grapevine Reveals the Possible Recombinant Origin of GCMV. Virus Genes 2015, 50, 165–171. [Google Scholar] [CrossRef]
- Qiao, W.; Falk, B.W. Efficient Protein Expression and Virus-Induced Gene Silencing in Plants Using a Crinivirus-Derived Vector. Viruses 2018, 10, 216. [Google Scholar] [CrossRef]
- Gopinath, K.; Wellink, J.; Porta, C.; Taylor, K.M.; Lomonossoff, G.P.; Van Kammen, A. Engineering Cowpea Mosaic Virus RNA-2 into a Vector to Express Heterologous Proteins in Plants. Virology 2000, 267, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; van Rij, R.; Silvera, D.; Andino, R. Toward a Poliovirus-Based Simian Immunodeficiency Virus Vaccine: Correlation between Genetic Stability and Immunogenicity. J. Virol. 1997, 71, 7841–7850. [Google Scholar] [CrossRef] [PubMed]
- Zarzyńska-Nowak, A.; Minicka, J.; Wieczorek, P.; Hasiów-Jaroszewska, B. Infekcyjne kopie TBRV sprzężone z białkiem zielonej fluorescencji jako narzędzie do analizy przebiegu patogenezy w tkankach roślinnych. In Proceedings of the 62 Sesja Naukowa Instytutu Ochrony Roślin—Państwowego Instytutu Badawczego, Poznań, Poland, 16 February 2022; pp. 49–50. [Google Scholar]
- Zhang, C.; Ghabrial, S.A. Development of Bean Pod Mottle Virus-Based Vectors for Stable Protein Expression and Sequence-Specific Virus-Induced Gene Silencing in Soybean. Virology 2006, 344, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Rapolu, M.; Kumar, S.; Gupta, M.; Liang, Z.; Han, Z.; Williams, P.; Su, W.W. Coordinated Protein Co-Expression in Plants by Harnessing the Synergy between an Intein and a Viral 2A Peptide. Plant Biotechnol. J. 2017, 15, 718–728. [Google Scholar] [CrossRef]
- Sempere, R.N.; Gómez, P.; Truniger, V.; Aranda, M.A. Development of Expression Vectors Based on Pepino Mosaic Virus. Plant Methods 2011, 7, 6. [Google Scholar] [CrossRef]
- Tavares-Esashika, M.L.; Campos, R.N.S.; Maeda, M.H.K.; Koyama, L.H.H.; Hamann, P.R.V.; Noronha, E.F.; Nagata, T. Development of a Heterologous Gene Expression Vector in Plants Based on an Infectious Clone of a Tobravirus, Pepper Ringspot Virus. Ann. Appl. Biol. 2022, 181, 107–116. [Google Scholar] [CrossRef]
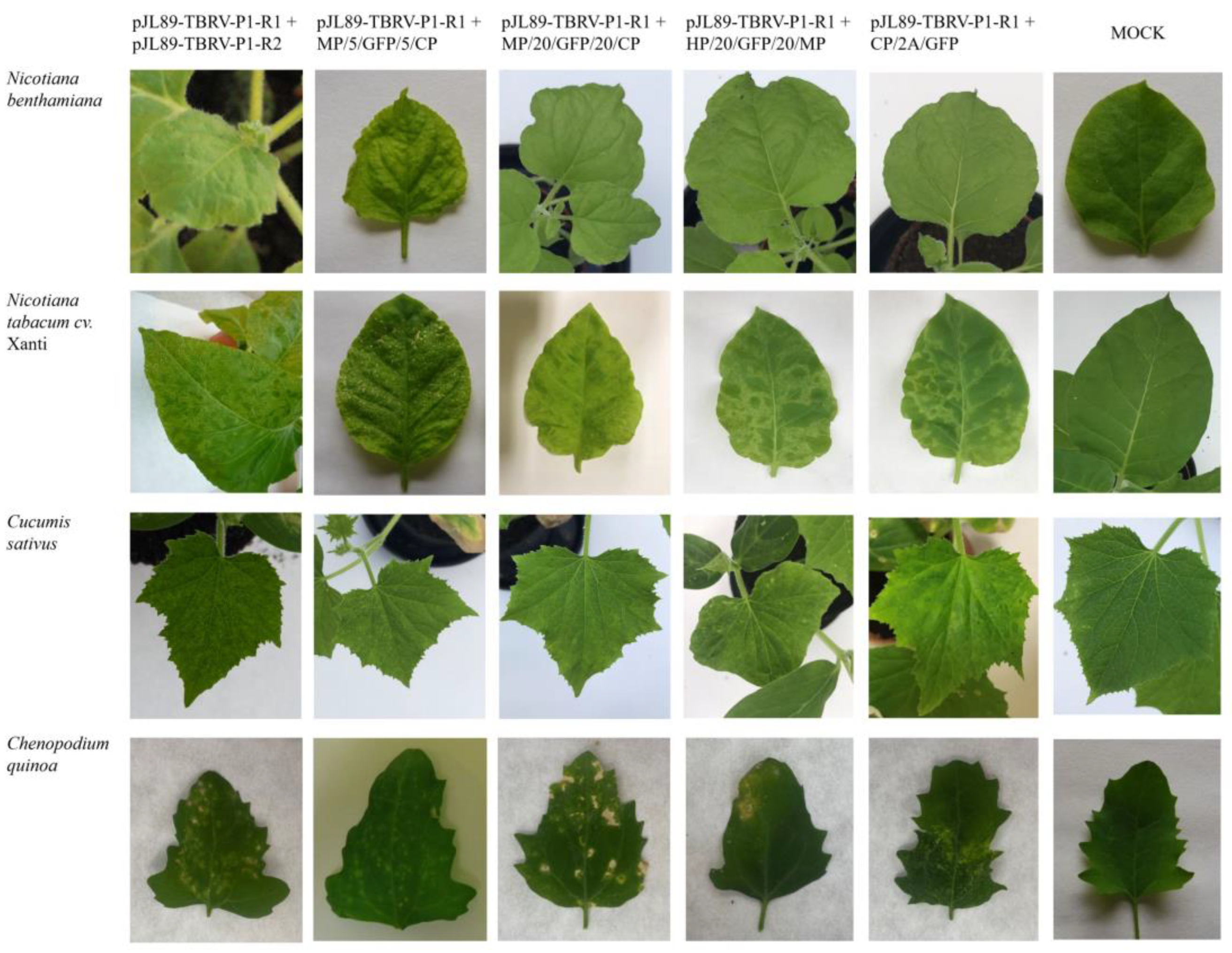


| Host/Clone | MP/5/GFP/5/CP | MP/20/GFP/20/CP | HP/20/GFP/20/MP | CP/2A/GFP |
|---|---|---|---|---|
| N. benthamiana | 1/4 | 4/4 | 3/4 | 4/4 |
| N. tabacum cv. Xanthi | 2/4 | 3/4 | 2/4 | 2/4 |
| C. sativus | 3/4 | 4/4 | 2/4 | 4/4 |
| C. quinoa | 3/4 | 3/4 | 2/4 | 4/4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarzyńska-Nowak, A.; Minicka, J.; Wieczorek, P.; Hasiów-Jaroszewska, B. Development of Stable Infectious cDNA Clones of Tomato Black Ring Virus Tagged with Green Fluorescent Protein. Viruses 2024, 16, 125. https://doi.org/10.3390/v16010125
Zarzyńska-Nowak A, Minicka J, Wieczorek P, Hasiów-Jaroszewska B. Development of Stable Infectious cDNA Clones of Tomato Black Ring Virus Tagged with Green Fluorescent Protein. Viruses. 2024; 16(1):125. https://doi.org/10.3390/v16010125
Chicago/Turabian StyleZarzyńska-Nowak, Aleksandra, Julia Minicka, Przemysław Wieczorek, and Beata Hasiów-Jaroszewska. 2024. "Development of Stable Infectious cDNA Clones of Tomato Black Ring Virus Tagged with Green Fluorescent Protein" Viruses 16, no. 1: 125. https://doi.org/10.3390/v16010125
APA StyleZarzyńska-Nowak, A., Minicka, J., Wieczorek, P., & Hasiów-Jaroszewska, B. (2024). Development of Stable Infectious cDNA Clones of Tomato Black Ring Virus Tagged with Green Fluorescent Protein. Viruses, 16(1), 125. https://doi.org/10.3390/v16010125








