Australian Cool-Season Pulse Seed-Borne Virus Research: 1. Alfalfa and Cucumber Mosaic Viruses and Less Important Viruses
Abstract
1. Introduction
2. Background Information
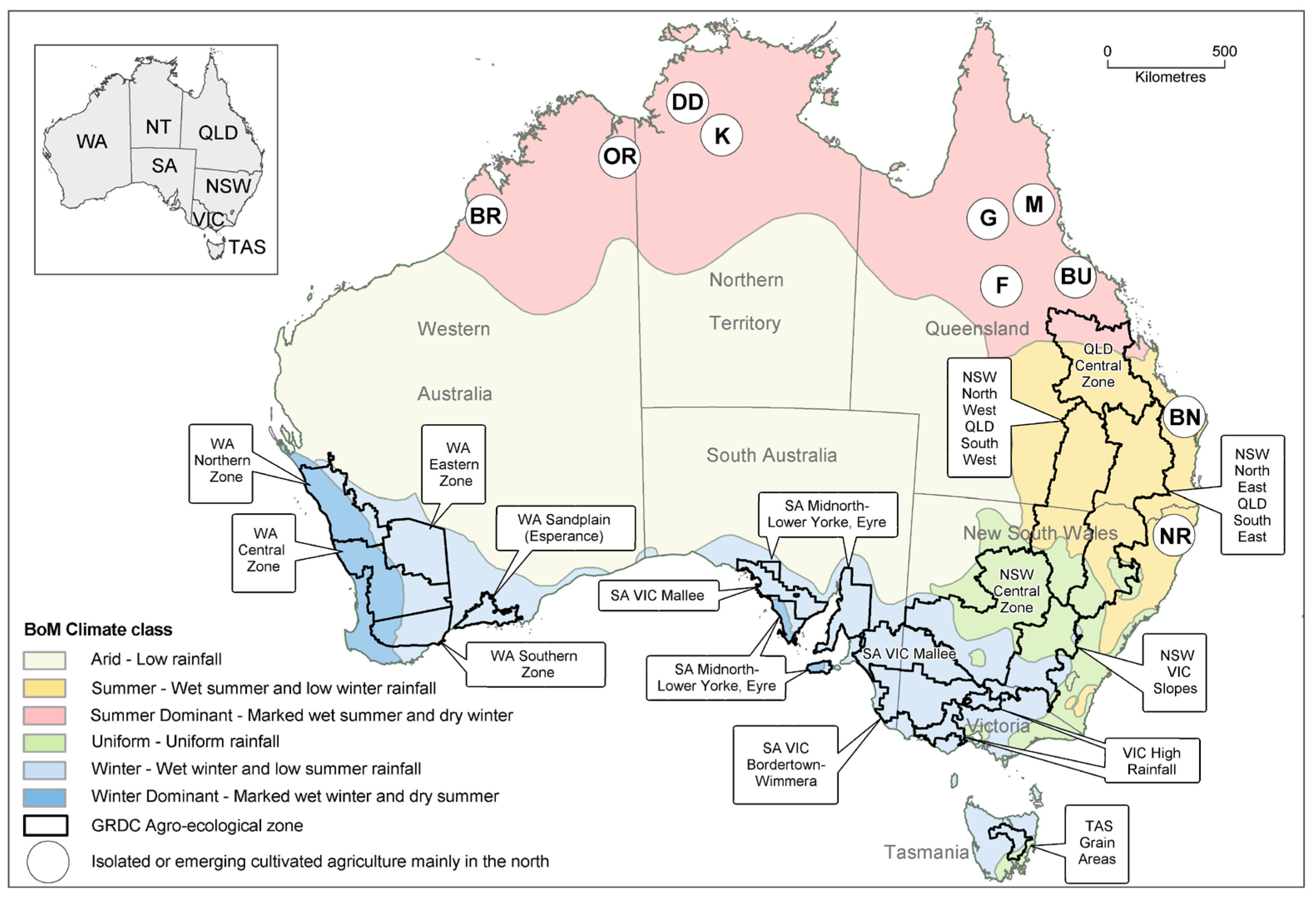
2.1. Australian Pulse Industry
2.2. Epidemiology and Management Principles
2.3. Future Threats to Effective Virus Management
2.4. Recent General Recommendations for Future Research
- As average temperatures increase and growing season rainfall rises or declines becoming more erratic, the climate of Australia’s grainbelt is changing fast. The weather is becoming increasingly unstable, causing a greater level of uncertainty about epidemics of virus diseases that increasingly compromises taking decisions over when control measures are required and, if so, which measures to deploy. Further research is therefore required to get ready for future pulse virus disease epidemics induced by climate change. For the viral pathogens identified as most likely to be affected, this will necessitate research addressing (a) the influence of climate change parameters (extreme weather events, insufficient rainfall, increased temperatures and wind speeds) upon their epidemiology and (b) the escalating difficulties in managing them effectively.
- Further research is required to establish how altering agricultural practices and technologies, and other alterations in prevailing circumstances unrelated to climate change, might influence virus disease epidemics occurring in pulse crops and, in turn, virus disease control strategies in the continent’s grainbelts. This includes (a) updating information on formerly better-studied pulse virus diseases and (b) addressing new virus disease problems of pulses likely to arise due to shifts in viral pathogen prevalence arising from farming system changes, cultivars or new virus strain and vector introductions.
- Ongoing annual surveillance for viruses infecting pulses and their vectors still needs to occur across the Australian grainbelt. This provides an understanding of their economic importance to prioritise research funding, an early warning of their occurrence across different growing seasons and rainfall zones, and the relative performances and robustness of any disease resistances where known.
- The use of historical data and new research to optimise modelling and decision support systems (DSSs) for virus diseases is crucial and needs to be expanded. With the better-researched pulse virus diseases, the principal focus should be on ensuring that all forecasting models and DSSs already available are updated to incorporate shifts in climatic, agronomic and pathosystem drivers since their initial development and delivery of DSSs derived from such models to the national pulse industry. For viral pathosystems without such models, further epidemiology and management research that complements existing findings is needed, leading to the development of improved, locally relevant forecasting models and DSSs and their delivery nationally.
- Molecular approaches to viral resistance in cool-season pulses warrant more support, especially those involving RNAi and CRISPR/Cas approaches.
- There is a clear need to ensure that the research capacity to respond to unforeseen circumstances involving pulse virus pathogens is maintained. This is necessary to ensure the pulse industry avoids being caught out by the absence of necessary expertise to address future unforeseen epidemics of virus diseases seriously affecting pulse crops.
- It is important to understand what biological and genotypic diversity is present within the economically important viruses of the different pulses that occur across Australia. This requires a combination of biological data derived from field and glasshouse experiments with whole genome sequencing of representative virus isolates from pulses followed by their phylogenetic analysis. Although much relevant biological data from Australia is available for AMV and CMV, information about the phylogenetics of a representative spectrum of complete isolate sequences of these two viruses from different pulses around Australia is lacking.
- It is important to keep up to date with new technologies that improve the efficiency of large-scale routine virus testing and diagnosis in pulse leaf and seed samples and the effectiveness of remote sensing technologies for virus disease and virus vector surveillance in the field using thermal, hyperspectral, multispectral and other remote sensing procedures.
2.5. Viruses Infecting Australian Pulse Crops
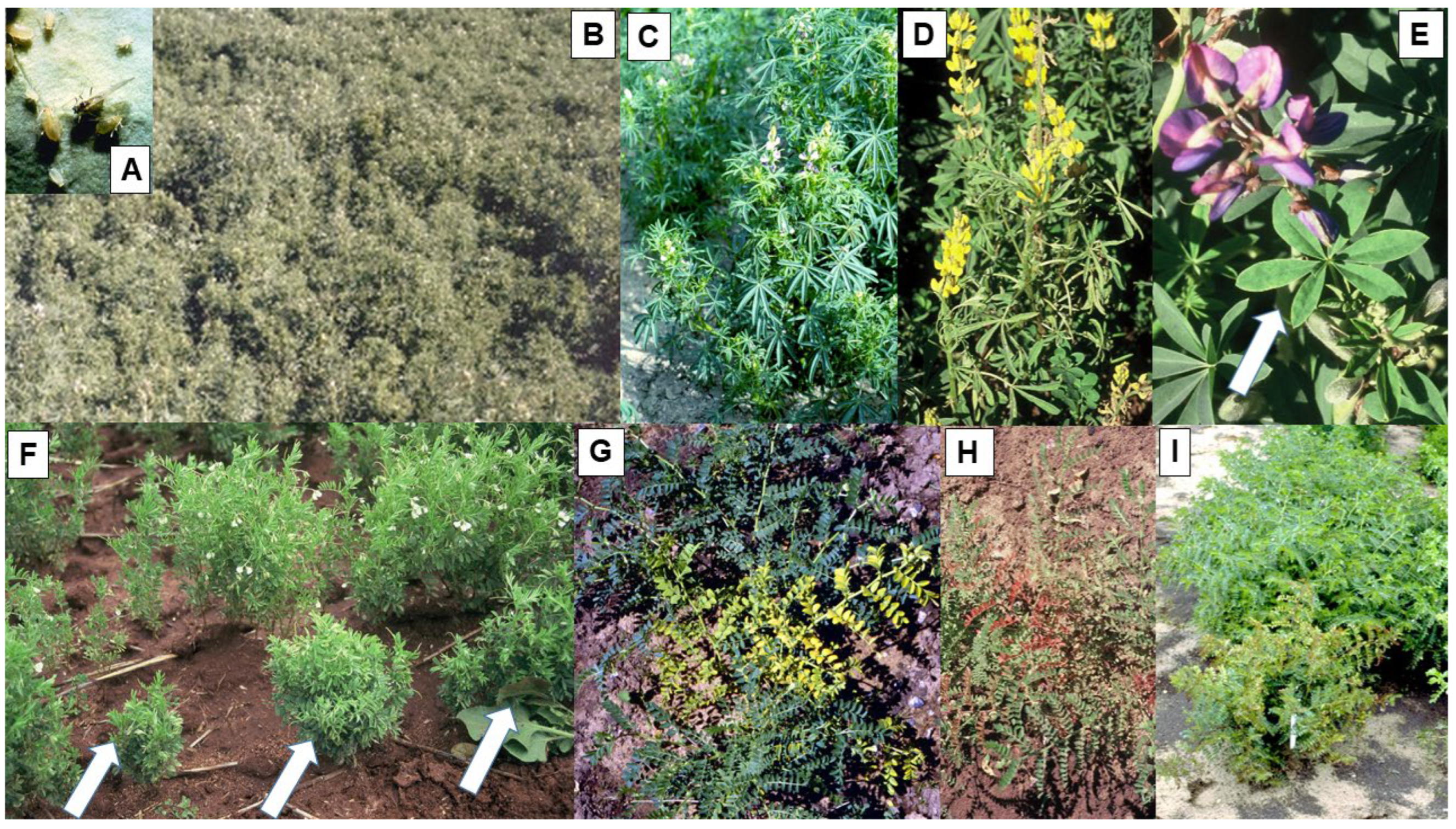
3. Cucumber Mosaic Virus
| Virus | Virus Genus | Pulse Crops Affected (Includes Non-Cool-Season Pulses) | Main Foliage Symptoms | Vector | Seed-Borne | Maximum Yield Loss | Seed Quality Defect | Region Found In a |
|---|---|---|---|---|---|---|---|---|
| Most important viruses | ||||||||
| Alfalfa mosaic virus (AMV) | Alfamovirus | Chickpea, field pea, faba bean, lentil, narrow-leafed lupin, yellow lupin, narbon bean, mung bean, common vetch, Adzuki bean, fenugreek, narbon bean, grass pea, dwarf chickling, purple vetch, Lathyrus ochrus | Mosaic, leaf deformation, dwarfing | Aphid | Yes | 98% | Reduced size | STE, MED, TE |
| Bean yellow mosaic virus (BYMV) | Potyvirus | Chickpea, common bean, faba bean, field pea, lentil, narrow-leafed lupin, white lupin, yellow lupin, sandplain lupin, pearl lupin, common vetch, bitter vetch, narbon bean, grass pea, dwarf chickling, fenugreek | Mosaic, leaf deformation, dwarfing | Aphids | Yes | 99% | Reduced size (necrosis and malformation in faba bean) | STE, MED, TE |
| Cucumber mosaic virus (CMV) | Cucumovirus | Chickpea, faba bean, field pea, lentil, narrow-leafed lupin, yellow lupin, pearl lupin, narbon bean, fenugreek, bitter vetch | Mosaic, chlorosis, leaf deformation, plant dwarfing | Aphids | Yes | 90% | Reduced size | STE, MED, TE |
| Pea seed-borne mosaic virus (PSbMV) | Potyvirus | Chickpea, faba bean, field pea, lentil, common vetch, fenugreek, narbon bean, dwarf chickling, grass pea, bitter vetch, purple vetch, L. clymenum, L. ochrus | Mosaic, mild dwarfing | Aphids | Yes | 96% | Necrotic rings, malformation, cracking, reduced size | STE, MED, TE |
| Less important viruses | ||||||||
| Broad bean stain virus (BBSV) | Comovirus | Faba bean, lentil, field pea, common vetch | Mottle, leaf deformation, necrosis | Beetle | Yes | 61% | Reduced seed size, necrosis, malformation | TE |
| Broad bean true mosaic virus (BBTV) | Comovirus | Faba bean, lentil, field pea, common vetch | Mottle, leaf deformation, necrosis | Beetle | Yes | 30% | Reduced seed size, necrosis, malformation | TE |
| Broadbean wilt virus 2 (BBWV-2) | Fabavirus | Faba bean, chickpea, field pea, narrow-leafed lupin, white lupin, cowpea, common bean | Vein clearing, mottle, leaf deformation, apical necrosis, ringspot, wilting | Aphid | Yes | 26% | Reduced seed size, necrosis, malformation | STE, TE |
| Cowpea mild mottle virus (CpMMV) | Calarvirus | Mungbean, common bean, lima bean, field pea, cowpea | Mosaic, leaf distortion, necrosis, dwarfing, pod distortion | Whitefly | Yes | 100% | Reduced seed size | STE |
| Peanut mottle virus (PMoV) | Potyvirus | Common bean, lima bean, Adzuki bean, cowpea, field pea, narrow-leafed lupin, white lupin | Mottle, necrosis | Aphid | Yes | 70% | Reduced size, malformation, discoloration | STE |
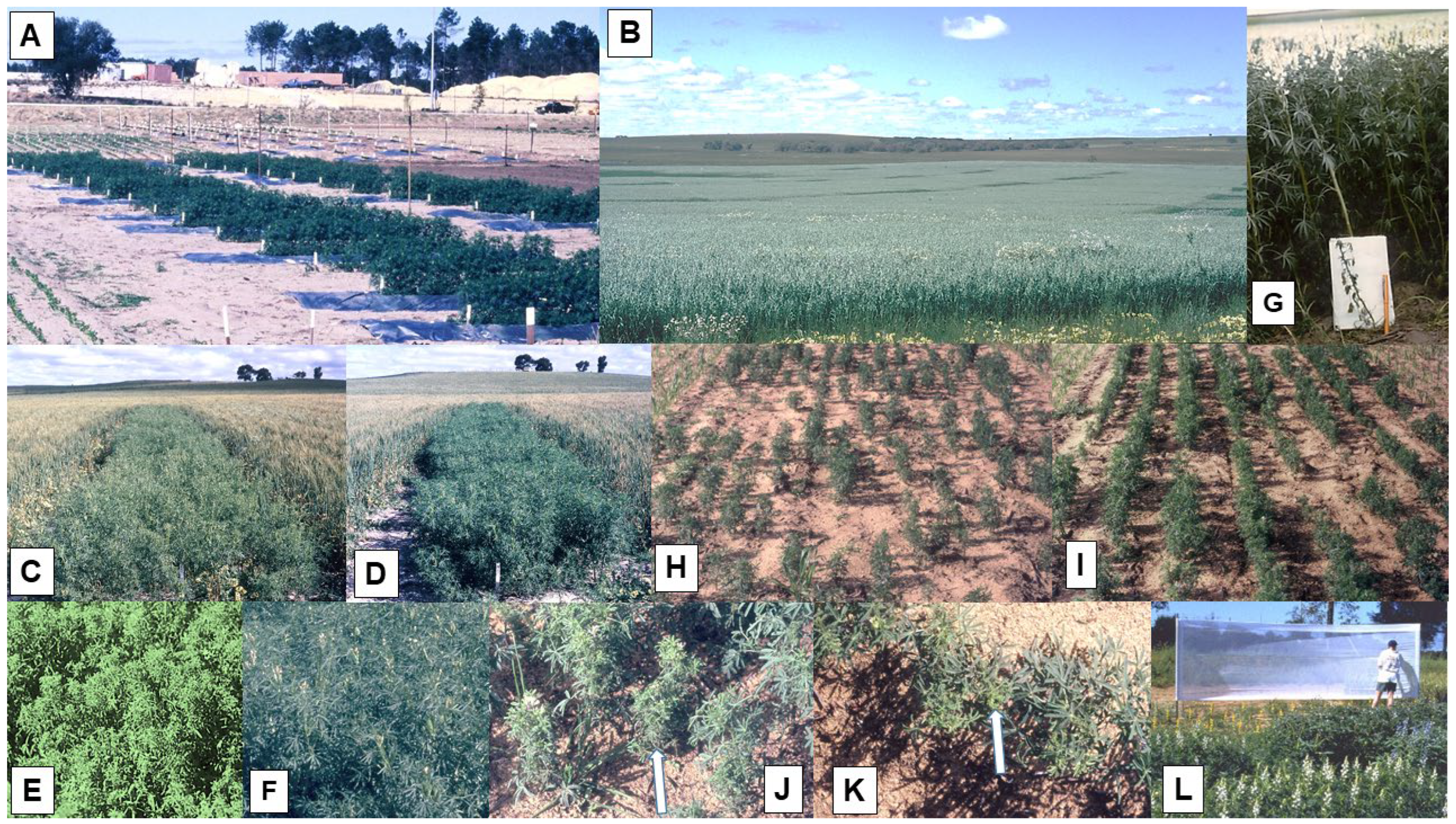
3.1. Lupins
3.1.1. Breeding Program and Commercial Seed Stock Contamination
3.1.2. Alternative Hosts
3.1.3. Cultural and Phytosanitary Control
3.1.4. Chemical Control
3.1.5. Host Resistance
- Deploy speed breeding to expedite the incorporation of CMV resistance into new lupin cultivars.
- Develop molecular markers suitable for use in streamlining the breeding of new narrow-leafed and yellow lupin cultivars with CMV resistance. This needs to include identifying quantitative trait loci (QTLs) for use as molecular markers to speed up breeding for polygenically inherited resistance, e.g., to seed transmission.
- Establish whether CMV infection resistance suitable for use in breeding lupin cultivars with this trait is present in narrow-leafed lupin germplasm.
- Incorporate CMV resistance gene Ncm-1 to provide CMV resistance during breeding yellow lupin cultivars for Australia.
- Employ accession P26956 as a parent to confer extreme CMV resistance when breeding new cultivars of pearl lupin suited to Australian growing conditions.
- Investigate the use of recently developed genetic modification procedures, especially genome editing and RNA silencing, to introduce stable resistance to CMV into new cultivars of yellow, narrow-leafed and pearl lupin.
3.1.6. Patterns of Spread
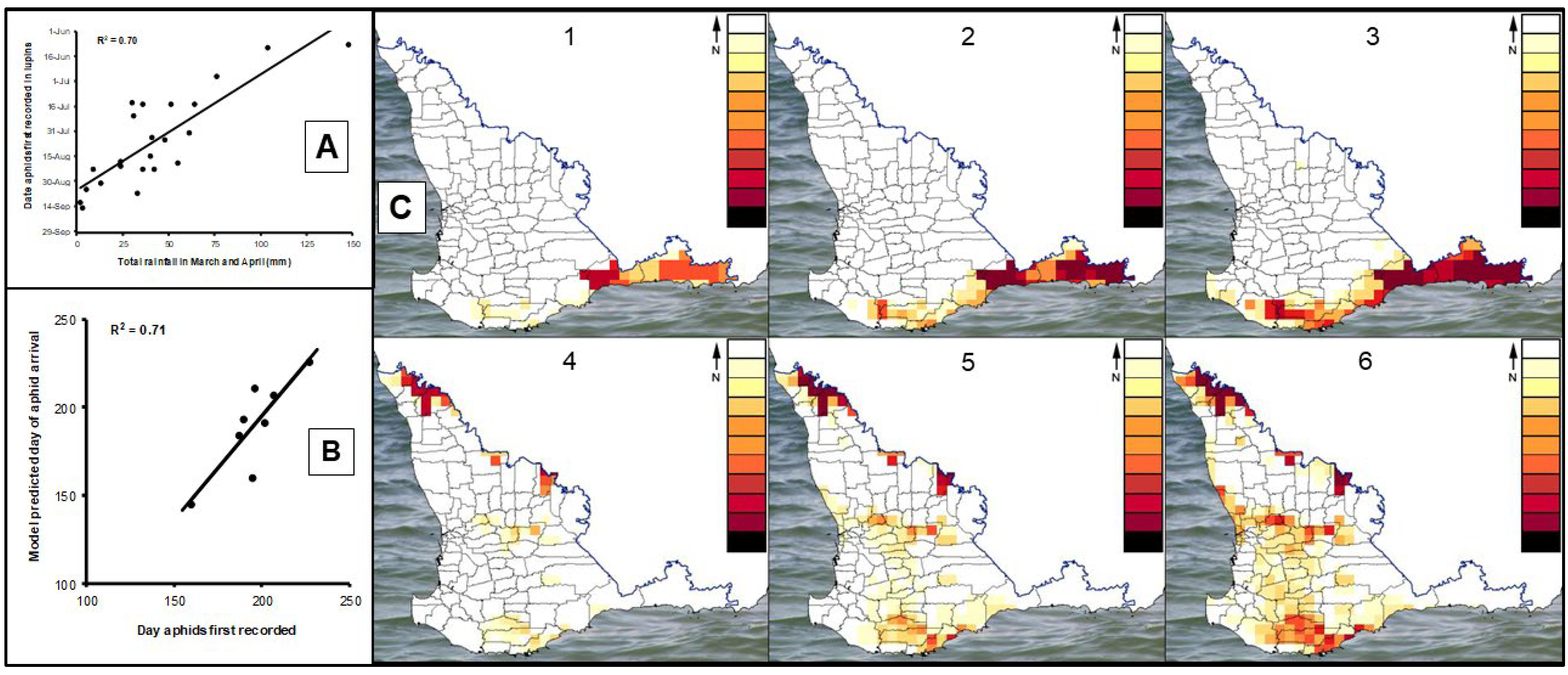
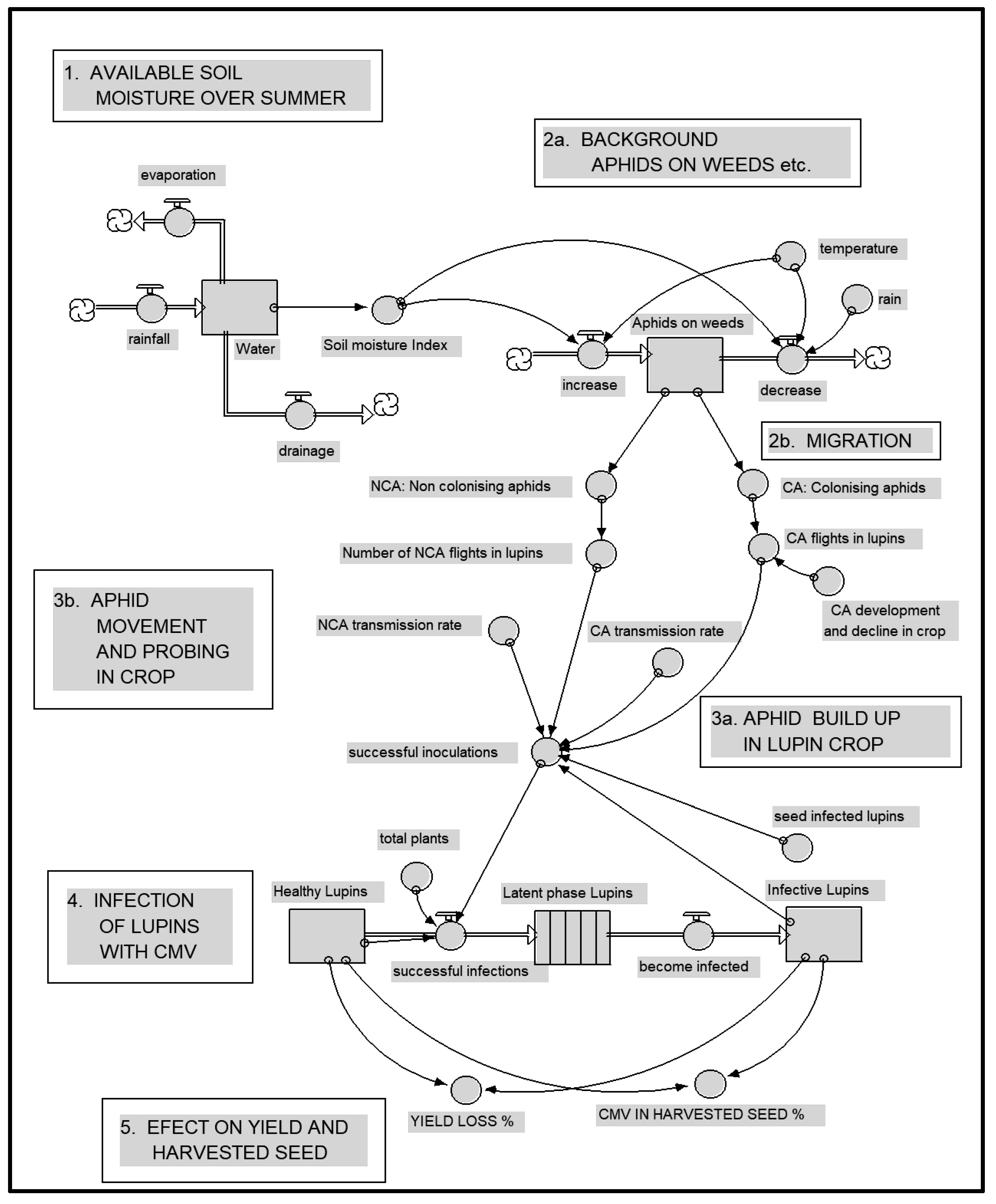
3.1.7. Epidemic Drivers and Forecasting
3.1.8. Integrated Disease Management
- In grainbelt zones at higher risk of losses from CMV infection, minimize the initial internal seed-borne infection source by sowing seed stocks with <0.1% CMV infection sourced from lower-risk zones (phytosanitary).
- In grainbelt zones at lower risk of losses from CMV infection, diminish the initial internal seed-borne infection source by sowing seed stocks with <0.5% CMV infection (phytosanitary).
- Sow cultivars with intrinsic CMV seed transmission rates that are low to help minimise the initial internal seed-borne CMV infection source, especially when retaining harvested seed for sowing in the following growing next season (host resistance).
- Sow seeds at high seeding rates to generate high plant densities and early canopy closure to (i) shade out seed-infected plants and early-current-season-infected plants, thereby minimising the early internal infection source for subsequent CMV spread by aphid vectors, and (ii) diminish aphid landing rates, thereby further diminishing CMV spread (cultural).
- Sow seeds at narrow row spacing to generate early canopy closure, thereby diminishing aphid landing rates and the extent of CMV spread before canopy closure (cultural).
- When sowing untested seeds at wide row spacing in lower CMV risk zones, ensure a high seeding rate is used to produce high plant densities within rows that shade out CMV seed-infected plants, thereby reducing the primary source of inoculum (cultural).
- Sow early maturing cultivars to diminish both late CMV spread by vector aphids and additional harvested seed infection in extended growing seasons (cultural).
- Maximise stubble groundcover using minimum tillage procedures that minimise soil cultivation to diminish aphid vector landing rates, thereby reducing CMV spread prior to canopy closure (cultural).
- Employ crop rotation to avoid volunteer seed-borne lupin infection sources growing within crops (cultural).
- Ensure isolation from neighbouring pulse (including lupin) crops or legume pastures to avoid any ingress of CMV from vector aphids flying from alternative external virus sources (cultural).
- Maximise weed control using selective herbicide to minimise potential weed infection sources of CMV within the crop (chemical).
- Apply insecticides solely to manage direct aphid feeding damage once threshold population numbers are reached (chemical).
- Sieve infected seed stocks before sowing to remove the small seed fraction before sowing, which helps reduce the number of seed-infected plants (phytosanitary).
- Mixed cropping with non-host (e.g., cereal) to diminish CMV spread to lupins gown for hay production (cultural).
- Spray high-value lupin seed crops regularly with a mixture of pyrethroid and neonicotinoid insecticides applied at high application rates to kill or repel incoming vector aphids (chemical). *
- Introduce a healthy seed pipeline by keeping parental and F1-generation plants inside insect-proof glasshouses/screenhouses where any initial sources of CMV infection can be identified and removed (phytosanitary).
- Maintain a healthy seed pipeline by growing F2 and later generations in isolation from other lupins to avoid CMV re-introduction (phytosanitary).
- Discard seed lots found to be CMV-infected by testing representative seed samples from selected breeding lines outside the growing season (phytosanitary).
- Employ rigorous roguing procedures to remove plants with symptoms of seed-borne infection from plots before aphid vectors spread CMV (phytosanitary).
- Destroy CMV-infected plots with herbicide sprays or by their physical removal to avoid its spread to other plots (chemical).
- Deploy reflective mulch to protect single-row plots to decrease aphid landing rates (cultural).
- Sow plots into retained stubble or add straw mulch to decrease aphid landing rates (cultural).
- Achieve early canopy cover by sowing plots at high seeding rates with narrow row spacing to decrease aphid landing rates and shade over plants with seed-borne CMV infection (cultural).
- Sow a non-host crop perimeter around plots to act as virus ‘cleansing barriers’ against CMV ingress from external sources (cultural).
- Spray plots regularly with a mixture of pyrethroid and neonicotinoid insecticides applied at high application rates to kill or repel incoming aphids (chemical). *
- Apply selective herbicides to remove potential CMV alternative hosts from between plots (chemical).
3.2. Pulses Other Than Lupins
3.2.1. Occurrence in Plots, Crops and Seed Stocks
3.2.2. Seed Yield Losses and Patterns of Spread
3.2.3. Host Resistance
3.2.4. Further Research
4. Alfalfa Mosaic Virus
4.1. Pulses Other Than Lupins
4.1.1. Occurrence in Plots, Crops and Seed Stocks
4.1.2. Seed Yield Losses
4.1.3. Host Resistance
4.1.4. Alternative Hosts
4.1.5. Further Research
4.2. Lupins
4.2.1. Research Findings
4.2.2. Further Research
5. Less Important Pulse Viruses
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thresh, J.M. The origins and epidemiology of some important plant virus diseases. Appl. Biol. 1980, 5, 1–65. [Google Scholar]
- Thresh, J.M. Cropping practices and virus spread. Annu. Rev. Phytopathol. 1982, 20, 193–218. [Google Scholar] [CrossRef]
- Thresh, J.M. Crop Viruses and Virus Diseases: A Global Perspective. In Virus Diseases and Crop Biosecurity; NATO Security through Science Series; Cooper, I., Kühne, T., Polishchuk, V.P., Eds.; Springer: Dordecht, The Netherlands, 2006; pp. 9–32. [Google Scholar]
- Bos, L. Crop losses caused by viruses. Crop Prot. 1982, 1, 263–282. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Naidu, R. Global dimensions of plant virus diseases: Current status and future perspectives. Ann. Rev. Virol. 2019, 6, 387–409. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A.C. Disease pandemics and major epidemics arising from new encounters between indigenous viruses and introduced crops. Viruses 2020, 12, 1388. [Google Scholar] [CrossRef]
- Jones, R.A.C. Global plant virus disease pandemics and epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef]
- Bos, L. New plant virus problems in developing countries: A corollary of agricultural modernization. Adv. Virus Res. 1992, 41, 349–407. [Google Scholar] [PubMed]
- Thresh, J.M. Control of tropical plant virus diseases. Adv. Virus Res. 2006, 67, 245–295. [Google Scholar]
- Jones, R.A.C. Control of Plant Virus Diseases. Adv. Virus Res. 2006, 67, 205–244. [Google Scholar]
- Jones, R.A.C. Plant virus emergence and evolution: Origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 2009, 141, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A.C. Plant virus ecology and epidemiology: Historical perspectives, recent progress and future prospects. Ann. Appl. Biol. 2014, 164, 320–347. [Google Scholar] [CrossRef]
- Jones, R.A.C. Future scenarios for plant virus pathogens as climate change progresses. Adv. Virus Res. 2016, 95, 87–147. [Google Scholar] [PubMed]
- Jones, R.A.C. Plant and insect viruses in managed and natural environments: Novel and neglected transmission pathways. Adv. Virus Res. 2018, 101, 149–187. [Google Scholar]
- Jones, R.A.C.; Barbetti, M.J. Influence of climate change on plant disease infections and epidemics caused by viruses and bacteria. CAB Rev. 2012, 7, 1–33. [Google Scholar] [CrossRef]
- Hull, R. Plant Virology, 5th ed.; Academic Press: San Diego, CA, USA, 2014. [Google Scholar]
- Trębicki, P. Climate change and plant virus epidemiology. Virus Res. 2020, 286, 198059. [Google Scholar] [CrossRef] [PubMed]
- Buchen-Osmond, C.; Crabtree, K.; Gibbs, A.J.; McLean, G.D. Viruses of Plants in Australia: Descriptions and Lists from the VIDE Database (No. 632.8 V821v); Australian National University: Canberra, Australia, 1988. [Google Scholar]
- Gibbs, A.J.; Mackenzie, A.; Wei, K.J.; Gibbs, M.J. The potyviruses of Australia. Arch. Virol. 2008, 153, 1411–1420. [Google Scholar] [CrossRef]
- Rodoni, B.C. The role of plant biosecurity in preventing and controlling emerging plant virus disease epidemics. Virus Res. 2009, 141, 150–157. [Google Scholar] [CrossRef]
- Gerritsen, R. Evidence for indigenous Australian agriculture. Australas. Sci. 2010, 31, 35–37. [Google Scholar]
- Davis, R.I.; Jones, L.M.; Pease, B.; Perkins, S.L.; Vala, H.R.; Kokoa, P.; Apa, M.; Dale, C.J. Plant virus and virus-like disease threats to Australia’s north targeted by the northern Australia quarantine strategy. Plants 2021, 10, 2175. [Google Scholar] [CrossRef]
- Whattam, M.; Dinsdale, A.; Elliott, C.E. Evolution of plant virus diagnostics used in Australian Post Entry Quarantine. Plants 2021, 10, 1430. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Sharman, M.; Trebicki, P.; Maina, S.; Congdon, B.S. Virus diseases of cereal and oilseed crops in Australia: Current position and future challenges. Viruses 2021, 13, 2051. [Google Scholar] [CrossRef] [PubMed]
- Maina, S.; Jones, R.A.C. Enhancing biosecurity against virus disease threats to Australian grain crops: Current situation and future prospects. Front. Hortic. 2023, 2, 1263604. [Google Scholar] [CrossRef]
- Boswell, K.F.; Gibbs, A.J. Viruses of Legumes 1983; Descriptions and Keys from VIDE; Research School of Biological Science, Australian National University: Canberra, Australia, 1983. [Google Scholar]
- Salam, M.U.; Davidson, J.A.; Thomas, G.J.; Ford, R.; Jones, R.A.C.; Lindbeck, K.D.; Macleod, W.J.; Kimber, R.B.; Galloway, J.; Mantri, N. Advances in winter pulse pathology research in Australia. Australas. Plant Pathol. 2011, 40, 549–567. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. Definition and Classification Commodities, 4. Pulses and Derived Products; Food and Agriculture Organization of the United Nations: Rome, Italy, 1994; Available online: http://www.fao.org/es/faodef/fdef04e.htm (accessed on 1 June 2023).
- Calles, T. The International Year of Pulses: What Are They and Why Are They Important? Food and Agriculture Organization, United Nations. 2016. Available online: https://www.fao.org/3/bl797e/BL797E.pdf. (accessed on 1 June 2023).
- Aykroyd, W.R.; Doughty, J.; Walker, A.F. Legumes in human nutrition. United Nations Food Agric. Organ. Food Nutr. Pap. 1982, 20, 1–152. [Google Scholar]
- Singh, N. Pulses: An overview. J. Food Sci. Technol. 2017, 54, 853–857. [Google Scholar] [CrossRef]
- Herridge, D.F.; Peoples, M.B.; Boddey, R.M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 2008, 311, 1–18. [Google Scholar] [CrossRef]
- Liu, C.; Plaza-Bonilla, D.; Coulter, J.A.; Kutcher, H.R.; Beckie, H.J.; Wang, L.; Floch, J.B.; Hamel, C.; Siddique, K.H.; Li, L.; et al. Diversifying crop rotations enhances agroecosystem services and resilience. Adv. Agron. 2022, 173, 299–335. [Google Scholar]
- Siddique, K.H.M.; Sykes, J. Pulse production in Australia past, present and future. Aust. J. Exp. Agric. 1997, 37, 103–111. [Google Scholar] [CrossRef]
- Pulse Australia. What Are Pulses? 2021. Available online: https://www.pulseaus.com.au/using-pulses/what-are-pulses (accessed on 7 June 2023).
- Gladstones, J.S. Lupins in Western Australia. 2. Cultivation methods. West. Aust. J. Agr. Fourth Ser. 1969, 10, 390–393. [Google Scholar]
- Siddique, K.H.M.; Loss, S.P.; Enneking, D. Narbon bean (Vicia narbonensis L.): A promising grain legume for low rainfall areas of south-western Australia. Aust. J. Exp. Agric. 1996, 36, 53–62. [Google Scholar] [CrossRef]
- Gladstones, J.S.; Atkins, C.A.; Hamblin, J. (Eds.) Lupins as Crop Plants: Biology, Production and Utilization; CAB International: Oxford, UK, 1998. [Google Scholar]
- French, R.J.; Sweetingham, M.W.; Shea, G.G. A comparison of the adaptation of yellow lupin (Lupinus luteus L.) and narrow-leafed lupin (L. angustifolius L.) to acid sandplain soils in low rainfall agricultural areas of Western Australia. Aust. J. Agric. Res. 2001, 52, 945–954. [Google Scholar] [CrossRef]
- Clements, J.C.; Buirchell, B.J.; Yang, H.; Smith, P.M.C.; Sweetingham, M.W.; Smith, C.G. Lupin. In Genetic Resources, Chromosome Engineering, and Crop Improvement. Grain Legumes; Singh, R.J., Jauhar, P.P.J., Eds.; Taylor and Francis: Boca Raton, FL, USA, 2005; Volume 1, pp. 231–323. [Google Scholar]
- Clements, J.C.; Sweetingham, M.S.; Smith, L.; Francis, G.; Thomas, G.; Sipsas, S. Crop Improvement in Lupinus mutabilis for Australian Agriculture—Progress and Prospects. In Lupins for Health and Wealth, Proceedings of the 12th International Lupin Conference, Fremantle, Australia, 14–18 September 2008; Palta, J.A., Berger, J.B., Eds.; International Lupin Association: Canterbury, New Zealand, 2008; pp. 244–250. [Google Scholar]
- Buirchell, B.J. Narrow-Leafed Lupin Breeding in Australia—Where to from Here? In Lupins for Health and Wealth, Proceedings of the 12th International Lupin Conference, Fremantle, Australia, 14–18 September 2008; Palta, J.A., Berger, J.B., Eds.; International Lupin Association: Canterbury, New Zealand, 2008; pp. 226–230. [Google Scholar]
- Jones, R.A.C. Virus Diseases of Australian Pastures. In Pasture and Forage Crop Pathology; Chackraborty, S.L.K., Skipp, R.A., Pederson, G.A., Bray, R., Latch, G.C.M., Nutter, F.W., Eds.; American Society of Agronomy: Madison, WI, USA, 1996; pp. 288–300. [Google Scholar]
- Jones, R.A.C. Developing integrated disease management strategies against non-persistently aphid-borne viruses: A model programme. Integr. Pest Manag. Rev. 2001, 6, 15–46. [Google Scholar] [CrossRef]
- Jones, R.A.C. Virus diseases of annual pasture legumes: Incidences, losses, epidemiology and management. Crop Pasture Sci. 2012, 63, 399–418. [Google Scholar] [CrossRef]
- Jones, R.A.C. Virus diseases of perennial pasture legumes: Incidences, losses, epidemiology and management. Crop Pasture Sci. 2013, 64, 199–215. [Google Scholar] [CrossRef]
- Jones, R.A.C. Alteration of plant species mixtures by virus infection: Managed pastures the forgotten dimension. Plant Pathol. 2022, 71, 1255–1281. [Google Scholar] [CrossRef]
- Brunt, A.; Crabtree, K.; Gibbs, A.J. Viruses of Tropical Plants: Descriptions and Lists from the VIDE Database; CAB International: Wallingford, UK, 1990. [Google Scholar]
- Jones, R.A.C.; McLean, G.D. Virus diseases of lupins. Ann. Appl. Biol. 1989, 114, 609–637. [Google Scholar] [CrossRef]
- Makkouk, K.M.; Kumari, S.G.; van Leur, J.A.G.; Jones, R.A.C. Control of plant virus diseases in cool-season legume crops. Adv. Virus Res. 2014, 90, 207–253. [Google Scholar] [PubMed]
- Jones, R.A.C. Host resistance to virus diseases provides a key enabler towards fast tracking gains in grain lupin breeding. Plants 2023, 12, 2521. [Google Scholar] [CrossRef]
- Persley, D.M.; Cooke, T.; House, S. (Eds.) Diseases of Vegetable Crops in Australia; CSIRO Publishing: Collingwood, Australia, 2010; 292p. [Google Scholar]
- Johnstone, G.R.; Barbetti, M.J. Impact of Fungal and Virus Diseases on Pasture. In Temperate Pastures: Their Production, Use and Management; Wheeler, J.L., Pearson, C.J., Roberts, G.E., Eds.; Australian Wool Corporation/CSIRO: Melbourne, Australia, 1987; pp. 235–248. [Google Scholar]
- Panetta, F.D.; Ridsdell-Smith, T.J.; Barbetti, M.J.; Jones, R.A.C. The Ecology of Weeds, Invertebrate Pests and Diseases of Australian Sheep Pastures. In Pests of Pastures: Weed, Invertebrate and Disease Pests of Australian Sheep Pastures; Delfosse, E.S., Ed.; Australian Wool Corporation/CSIRO: Melbourne, Australia, 1993; pp. 87–114. [Google Scholar]
- Barbetti, M.J.; Jones, R.A.C.; Riley, I.T. Problems and Progress in Assessing Direct and Indirect Yield Losses Caused by Pathogens in Pasture Species. In Pasture and Forage Crop Pathology; Chakraborty, S., Leath, K.T.L., Skipp, R.A., Pederson, G.A., Bray, R.A., Latch, G.C.M., Nutter, F.W., Eds.; American Society of Agronomy: Madison, WI, USA, 1996. [Google Scholar]
- Barbetti, M.J.; Pei You, M.P.; Jones, R.A.C. Medicago Truncatula and Other Annual Medicago spp.—Interactions with Root and Foliar Fungal, Oomycete, and Viral Pathogens. In The Model Legume Medicago truncatula; de Brujin, F., Ed.; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2020; pp. 293–306. [Google Scholar]
- Nichols, P.G.H.; Jones, R.A.C.; Ridsdill-Smith, T.J.; Barbetti, M.J. Genetic improvement of subterranean clover (Trifolium subterraneum L.). 2. Breeding for disease and pest resistance. Crop Pasture Sci. 2014, 65, 1207–1229. [Google Scholar] [CrossRef]
- Johnstone, G.R.; McLean, G.D. Virus diseases of subterranean clover. Ann. Appl. Biol. 1987, 110, 421–440. [Google Scholar] [CrossRef]
- Bos, L.; Hampton, R.O.; Makkouk, K.M. Viruses and Virus Diseases of Pea, Lentil, Faba Bean and Chickpea. In World Crops: Cool Season Food Legumes; Summerfield, R.J., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1988; pp. 591–615. [Google Scholar]
- Edwardson, J.R.; Christie, R.G. CRC Handbook of Viruses Infecting Legumes; CRC Press: Boca Raton, FL, USA, 1991. [Google Scholar]
- Jones, R.A.C. Determining threshold levels for seed-borne virus infection in seed stocks. Virus Res. 2000, 71, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A.C. Using epidemiological information to develop effective integrated virus disease management strategies. Virus Res. 2004, 100, 5–30. [Google Scholar] [CrossRef]
- Bos, L. Legume Virology. In Encyclopedia of Virology, 3rd ed.; Mahy, W.J., Van Regenmortel, M.H.N., Eds.; Academic Press: New York, NY, USA, 2008; pp. 212–220. [Google Scholar]
- Sastry, K.S. Seed-Borne Plant Virus Diseases; Springer: New York, NY, USA, 2013. [Google Scholar]
- Persley, D.M.; Steele, V.; Sharman, M.; Campbell, P.R.; Geering, A.D.; Gambley, C. First report of a carlavirus infecting plants in the Fabaceae in Australia. New Dis. Rep. 2020, 41, 26. [Google Scholar] [CrossRef]
- Gambley, C.; Nimmo, P.; Persley, D.; Steele, V.; Sharman, M.; Campbell, P. Cowpea mild mottle virus, a sometimes problem for French bean crops. Australas. Plant Pathol. 2022, 51, 565–576. [Google Scholar] [CrossRef]
- Henzell, T. Australian Agriculture: Its History and Challenges; CSIRO Publishing: Melbourne, Australia, 2007. [Google Scholar]
- Brown, A.; De Costa, C.; Guo, F. Our Food Future: Trends and Opportunities; Research Report 20.1; Australian Bureau of Agricultural and Resource Economics and Sciences: Canberra, Australia, 2020. [Google Scholar]
- Pratley, J.; Kirkegaard, J. (Eds.) Australian Agriculture in 2020: From Conservation to Automation; Agronomy Australia and Charles Sturt University: Wagga Wagga, Australia, 2020; Available online: http://www.agronomyaustraliaproceedings.org/images/sampledata/specialpublications/Australian%20Agriculture%20in%202020.pdf (accessed on 1 September 2023).
- Freund, M.; Henley, B.J.; Karoly, D.J.; Allen, K.J.; Baker, P.J. Multi-century cool- and warm-season rainfall reconstructions for Australia’s major climatic regions. Clim. Past 2017, 13, 1751–1770. [Google Scholar] [CrossRef]
- Stephens, D.J.; Lyons, T.J. Rainfall-yield relationships across the Australian wheatbelt. Aust. J. Agric. Res. 1998, 49, 211–224. [Google Scholar] [CrossRef]
- Turner, N.C.; Wright, G.C.; Siddique, K.H.M. Adaptation of grain legumes (pulses) to water-limited environments. Adv. Agron. 2001, 71, 193–231. [Google Scholar]
- Turner, N.C.; Asseng, S. Productivity, sustainability, and rainfall-use efficiency in Australian rainfed Mediterranean agricultural systems. Aust. J. Agric. Res. 2005, 56, 1123–1136. [Google Scholar] [CrossRef]
- Australian Export Grains Innovation Centre. Australian Pulses. In Australian Export Grains Innovation Centre, Sydney and Perth, Australia; 2022; Available online: https://www.aegic.org.au/australian-grains/pulses/ (accessed on 20 January 2023).
- Australian Export Grains Innovation Centre. What Grows Where? In Australian Grain Production—A Snapshot. Australian Grain Note; AEGIC: Sydney/Perth, Australia, 2020; Available online: https://www.aegic.org.au/australian-grain-production-a-snapshot/ (accessed on 1 September 2023).
- Kimberley Development Commission. Primary Industries: The Ord River Irrigation Area. 2020. Available online: https://kdc.wa.gov.au/economic-profile/primaryindustries/#:~:text=The%20Ord%20Valley%20produces%20a,stages%20over%20the%20last%20years (accessed on 8 September 2021).
- Northern Territory Government. Agriculture, Forestry and Fishing. Northern Territory Economy. 2017. Available online: https://nteconomy.nt.gov.au/industry-analysis/agriculture,-foresty-and-fishing (accessed on 8 September 2021).
- Cooperative Research Centre for Northern Australia. Northern Australian Broadacre Cropping Situational Analysis. 2020. Available online: https://www.crcna.com.au/resources/publications/northern-australian-broadacre-cropping-situationalanalysis (accessed on 8 September 2021).
- Grains Research and Development Corporation. Australian Grains Focus 2010–2011; Grains Research and Development Corporation: Kingston, Australia, 2010; Available online: https://grdc.com.au/__data/assets/pdf_file/0025/208735/grdc-australian-grains-focus-20102011.pdf (accessed on 9 September 2021).
- Jones, R.A.C.; Salam, M.U.; Maling, T.J.; Diggle, A.J.; Thackray, D.J. Principles of predicting plant virus disease epidemics. Ann. Rev. Phytopathol. 2010, 48, 179–203. [Google Scholar] [CrossRef]
- Jones, R.A.C. Trends in plant virus epidemiology: Opportunities from new or improved technologies. Virus Res. 2014, 186, 3–19. [Google Scholar] [CrossRef]
- Oerke, E.C. Remote sensing of diseases. Annu. Rev. Phytopathol. 2020, 58, 225–252. [Google Scholar] [CrossRef]
- Neupane, K.; Baysal-Gurel, F. Automatic identification and monitoring of plant diseases using unmanned aerial vehicles: A review. Remote Sens. 2021, 13, 3841. [Google Scholar] [CrossRef]
- Rhodes, M.W.; Bennie, J.J.; Spalding, A.; Ffrench-Constant, R.H.; Maclean, I.M. Recent advances in the remote sensing of insects. Biol. Rev. 2022, 97, 343–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Ostendorf, B.; Gautam, D.; Habili, N.; Pagay, V. Plant viral disease detection: From molecular diagnosis to optical sensing technology—A multidisciplinary review. Remote Sens. 2022, 14, 1542. [Google Scholar] [CrossRef]
- Croser, J.S.; Pazos-Navarro, M.; Bennett, R.G.; Tschirren, S.; Edwards, K.; Erskine, W.; Creasy, R.; Ribalta, F.M. Time to flowering of temperate pulses in vivo and generation turnover in vivo–in vitro of narrow-leaf lupin accelerated by low red to far-red ratio and high intensity in the far-red region. Plant Cell Tissue Org. Cult. 2016, 127, 591–599. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, X.; Zhou, G.; Zhang, T. Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 2020, 18, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-based technologies for engineering plant virus resistance. Plants 2021, 10, 82. [Google Scholar] [CrossRef]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N. RNA Interference and CRISPR/Cas9 Applications for Virus Resistance. In CRISPR and RNAi Systems; Abd-Elsalam, K.A., Lim, K.-T., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 163–182. [Google Scholar]
- Ding, S.W. Transgene silencing, RNA interference, and the antiviral defense mechanism directed by small interfering RNAs. Phytopathology 2023, 113, 616–625. [Google Scholar] [CrossRef]
- Jones, R.A.C. Progress in Understanding Pulse Disease Epidemiology and Management in Western Australia; Department of Primary Industries and Regional Development, Western Australia: Perth, Australia, 2018; 62p. [Google Scholar]
- Jones, R.A.C.; Proudlove, W. Further studies on cucumber mosaic virus infection of narrow-leafed lupin (Lupinus angustifolius): Seed-borne infection, aphid transmission, spread and effects on grain yield. Ann. Appl. Biol. 1991, 118, 319–329. [Google Scholar] [CrossRef]
- Bwye, A.M.; Jones, R.A.C.; Proudlove, W. Effects of sowing seed with different levels of infection, plant density and the growth stage at which plants first develop symptoms on cucumber mosaic-virus infection of narrow-leafed lupins (Lupinus angustifolius). Aust. J. Agric. Res. 1994, 45, 1395–1412. [Google Scholar] [CrossRef]
- Bwye, A.M.; Proudlove, W.; Berlandier, F.A.; Jones, R.A.C. Effects of applying insecticides to control aphid vectors and cucumber mosaic virus in narrow-leafed lupins (Lupinus angustifolius). Aust. J. Exper. Agric. 1997, 37, 93–102. [Google Scholar] [CrossRef]
- Bwye, A.M.; Jones, R.A.C.; Proudlove, W. Effects of different cultural practices on spread of cucumber mosaic virus in narrow-leafed lupins (Lupinus angustifolius). Aust. J. Agric. Res. 1999, 50, 985–996. [Google Scholar] [CrossRef]
- Thackray, D.J.; Jones, R.A.C.; Bwye, A.M.; Coutts, B.A. Further studies on the effects of insecticides on aphid vector numbers and spread of cucumber mosaic virus in narrow-leafed lupins (Lupinus angustifolius). Crop Prot. 2000, 19, 121–139. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Coutts, B.A.; Latham, L.J.; McKirdy, S.J. Cucumber mosaic virus infection of chickpea stands: Temporal and spatial patterns of spread and yield-limiting potential. Plant Pathol. 2008, 57, 842–853. [Google Scholar] [CrossRef]
- Jones, R.A.C. Effects of cereal borders, admixture with cereals and plant density on the spread of bean yellow mosaic potyvirus into narrow-leafed lupins (Lupinus angustifolius). Ann. Appl. Biol. 1993, 122, 501–518. [Google Scholar] [CrossRef]
- Jones, R.A.C. Virus Diseases of Lupins; Western Australian Department of Agriculture Bulletin No. 4294; Department of Primary Industries and Regional Development: Perth, Australia, 1994; 14p. [Google Scholar]
- Jones, R.A.C.; Coutts, B.A.; Cheng, Y. Yield limiting potential of necrotic and non-necrotic strains of bean yellow mosaic virus in narrow-leafed lupin (Lupinus angustifolius). Aust. J. Agric. Res. 2003, 54, 849–859. [Google Scholar] [CrossRef]
- Coutts, B.A.; Prince, R.T.; Jones, R.A.C. Quantifying effects of seed-borne virus inoculum on virus spread, yield losses and seed infection in the pea seed-borne mosaic virus—Field pea pathosystem. Phytopathology 2009, 99, 1156–1167. [Google Scholar] [CrossRef]
- Palukaitis, P.; Garcia-Arenal, F. Cucumber Mosaic Virus. In Descriptions of Plant Viruses No. 400; Association of Applied Biologists: Wellesbourne, UK, 2003. [Google Scholar]
- Crowley, N.C. Some variables affecting the use of cowpea as an assay host for cucumber mosaic virus. Aust. J. BioI. Sci. 1954, 7, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Harvey, H.L. Plant disease—Cucumber mosaic virus. West. Aust. J. Agric. Third Ser. 1954, 3, 39–42. [Google Scholar]
- Latham, L.J.; Jones, R.A.C. Incidence of virus infection in experimental plots, commercial crops and seed stocks of cool season crop legumes. Aust. J. Agric. Res. 2001, 52, 397–413. [Google Scholar] [CrossRef]
- Latham, L.J.; Jones, R.A.C.; McKirdy, S.J. Cucumber mosaic cucumovirus infection of cool-season crop, annual pasture, and forage legumes: Susceptibility, sensitivity and seed transmission. Aust. J. Agric. Res. 2001, 52, 683–697. [Google Scholar] [CrossRef]
- Aftab, M.; Freeman, A.; Davidson, J. Pulse virus surveys in south eastern Australia (2003–2004). In Proceedings of the Australasian Plant Pathology Society 15th Biennial Conference, Geelong, Australia, 24–29 September 2005; p. 338. Available online: https://catalogue.nla.gov.au/catalog/4494494 (accessed on 3 August 2023).
- Aftab, M.; Nancarrow, N.; Freeman, A.; Davidson, J.; Rodoni, B.; Trębicki, P. Natural infection of cucumber mosaic virus, pea seed-borne mosaic virus and turnip yellows virus in a fenugreek crop (Trigonella foenum-graecum). Australas. Plant Dis. Notes 2018, 13, 2. [Google Scholar] [CrossRef]
- Association of Applied Biologists. Descriptions of Plant Viruses; Association of Applied Biologists: Warwick, UK, 2023; Available online: https://www.dpvweb.net/ (accessed on 3 August 2023).
- Centre for Agriculture and Biosciences International. Data Sheets. In Invasive Species Compendium; Centre for Agriculture and Biosciences International: Wallingford/Oxfordshire, UK, 2022; Available online: https://www.cabi.org/Uploads/CABI/publishing/promotional-materials/insert/IS%20Compendium%20-%20A4%20Insert.pdf (accessed on 3 August 2023).
- Gladstones, J.S. Uniwhite—A new lupin variety. West. Aust. J. Agric. Fourth Ser. 1967, 8, 190–197. [Google Scholar]
- Jones, R.A.C. Seed-borne cucumber mosaic virus infection of narrow-leafed lupin (Lupinus angustifolius) in Western Australia. Ann. Appl. Biol. 1988, 113, 507–518. [Google Scholar] [CrossRef]
- McLean, G.D.; Price, L.K. Virus, Viroid, Mycoplasma and Rickettsial Diseases of Plants in Western Australia; Western Australian Department of Agriculture Technical Bulletin No. 68; Department of Primary Industries and Regional Development: Perth, Australia, 1984; 22p. [Google Scholar]
- Bowyer, J.W.; Keirnan, E. Cucumber mosaic virus in lupin. Australas. Plant Pathol. 1981, 10, 28–29. [Google Scholar] [CrossRef]
- Alberts, E.; Hannay, J.; Randles, J.W. An epidemic of cucumber mosaic virus in South Australian lupins. Aust. J. Agric. Res. 1985, 36, 267–273. [Google Scholar] [CrossRef]
- Jones, R.A.C. Cucumber mosaic virus in lupins. West Aust. J. Agric. Fourth Ser. 1987, 8, 190–197. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Coutts, B.A.; Kehoe, M.A. Cucumber Mosaic Virus in Lupins; Western Australian Department of Agriculture and Food, Farmnote No. 402; Department of Primary Industries and Regional Development: Perth, Australia, 2010. [Google Scholar]
- Wells, H.D.; Corbett, M.K.; Forbes, I. Cucumber mosaic virus is seed-borne in blue lupines. Phytopathology 1964, 54, 627. [Google Scholar]
- Lupuwana, P.; Rybicki, E.P.; von Wechmar, M.B. Cucumber mosaic virus in lupins. In Proceedings of the 23rd Congress South African Society for Plant Pathology, Ciskei, South Africa, 21 January 1985; p. 54. [Google Scholar]
- Jones, R.A.C. Seed-borne virus diseases in the Western Australian lupin, subterranean clover and annual medic seed collections and breeding/evaluation programmes. In Proceedings of the Workshop on Production, Maintenance and Exchange of Healthy Legume Germplasm, Melbourne, Australia, 1–2 April 1990; Sward, R.J., Ed.; Plant Research Institute, Department of Agriculture and Rural Affairs: Melbourne, Australia; p. 31. [Google Scholar]
- Geering, A.D.W.; Randles, J.W. Interactions between a seed-borne strain of cucumber mosaic cucumovirus and its lupin host. Ann. Appl. Biol. 1994. 124, 301–314. [CrossRef]
- Wahyuni, W.S.; Francki, R.I.B. Responses of some grain and pasture legumes to 16 strains of cucumber mosaic virus (CMV). Aust. J. Agric. Res. 1992, 43, 465477. [Google Scholar] [CrossRef]
- Wahyuni, W.S.; Dietzgen, R.G.; Hanada, K.; Francki, R.I.B. Serological and biological variation between and within subgroup I and II strains of cucumber mosaic virus. Plant Pathol. 1992, 41, 282–297. [Google Scholar] [CrossRef]
- McKirdy, S.J.; Jones, R.A.C. Infection of alternative hosts associated with narrow-leafed lupin (Lupinus angustifolius) and subterranean clover (Trifolium subterraneum) by cucumber mosaic virus and its persistence between growing seasons. Aust. J. Agric. Res. 1994, 45, 1035–1049. [Google Scholar] [CrossRef]
- Jones, R.A.C.; McKirdy, S.J. Seed-borne cucumber mosaic virus infection of subterranean clover in Western Australia. Ann. Appl. Biol. 1990, 116, 73–86. [Google Scholar] [CrossRef]
- Jones, R.A.C. Losses in productivity of subterranean clover swards caused by sowing cucumber mosaic virus-infected seed. Ann. Appl. Biol. 1991, 119, 273–288. [Google Scholar] [CrossRef]
- Pathipanawat, W.; Jones, R.A.C.; Sivasithamparam, K. Studies on seed and pollen transmission of alfalfa mosaic, cucumber mosaic and bean yellow mosaic viruses in cultivars and accessions of annual Medicago species. Aust. J. Agric. Res. 1995, 46, 153–165. [Google Scholar] [CrossRef]
- Cowling, W.A.; Gladstones, J.S. Lupin Breeding in Australia. In Linking Research and Marketing Opportunities for Pulses in the 21st Century, Proceedings of the Third International Food Legumes Research Conference; Springer: Dordrecht, The Netherlands, 2000; pp. 541–547. [Google Scholar]
- Jones, R.A.C. Reflective mulch decreases the spread of two non-persistently aphid transmitted viruses to narrow-leafed lupin (Lupinus angustifolius). Ann. Appl. Biol. 1991, 118, 79–85. [Google Scholar] [CrossRef]
- Berlandier, F.A.; Thackray, D.J.; Jones, R.A.C.; Latham, L.J.; Cartwright, L. Determining the relative roles of different aphid species as vectors of cucumber mosaic and bean yellow mosaic viruses in lupins. Ann. Appl. Biol. 1997, 131, 297–314. [Google Scholar] [CrossRef]
- Bwye, A.M.; Jones, R.A.C.; Proudlove, W. Management of cucumber mosaic virus in lupins. West. Aust. J. Agric. Fourth Ser. 1995, 36, 124–130. [Google Scholar]
- Wylie, S.; Wilson, C.R.; Jones, R.A.C.; Jones, M.G.K. A polymerase chain reaction assay for cucumber mosaic virus in lupin seeds. Aust. J. Agric. Res. 1993, 44, 41–51. [Google Scholar] [CrossRef]
- O’Keefe, D.C.; Berryman, D.I.; Coutts, B.A.; Jones, R.A.C. Absence of seed coat contamination with cucumber mosaic virus allows large-scale detection of seed transmission in lupin seed samples. In Proceedings of the 7th Australasian Plant Virology Workshop, Rottnest Island, Perth, Australia, 9–12 November 2006; Murdoch University: Perth, Australia, 2006; p. 57. [Google Scholar]
- O’Keefe, D.C.; Berryman, D.I.; Coutts, B.A.; Jones, R.A.C. Lack of seed coat contamination with cucumber mosaic virus in lupin permits reliable, large-scale detection of seed transmission in seed samples. Plant Dis. 2007, 91, 504–508. [Google Scholar] [CrossRef][Green Version]
- Jones, R.A.C. Patterns of spread of two non-persistently aphid-borne viruses in lupin stands under four different infection scenarios. Ann. Appl. Biol. 2005, 146, 337–350. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Cowling, W.A. Screening for resistance to seed transmission of cucumber mosaic virus in Lupinus angustifolius. In Advances in Lupin Research; Neves Martins, J.M., Reiraio da Costa, M.L., Eds.; Instituto Superior de Agronomia Press: Lisbon, Portugal, 1994; pp. 120–122. [Google Scholar]
- Jones, R.A.C.; Cowling, W.A. Resistance to seed transmission of cucumber mosaic virus in narrow-leafed lupins (Lupinus angustifolius). Aust. J. Agric. Res. 1995, 46, 1339–1352. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Coutts, B.A.; Reeve, N.; Cowling, W.A.; Buirchell, B.J. Screening for Resistance to Cucumber Mosaic Virus in Lupins. In Crop Update Conference, Lupin 1999; Agriculture Western Australia: Perth, Australia, 1999; pp. 27–28. [Google Scholar]
- Jones, R.A.C.; Latham, L.J. Natural resistance to cucumber mosaic virus in broad-leafed lupins. In Proceedings of the First Australian Lupin Technical Symposium, Perth, Australia, 17–21 October 1994; Department of Agriculture: South Perth, Australia, 1994; p. 278. [Google Scholar]
- Jones, R.A.C.; Latham, L.J. Natural resistance to cucumber mosaic virus in lupin species. Ann. Appl. Biol. 1996, 129, 523–542. [Google Scholar] [CrossRef]
- Garcıa-Arenal, F.; Palukaitis, P. Cucumber Mosaic Virus. In Desk Encyclopedia of Plant and Fungal Virology; Mahy, B.J., van Regenmortel, M.H.V., Eds.; Elsevier: Amsterdam, The Netherlands, 2008; pp. 171–176. [Google Scholar]
- Jones, R.A.C.; Buirchell, G.M. Resistance to cucumber mosaic virus in Lupinus mutabilis (pearl lupin). Australas. Plant Pathol. 2004, 33, 591–593. [Google Scholar] [CrossRef]
- Singh, Z.; Jones, M.G.K.; Jones, R.A.C. Effectiveness of coat protein and defective replicase gene-mediated resistance against Australian isolates of cucumber mosaic virus. Aust. J. Exp. Agric. 1998, 38, 375–383. [Google Scholar] [CrossRef]
- Liu, L.; Yang, R.; Wylie, S.J.; Li, H.; Jones, M.G.K. Protection conferred by the cucumber mosaic virus replicase gene in transgenic narrow-leafed lupin. In Proceedings of the 15th Meeting International Working Group on Legume Viruses, Fremantle, Australia, 15–17 August 1999; Centre for Legumes in Mediterranean Agriculture: Perth, Australia, 1999; p. 65. [Google Scholar]
- Yang, R.; Liu, L.; Dwyer, G.I.; Wylie, S.J.; Jones, R.A.C.; Jones, M.G.K. Engineering cucumber mosaic virus resistance in narrow-leafed lupins (Lupinus angustifolius) based on a defective replicase gene. In Proceedings of the 15th Meeting International Working Group on Legume Viruses, Fremantle, Australia, 15–17 August 1999; Centre for Legumes in Mediterranean Agriculture: Perth, Australia, 1999; p. 42. [Google Scholar]
- Jones, M.G.K.; Wylie, S.J.; Berryman, D.; Selladurai, S.; Brien, S.J.; Li, D.; Yang, R.; Ryan, K.; Li, H.; Li, L. Biotechnological Tools in Lupin Breeding and Disease Control. In Lupin, an Ancient Crop for the New Millennium, Proceedings of the 9th International Lupin Conference, Klink/Muritz, Germany, 20–24 June 1999; van Santen, E., Wink, M., Weissmann, S., Romer, P., Eds.; International Lupin Association: Canterbury, New Zealand, 2000; pp. 111–114. Available online: https://researchrepository.murdoch.edu.au/id/eprint/19710/1/WYLIE-20040624162812.pdf (accessed on 3 August 2023).
- Yang, R. Towards Genetic Engineering Cucumber Mosaic Virus (CMV) Resistance in Lupins. Ph.D. Thesis, Murdoch University, Perth, Australia, 2000. Available online: https://researchportal.murdoch.edu.au/esploro/outputs/doctoral/Towards-geneticengineering-cucumber-mosaic-virus/991005541812107891 (accessed on 10 May 2023).
- Perry, J.N.; Bell, E.D.; Smith, R.H.; Woiwod, I.P. SADIE: Software to measure and model spatial pattern. Asp. Appl. Biol. 1996, 46, 95–102. [Google Scholar]
- Perry, J.N.; Winder, L.; Holland, J.M.; Alston, R.D. Red-blue plots for detecting clusters in count data. Ecol. Lett. 1999, 2, 106–113. [Google Scholar] [CrossRef]
- Thackray, D.J.; Jones, R.A.C. Forecasting Aphid and Virus Risk in Lupins. In Crop Update Conference, Lupin 1999; Agriculture Western Australia: Perth, Australia, 1999; pp. 24–26. [Google Scholar]
- Thackray, D.J.; Hawkes, J.; Jones, R.A.C. Forecasting Aphid and Virus Risk in Lupins. In Crop Update Conference, Lupin 2000; O’Neill, B., Ed.; Agriculture Western Australia: Perth, Australia, 2000; pp. 19–21. [Google Scholar]
- Thackray, D.J.; Diggle, A.J.; Berlandier, F.A.; Jones, R.A.C. Development of a Simulation Model Forecasting Aphid Outbreaks and Spread of Cucumber Mosaic Virus in Lupins. In Pest Management—Future Challenges, Proceedings of the 6th Australasian Applied Entomological Research Conference, Brisbane, Australia, 29 September–2 October 1998; Zalucki, M.P., Drew, R.A.I., White, G.C., Eds.; University of Queensland Press: Brisbane, Australia, 1998; Volume 2, pp. 99–106. [Google Scholar]
- Thackray, D.J.; Diggle, A.J.; Berlandier, F.A.; Jones, R.A.C. Forecasting aphid outbreaks and epidemics of cucumber mosaic virus in lupin crops in a Mediterranean-type environment. Virus Res. 2004, 100, 67–82. [Google Scholar] [CrossRef]
- Thackray, D.J. Implications of the Green Bridge for Viral and Fungal Disease Carry-Over between Seasons. In Crop Update Conference, Lupin 2000; O’Neill, B., Ed.; Agriculture Western Australia: Perth, Australia, 2000; pp. 17–19. [Google Scholar]
- Hawkes, J.R.; Jones, R.A.C. Incidence and distribution of barley yellow dwarf virus and cereal yellow dwarf virus in oversummering grasses in a Mediterranean-type environment. Aust. J. Agric. Res. 2005, 56, 257–270. [Google Scholar] [CrossRef]
- Coutts, B.A.; Hawkes, J.R.; Jones, R.A.C. Occurrence of Beet western yellows virus and its aphid vectors in over-summering broad-leafed weeds and volunteer crop plants in the grainbelt region of south-western Australia. Aust. J. Agric. Res. 2006, 57, 975–982. [Google Scholar] [CrossRef]
- Maling, T.; Diggle, A.J.; Thackray, D.J.; Siddique, K.H.M.; Jones, R.A.C. An epidemiological model for externally sourced vector-borne viruses applied to bean yellow mosaic virus in lupin crops in a Mediterranean type environment. Phytopathology 2008, 98, 1280–1290. [Google Scholar] [CrossRef]
- Department of Primary Industries and Regional Development. Western Australian Lupin Industry, Department of Primary Industries and Regional Development, South Perth, Western Australia. 2023. Available online: https://www.agric.wa.gov.au/investment/western-australian-lupin-industry (accessed on 1 July 2023).
- Fletcher, A.; Lawes, R.; Weeks, C. Crop area increases drive earlier and dry sowing in Western Australia: Implications for farming systems. Crop Pasture Sci. 2016, 67, 1268–1280. [Google Scholar] [CrossRef]
- Australian Bureau of Agricultural Resource Economics and Sciences. Australian Crop Report, Western Australia; Department of Agriculture, Fisheries and Forestry: Canberra, Australia, 2022. Available online: https://www.agriculture.gov.au/abares/research-topics/agricultural-outlook/australian-crop-report/western-australia (accessed on 20 January 2023).
- Jones, R.A.C.; Coutts, B.A. Alfalfa mosaic and cucumber mosaic virus infection in chickpea and lentil: Incidence and seed transmission. Ann. Appl. Biol. 1996, 129, 491–506. [Google Scholar] [CrossRef]
- Freeman, A.J.; Spackman, M.E.; Aftab, M.; McQueen, V.; King, S.; van Leur, J.A.; Loh, M.H.; Rodoni, B. Comparison of tissue blot immunoassay and reverse transcription polymerase chain reaction assay for virus-testing pulse crops from a South-Eastern Australia survey. Australas. Plant Pathol. 2013, 42, 675–683. [Google Scholar] [CrossRef]
- van Leur, J.A.G.; Aftab, M.; Manning, W.; Bowring, A.; Riley, M.J. A severe outbreak of chickpea viruses in northern New South Wales, Australia, during 2012. Australas. Plant Dis. Notes 2013, 8, 49–53. [Google Scholar] [CrossRef][Green Version]
- Sharman, M.; Moore, M.; Van Leur, J.; Aftab, M.; Verrell, A. Viral Diseases of Chickpea—Impact and Management; GRDC Crop Update: Mungundi, Australia, 2014; pp. 110–114. [Google Scholar]
- Aftab, M.; Freeman, A.; Davidson, J. Virus diseases in South Australian pulse crops (2005–2006). In Proceedings of the 16th Biennial Australasian Plant Pathology Society Conference, Adelaide, Australia, 26–29 September 2007; pp. 24–27. [Google Scholar]
- Freeman, A.J.; Aftab, M. Surveying for and mapping of viruses in pulse crops in south-eastern Australia. In Proceedings of the 13th Biennial Conference of the Australasian Plant Pathology Society, Cairns, Australia, 24–27 September 2001; p. 149. [Google Scholar]
- Freeman, A.J.; Aftab, M.; Dobson, V. Surveying for viruses in pulse crops in Victoria. In Proceedings of the 8th International Congress of Plant Pathology, Christchurch, New Zealand, 2–7 February 2003; Volume 2, p. 261. [Google Scholar]
- Freeman, A.J.; Aftab, M.; McQueen, V.; Davidson, J. The occurrence of common viruses in pulse crops in south eastern Australia. In Proceedings of the 15th Biennial Australasian Plant Pathology Society Conference, Geelong, Australia, 26–30 September 2005; p. 339. [Google Scholar]
- Maina, S.; Zheng, L.; Rodoni, B.C. Targeted Genome Sequencing (TG-Seq) Approaches to Detect Plant Viruses. Viruses 2021, 13, 583. [Google Scholar] [CrossRef] [PubMed]
- Latham, L.J.; Jones, R.A.C.; Coutts, B.A. Yield losses caused by virus infection in four combinations of non-persistently aphid-transmitted virus and cool-season crop legume. Aust. J. Exp. Agric. 2004, 44, 57–63. [Google Scholar] [CrossRef]
- Freeman, A.; Aftab, M. Effective management of viruses in pulse crops in south eastern Australia should include management of weeds. Australas. Plant Pathol. 2011, 40, 430–441. [Google Scholar] [CrossRef]
- Jones, R.A.C. Seed-borne virus diseases in annual pasture legumes. J. Agric. West. Aust. Fourth Ser. 1988, 29, 58–61. [Google Scholar]
- Swenson, K.G.; Venables, D.G. Detection of two legume viruses in Australia. Aust. J. Exp. Agric. 1961, 1, 116–118. [Google Scholar] [CrossRef]
- Garran, J.; Gibbs, A.J. Studies on alfalfa mosaic virus and alfalfa aphids. Aust. J. Agric. Res. 1982, 33, 657–664. [Google Scholar] [CrossRef]
- Jaspars, E.M.J.; Bos, L. Alfalfa Mosaic Virus. In Descriptions of Plant Viruses No. 229; Association of Applied Biologists: Wellesbourne, UK, 1980. [Google Scholar]
- Jones, R.A.C.; Pathipanawat, W. Seed-borne alfalfa mosaic virus infecting annual medics (Medicago spp.) in Western Australia. Ann. Appl. Biol. 1989, 115, 263–277. [Google Scholar] [CrossRef]
- Jones, R.A.C. Report on Survey for Virus Diseases in crops in the Ord River Irrigation Area, 12–14 August 1996. In Northern Australian Quarantine Strategy (NAQS); Agriculture Western Australia: Perth, Australia, 1996. [Google Scholar]
- van Leur, J.A.G.; Makkouk, K.M.; Freeman, A.; Schilg, M.A. Occurrence of viruses in faba bean on the Liverpool Plains, northern New South Wales. In Proceedings of the 8th International Congress of Plant Pathology, Christchurch, New Zealand, 2–7 February 2003; p. 265. [Google Scholar]
- Maina, S.; Zheng, L.; Kinoti, W.; Aftab, M.; Nancarrow, N.; Trębicki, P.; King, S.; Constable, F.; Rodoni, B. Metagenomic analysis reveals a nearly complete genome sequence of alfalfa mosaic virus from a field pea in Australia. Microbiol. Res. Announc. 2019, 8, 10–128. [Google Scholar] [CrossRef]
- Latham, L.J.; Jones, R.A.C. Alfalfa mosaic and pea seed-borne mosaic viruses in cool season crop, annual pasture, and forage legumes: Susceptibility, sensitivity, and seed transmission. Crop Past. Sci. 2001, 52, 771–790. [Google Scholar] [CrossRef]
- McKirdy, S.J.; Jones, R.A.C. Infection of alternative hosts associated with annual medics (Medicago spp.) by alfalfa mosaic virus and its persistence between growing seasons. Aust. J. Agric. Res. 1994, 45, 1413–1426. [Google Scholar] [CrossRef]
- Jones, R.A.C. Occurrence of virus infection in seed stocks and 3-year-old pastures of lucerne (Medicago sativa). Aust. J. Agric. Res. 2004, 55, 757–764. [Google Scholar] [CrossRef]
- Nutter, F.W.; Jones, R.A.C.; Geering, A.D.W.; Randles, J.W.; Graetz, D.; Alberts, E.V. Quantification of alfalfa mosaic virus and cucumber mosaic virus epidemics in the lupin-medic pathosystem. Phytopathology 1996, 86, S14. [Google Scholar]
- Jones, R.A.C.; Pearce, A.R.; Prince, R.T.; Coutts, B.A. Natural resistance to alfalfa mosaic virus in different lupin species. Australas. Plant Pathol. 2008, 37, 112–116. [Google Scholar] [CrossRef]
- Stubbs, L.L. A destructive vascular wilt virus disease of broad bean (Vicia faba L.) in Victoria. J. Dep. Agric. Vic. 1947, 46, 323–332. [Google Scholar]
- Taylor, R.H.; Stubbs, L.L. Broad Bean Wilt Virus. In Descriptions of Plant Viruses; No. 81; Association of Applied Biologists: Wellesbourne, Australia, 1972. [Google Scholar]
- Zhou, X. Broad Bean Wilt Virus 2. In Descriptions of Plant Viruses; No. 392; Association of Applied Biologists: Wellesbourne, Australia, 2002. [Google Scholar]
- Makkouk, K.M.; Kumari, S.G.; Bos, L. Broad bean wilt virus: Host range, purification, serology, transmission characteristics, and occurrence in faba bean in West Asia and North Africa. Neth. J. Plant Pathol. 1990, 96, 291–300. [Google Scholar] [CrossRef]
- Moghal, S.M.; Francki, R.I.B. Occurrence and properties of broad bean stain virus in South Australia. Aust. J. Biol. Sci. 1974, 27, 341–348. [Google Scholar] [CrossRef][Green Version]
- Randles, J.W.; Dube, A.J. Three seedborne pathogens isolated from Vicia faba seed imported from the United Kingdom. Australas. Plant Pathol. Soc. Newsl. 1977, 6, 37–38. [Google Scholar] [CrossRef]
- Gibbs, A.J.; Smith, H.G. Broad Bean True Mosaic Virus. In Descriptions of Plant Viruses; No. 20; Association of Applied Biologists: Wellesbourne, Australia, 1970. [Google Scholar]
- Gibbs, A.J.; Smith, H.G. Broad Bean Stain Virus. In Descriptions of Plant Viruses; No. 29; Association of Applied Biologists: Wellesbourne, UK, 1970. [Google Scholar]
- Behncken, G.M. The occurrence of peanut mottle virus in Queensland. Aust. J. Agric. Res. 1970, 21, 465–472. [Google Scholar] [CrossRef]
- Behncken, G.M.; McCarthy, G.J. Peanut mottle virus in peanuts, navy beans and soybeans. Qld. Agric. J. 1973, 99, 635–637. [Google Scholar]
- Zanardo, L.G.; Carvalho, C.M. Cowpea mild mottle virus (Carlavirus, Betaflexiviridae): A review. Tropical Plant Pathol. 2017, 42, 417–430. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jones, R.A.C.; Congdon, B.S. Australian Cool-Season Pulse Seed-Borne Virus Research: 1. Alfalfa and Cucumber Mosaic Viruses and Less Important Viruses. Viruses 2024, 16, 144. https://doi.org/10.3390/v16010144
Jones RAC, Congdon BS. Australian Cool-Season Pulse Seed-Borne Virus Research: 1. Alfalfa and Cucumber Mosaic Viruses and Less Important Viruses. Viruses. 2024; 16(1):144. https://doi.org/10.3390/v16010144
Chicago/Turabian StyleJones, Roger A. C., and Benjamin S. Congdon. 2024. "Australian Cool-Season Pulse Seed-Borne Virus Research: 1. Alfalfa and Cucumber Mosaic Viruses and Less Important Viruses" Viruses 16, no. 1: 144. https://doi.org/10.3390/v16010144
APA StyleJones, R. A. C., & Congdon, B. S. (2024). Australian Cool-Season Pulse Seed-Borne Virus Research: 1. Alfalfa and Cucumber Mosaic Viruses and Less Important Viruses. Viruses, 16(1), 144. https://doi.org/10.3390/v16010144








