Chloride Intracellular Channel Protein 1 (CLIC1) Is a Critical Host Cellular Factor for Influenza A Virus Replication
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Infection and Plaque Assay
2.3. Cell Viability
2.4. siRNA Transfection
2.5. Protein Extraction and Quantification
2.6. SomaScan Analyses
2.7. Immunoblotting
2.8. RNA Extraction and Real-Time PCR
2.9. Impact of CLIC1 Inhibitors on IAV Replication
2.10. Photomicrography
2.11. Immunofluorescent Microscopy
2.12. Statistical and Bioinformatics Analyses
3. Results
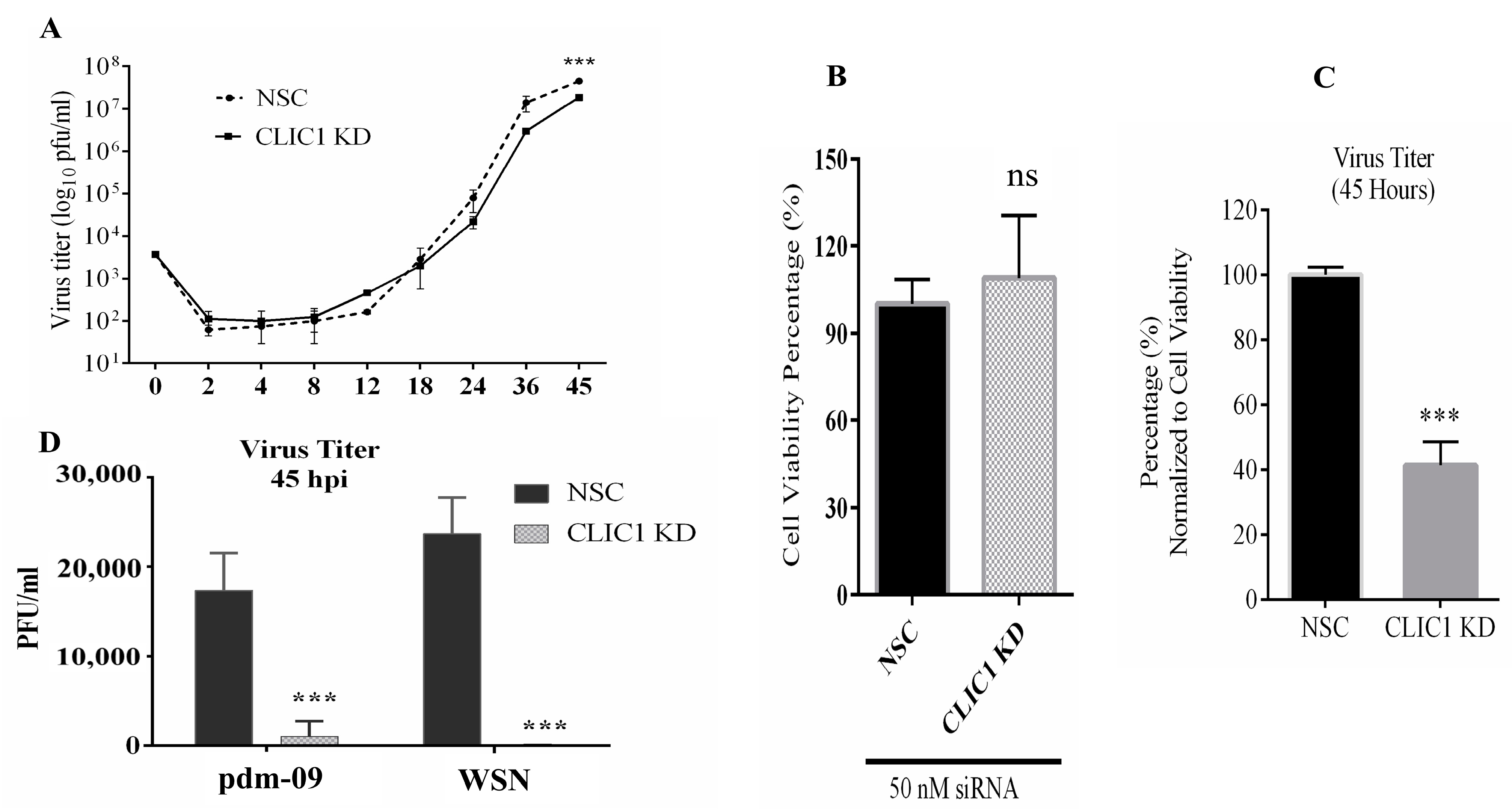
3.1. Optimization of CLIC1 Knockdown by siRNA Treatment
3.2. Impact of CLIC1 KD on IAV Replication
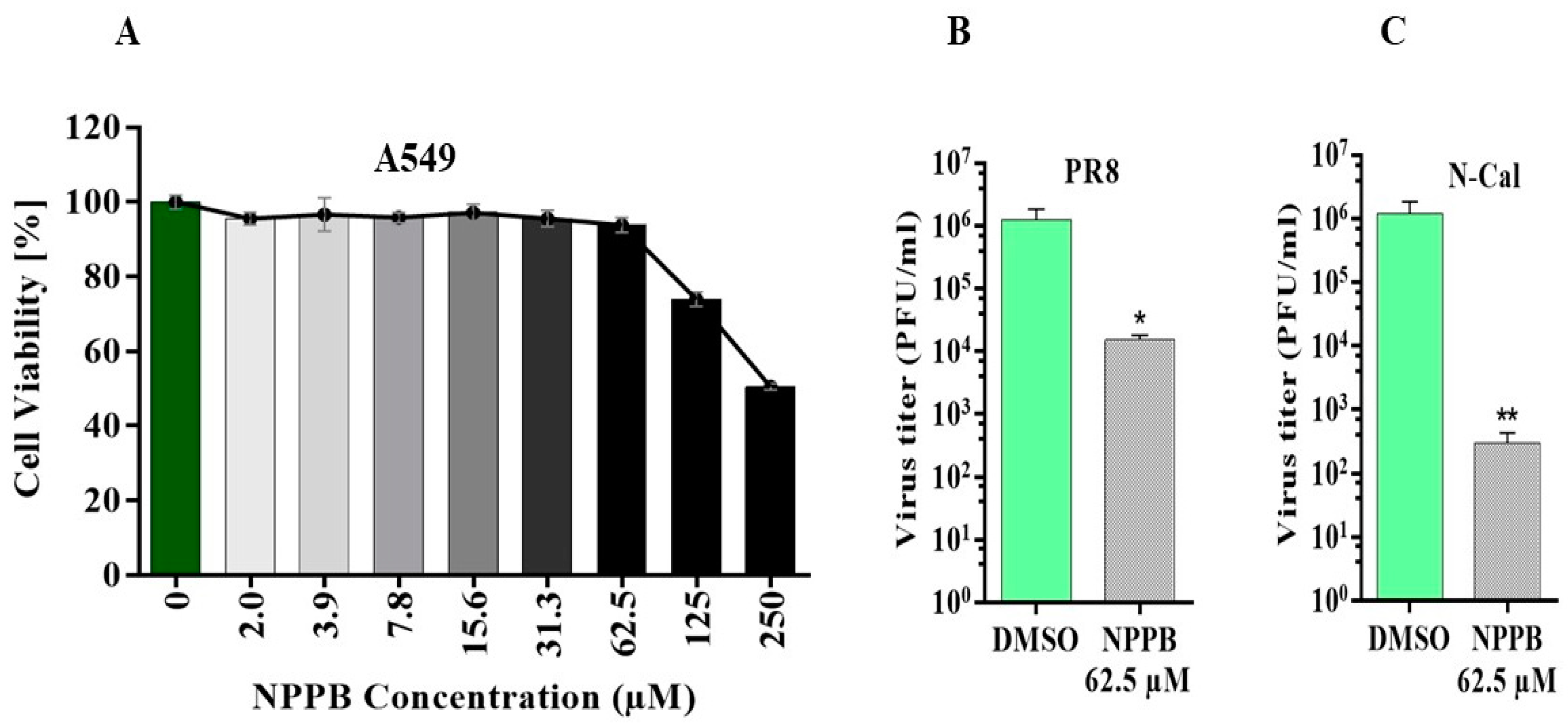
3.3. The Chloride Channel Inhibitor NPPB Suppresses the Replication of IAV Virus
3.4. Impact of CLIC1 KD on Viral Protein and RNA Expression
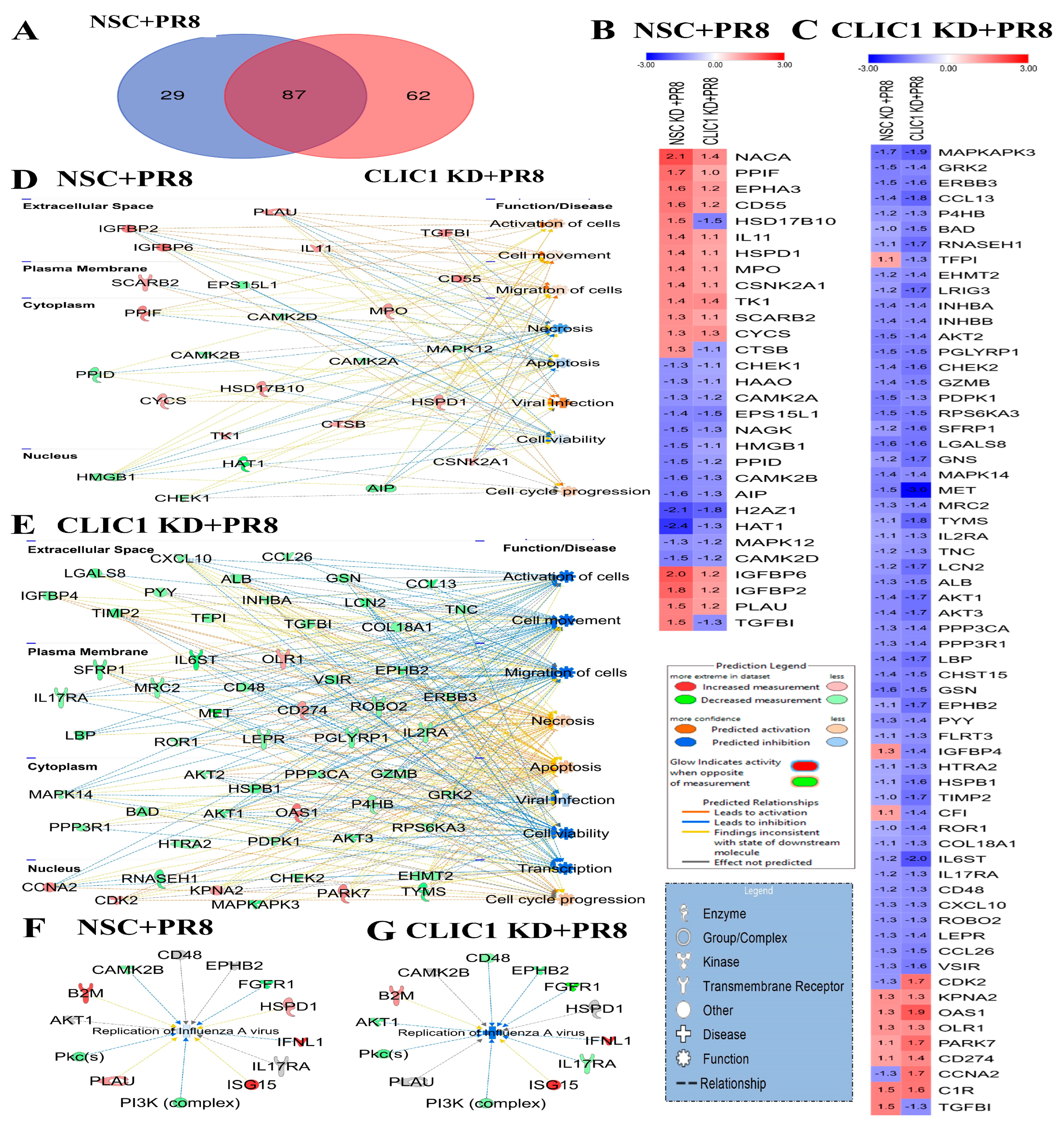
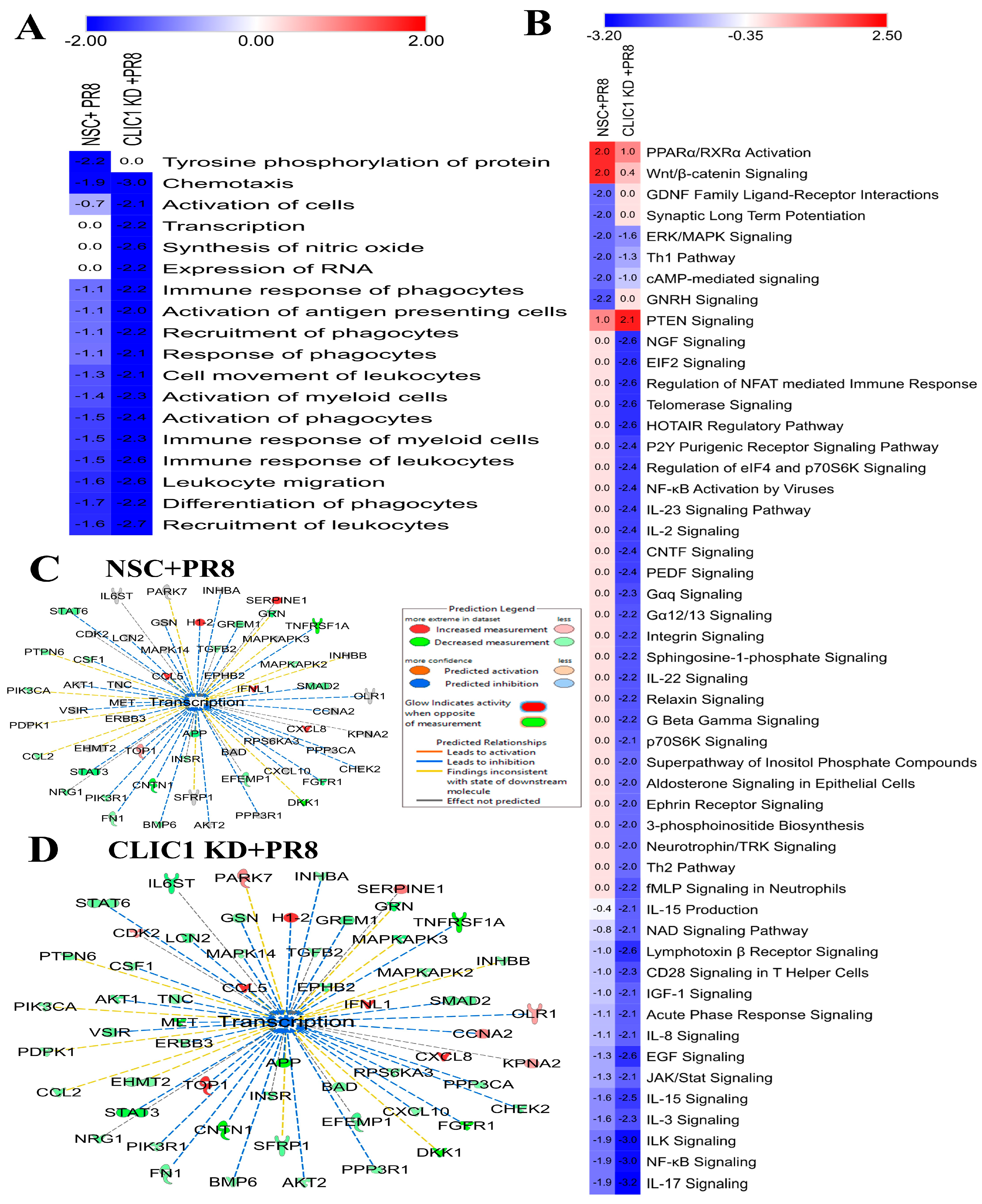
3.5. Proteomic Dysregulation Caused by CLIC1 KD during IAV Infection
4. Discussion
4.1. CLIC1 Knockdown Alters IAV-Mediated Host Proteomic Responses
4.2. CLIC1 Knockdown Cause Differential Regulation of Cellular Functions and Signaling Pathways Dysregulated by IAV Infection
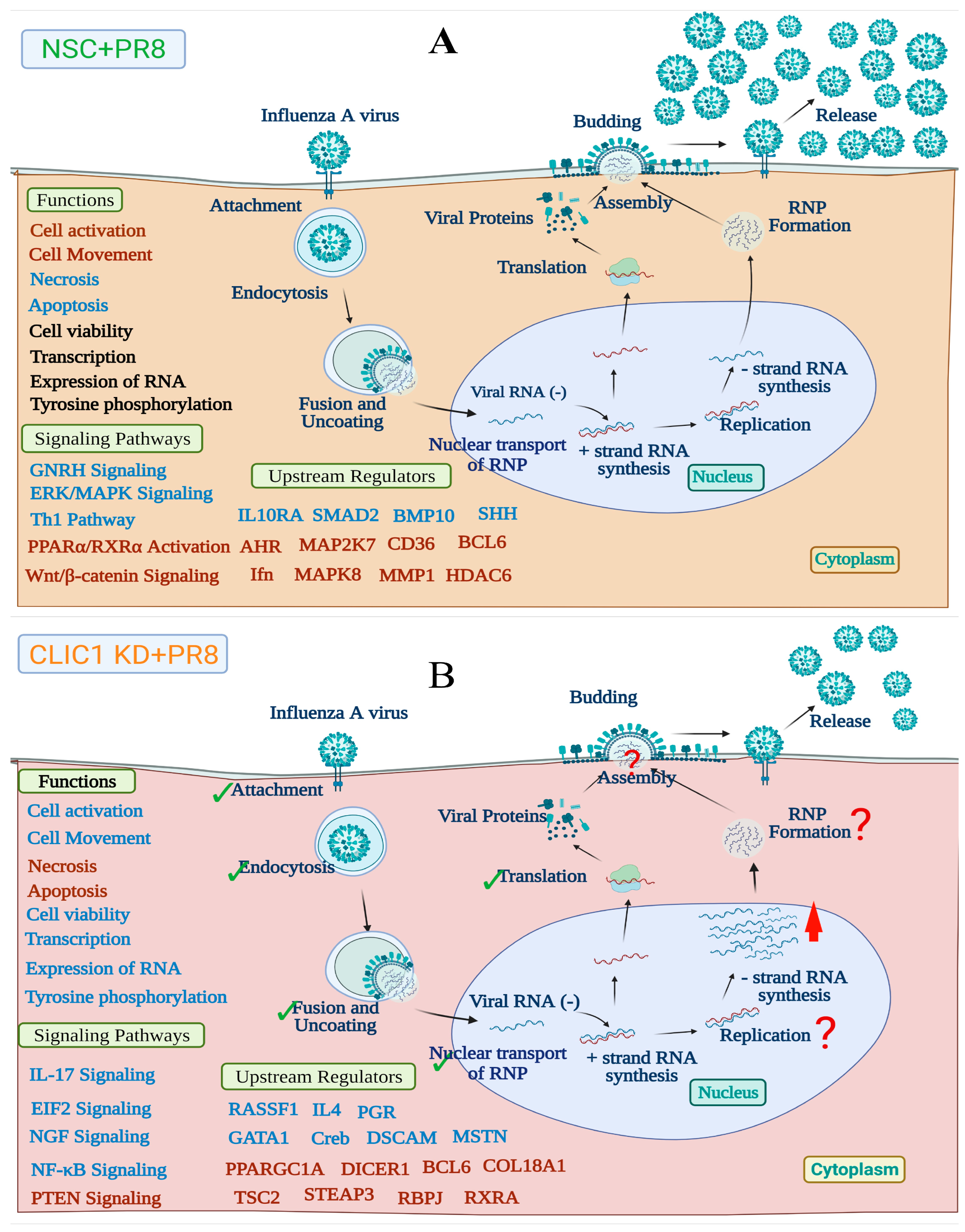
4.3. CLIC1 Is Important for Later Stages of IAV Replication
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Glezen, W.P. Prevention and Treatment of Seasonal Influenza. N. Engl. J. Med. 2008, 359, 2579–2585. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. Influenza: The once and future pandemic. Public Health Rep. 2010, 125 (Suppl. S3), 15–26. [Google Scholar] [CrossRef]
- Johnson, N.P.A.S.; Mueller, J. Updating the accounts: Global mortality of the 1918–1920 ‘Spanish’ influenza pandemic. Bull. Hist. Med. 2002, 76, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.L.; Palese, P. Orthomyxoviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Cohen, J.I., Griffin, D.E., Lamb, R.A., Martin, M.A., Racaniello, V.R., Roizman, B., Eds.; One. Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef]
- Sorrell, E.M.; Ramirez-Nieto, G.C.; Gomez-Osorio, I.G.; Perez, D.R. Genesis of pandemic influenza. Cytogenet. Genome Res. 2007, 117, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Simon, P.F.; McCorrister, S.; Hu, P.; Chong, P.; Silaghi, A.; Westmacott, G.; Coombs, K.M.; Kobasa, D. Highly pathogenic H5N1 and novel H7N9 influenza A viruses induce more profound proteomic host responses than seasonal and pandemic H1N1 strains. J. Proteome Res. 2015, 14, 4511–4523. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Gao, A.; Coombs, K.M. Influenza A Virus Uses PSMA2 for Downregulation of the NRF2-Mediated Oxidative Stress Response. J. Virol. 2022, 96, e0199021. [Google Scholar] [CrossRef]
- Rousseau, E.; Michaud, C.; Lefebvre, D.; Proteau, S.; Decrouy, A. Reconstitution of ionic channels from inner and outer membranes of mammalian cardiac nuclei. Biophys. J. 1996, 70, 703–714. [Google Scholar] [CrossRef]
- Peretti, M.; Angelini, M.; Savalli, N.; Florio, T.; Yuspa, S.H.; Mazzanti, M. Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim. Biophys. Acta 2015, 1848 Pt B, 2523–2531. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, C.; Yu, P.; Tang, B.; Liu, T.; Cui, H.; Xu, J. Regulation of colon cancer cell migration and invasion by CLIC1-mediated RVD. Mol. Cell. Biochem. 2012, 365, 313–321. [Google Scholar] [CrossRef]
- Tulk, B.M.; Schlesinger, P.H.; Kapadia, S.A.; Edwards, J.C. CLIC-1 functions as a chloride channel when expressed and purified from bacteria. J. Biol. Chem. 2000, 275, 26986–26993. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, S.M.; Martin, D.K.; Por, S.B.; Robbins, J.M.; Warton, K.; Bootcov, M.R.; Schofield, P.R.; Campbell, T.J.; Breit, S.N. Molecular cloning and expression of a chloride ion channel of cell nuclei. J. Biol. Chem. 1997, 272, 12575–12582. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Wu, C.C.; Chang, K.P.; Yu, J.S.; Chang, Y.C.; Liao, P.C. Cell secretome analysis using hollow fiber culture system leads to the discovery of CLIC1 protein as a novel plasma marker for nasopharyngeal carcinoma. J. Proteome Res. 2009, 8, 5465–5474. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, X.; Wang, W.; Shao, W.; Li, L.; Yin, W.; Xiu, L.; Mo, M.; Zhao, J.; He, Q.; et al. The expression and clinical significance of CLIC1 and HSP27 in lung adenocarcinoma. Tumour. Biol. 2011, 32, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.G.; Patel, N.J.; Singh, H. Intracellular Chloride Channels: Novel Biomarkers in Diseases. Front. Physiol. 2020, 11, 96. [Google Scholar] [CrossRef]
- Stakaityte, G.; Nwogu, N.; Lippiat, J.D.; Blair, G.E.; Poterlowicz, K.; Boyne, J.R.; Macdonald, A.; Mankouri, J.; Whitehouse, A. The cellular chloride channels CLIC1 and CLIC4 contribute to virus-mediated cell motility. J. Biol. Chem. 2018, 293, 4582–4590. [Google Scholar] [CrossRef]
- Krishnan, M.N.; Ng, A.; Sukumaran, B.; Gilfoy, F.D.; Uchil, P.D.; Sultana, H.; Brass, A.L.; Adametz, R.; Tsui, M.; Qian, F.; et al. RNA interference screen for human genes associated with West Nile virus infection. Nature 2008, 455, 242–245. [Google Scholar] [CrossRef]
- Sivan, G.; Martin, S.E.; Myers, T.G.; Buehler, E.; Szymczyk, K.H.; Ormanoglu, P.; Moss, B. Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3519–3524. [Google Scholar] [CrossRef]
- Coombs, K.M.; Berard, A.; Xu, W.; Krokhin, O.; Meng, X.; Cortens, J.P.; Kobasa, D.; Wilkins, J.; Brown, E.G. Quantitative Proteomic Analyses of Influenza Virus-Infected Cultured Human Lung Cells. J. Virol. 2010, 84, 10888–10906. [Google Scholar] [CrossRef]
- ur Rashid, M.; Coombs, K.M. Serum-reduced media impacts on cell viability and protein expression in human lung epithelial cells. J. Cell Physiol. 2019, 234, 7718–7724. [Google Scholar] [CrossRef]
- Candia, J.; Cheung, F.; Kotliarov, Y.; Fantoni, G.; Sellers, B.; Griesman, T.; Huang, J.; Stuccio, S.; Zingone, A.; Ryan, B.M.; et al. Assessment of Variability in the SOMAscan Assay. Sci. Rep. 2017, 7, 14248. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.; Carter, J.; Cunningham, V.; Dalby, A.; Eaton, B.; et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef]
- Brody, E.N.; Gold, L.; Lawn, R.M.; Walker, J.J.; Zichi, D. High-content affinity-based proteomics: Unlocking protein biomarker discovery. Expert Rev. Mol. Diagn. 2010, 10, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.-U.; Zahedi-Amiri, A.; Glover, K.K.; Gao, A.; Nickol, M.E.; Kindrachuk, J.; Wilkins, J.A.; Coombs, K.M. Zika virus dysregulates human sertoli cell proteins involved in spermatogenesis with little effect on tight junctions. PLoS Negl. Trop. Dis. 2020, 14, e0008335. [Google Scholar] [CrossRef]
- Rahim, M.N.; Selman, M.; Sauder, P.J.; Forbes, N.E.; Stecho, W.; Xu, W.; Lebar, M.; Brown, E.G.; Coombs, K.M. Generation and characterization of a new panel of broadly reactive anti-NS1 mabs for detection of influenza A virus. J. Gen. Virol. 2013, 94, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, I.; Fernando, J.; Mainez, J.; Sancho, P. TGF-beta signaling in cancer treatment. Curr. Pharm. Des. 2014, 20, 2934–2947. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, S.; Wang, N.; Guan, L.; Shao, C.; Lin, Y.; Liu, J.; Li, Y. High Expression of TGF-β1 Contributes to Hepatocellular Carcinoma Prognosis via Regulating Tumor Immunity. Front. Oncol. 2022, 12, 861601. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, H.; Faghihloo, E. Viruses as key modulators of the TGF-β pathway; a double-edged sword involved in cancer. Rev. Med. Virol. 2018, 28, e1967. [Google Scholar] [CrossRef]
- Li, N.; Ren, A.; Wang, X.; Fan, X.; Zhao, Y.; Gao, G.F.; Cleary, P.; Wang, B. Influenza viral neuraminidase primes bacterial coinfection through TGF-β–mediated expression of host cell receptors. Proc. Natl. Acad. Sci. USA 2015, 112, 238–243. [Google Scholar] [CrossRef]
- Jolly, L.; Stavrou, A.; Vanderstoken, G.; Meliopoulos, V.A.; Habgood, A.; Tatler, A.L.; Porte, J.; Knox, A.; Weinreb, P.; Violette, S.; et al. Influenza Promotes Collagen Deposition via αvβ6 Integrin-mediated Transforming Growth Factor β Activation. J. Biol. Chem. 2014, 289, 35246. [Google Scholar] [CrossRef]
- Li, D.; Xiao, Z.W.; Ding, J.; Yu, J.P. NACA as a Potential Cellular Target of Hepatitis B Virus PreS1 Protein. Dig. Dis. Sci. 2005, 50, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Feng, J.; Liu, Y.; Zhao, M.; Yuan, Y.; Yuan, H.; Yun, H.; Sun, M.; Bu, Y.; Liu, L.; et al. HAT1 signaling confers to assembly and epigenetic regulation of HBV cccDNA minichromosome. Theranostics 2019, 9, 7345. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Wang, Y.; Chen, D.; Wang, Y.; Wang, R.; Gong, D.; He, H.; Rock, D.L.; Hao, W.; Luo, S. Identification of host cellular proteins LAGE3 and IGFBP6 that interact with orf virus protein ORFV024. Gene 2018, 661, 60–67. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Q.; Jin, Y.; Yang, Q. Antiviral activity of interleukin-11 as a response to porcine epidemic diarrhea virus infection. Vet. Res. 2019, 50, 111. [Google Scholar] [CrossRef] [PubMed]
- König, R.; Stertz, S.; Zhou, Y.; Inoue, A.; Hoffmann, H.H.; Bhattacharyya, S.; Alamares, J.G.; Tscherne, D.M.; Ortigoza, M.B.; Liang, Y.; et al. Human host factors required for influenza virus replication. Nature 2010, 463, 813–817. [Google Scholar] [CrossRef]
- Ween, M.P.; Oehler, M.K.; Ricciardelli, C. Transforming growth factor-beta-induced protein (TGFBI)/(βig-H3): A matrix protein with dual functions in ovarian cancer. Int. J. Mol. Sci. 2012, 13, 10461–10477. [Google Scholar] [CrossRef]
- Wickenhagen, A.; Sugrue, E.; Lytras, S.; Kuchi, S.; Noerenberg, M.; Turnbull, M.L.; Loney, C.; Herder, V.; Allan, J.; Jarmson, I.; et al. A prenylated dsRNA sensor protects against severe COVID-19. Science 2021, 374, eabj3624. [Google Scholar] [CrossRef]
- Herrmann, C.H.; Carroll, R.G.; Wei, P.; Jones, K.A.; Rice, A.P. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 1998, 72, 9881–9888. [Google Scholar] [CrossRef]
- Yan, Y.; Tang, Y.D.; Zheng, C. When cyclin-dependent kinases meet viral infections, including SARS-CoV-2. J. Med. Virol. 2022, 94, 2962–2968. [Google Scholar] [CrossRef]
- Su, M.; Chen, Y.; Qi, S.; Shi, D.; Feng, L.; Sun, D. A Mini-Review on Cell Cycle Regulation of Coronavirus Infection. Front. Vet. Sci. 2020, 7, 943. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Marrero, M.C.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Salinas, L.; Verdugo-Rodriguez, A.; Rodriguez, L.L.; Borca, M.V. The role of interleukin 6 during viral infections. Front. Microbiol. 2019, 10, 1057. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Salinas, L.; Pauszek, S.J.; Stenfeldt, C.; O’Hearn, E.S.; Pacheco, J.M.; Borca, M.V.; Verdugo-Rodriguez, A.; Arzt, J.; Rodriguez, L.L. Increased virulence of an epidemic strain of vesicular stomatitis virus is associated with interference of the innate response in pigs. Front. Microbiol. 2018, 9, 1891. [Google Scholar] [CrossRef] [PubMed]
- Mamidi, P.; Nayak, T.K.; Kumar, A.; Kumar, S.; Chatterjee, S.; De, S.; Datey, A.; Ghosh, S.; Keshry, S.S.; Singh, S.; et al. MK2a inhibitor CMPD1 abrogates chikungunya virus infection by modulating actin remodeling pathway. PLoS Pathog. 2021, 17, e1009667. [Google Scholar] [CrossRef]
- Mosavi, S.Z.; Shahsavandi, S.; Ebrahimi, M.M.; Hatami, A.R.; Sadeghi, K.; Shahivandi, H. Necrotic Response to Low Pathogenic H9N2 Influenza Virus in Chicken Hepatoma Cells. Jundishapur. J. Microbiol. 2015, 8, e13770. [Google Scholar] [CrossRef]
- Teifke, J.P.; Klopfleisch, R.; Globig, A.; Starick, E.; Hoffmann, B.; Wolf, P.U.; Beer, M.; Mettenleiter, T.C.; Harder, T.C. Pathology of natural infections by H5N1 highly pathogenic avian influenza virus in mute (Cygnus olor) and whooper (Cygnus cygnus) swans. Vet. Pathol. 2007, 44, 137–143. [Google Scholar] [CrossRef]
- Chen, W.; Calvo, P.A.; Malide, D.; Gibbs, J.; Schubert, U.; Bacik, I.; Basta, S.; O’Neill, R.; Schickli, J.; Palese, P.; et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001, 7, 1306–1312. [Google Scholar] [CrossRef]
- Schultz-Cherry, S.; Dybdahl-Sissoko, N.; Neumann, G.; Kawaoka, Y.; Hinshaw, V.S. Influenza Virus NS1 Protein Induces Apoptosis in Cultured Cells. J. Virol. 2001, 75, 7875. [Google Scholar] [CrossRef]
- Taylor, H.; Laurence, A.D.J.; Uhlig, H.H. The Role of PTEN in Innate and Adaptive Immunity. Cold Spring Harb. Perspect. Med. 2019, 9, a036996. [Google Scholar] [CrossRef]
- Sahu, U.; Biswas, D.; Prajapati, V.K.; Singh, A.K.; Samant, M.; Khare, P. Interleukin-17-A multifaceted cytokine in viral infections. J. Cell. Physiol. 2021, 236, 8000–8019. [Google Scholar] [CrossRef] [PubMed]
- Verbist, K.C.; Klonowski, K.D. Functions of IL-15 in Anti-Viral Immunity: Multiplicity and Variety. Cytokine 2012, 59, 467. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.-L.; Ziltenert, H.J.; Liew, F.Y. Interleukin-3 protects mice from acute herpes simplex virus infection. Immunology 1990, 71, 358. [Google Scholar] [PubMed]
- Brias, S.G.; Stack, G.; Stacey, M.A.; Redwood, A.J.; Humphreys, I.R. The role of IL-22 in viral infections: Paradigms and paradoxes. Front. Immunol. 2016, 7, 211. [Google Scholar] [CrossRef]
- Murayama, T.; Kuno, K.; Jisaki, F.; Obuchi, M.; Sakamuro, D.; Furukawa, T.; Mukaida, N.; Matsushima, K. Enhancement human cytomegalovirus replication in a human lung fibroblast cell line by interleukin-8. J. Virol. 1994, 68, 7582–7585. [Google Scholar] [CrossRef]
- Khabar, K.S.A.; Al-Zoghaibi, F.; Murayama, T.; Matsushima, K.; Mukaida, N.; Siddiqui, Y.; Dhalla, M.; Al-Ahdal, M.N. Interleukin-8 selectively enhances cytopathic effect (CPE) induced by positive-strand RNA viruses in the human WISH cell line. Biochem. Biophys. Res. Commun. 1997, 235, 774–778. [Google Scholar] [CrossRef]
- Choi, A.M.K.; Jacoby, D.B. Influenza virus A infection induces interleukin-8 gene expression in human airway epithelial cells. FEBS Lett. 1992, 309, 327–329. [Google Scholar] [CrossRef]
- Fiedler, M.A.; Wernke-Dollries, K.; Stark, J.M. Respiratory syncytial virus increases IL-8 gene expression and protein release in A549 cells. Am. J. Physiol. 1995, 269 Pt 1, L865–L872. [Google Scholar] [CrossRef]
- Sheth, R.; Anderson, J.; Sato, T.; Oh, B.; Hempson, S.J.; Rollo, E.; Mackow, E.R.; Shaw, R.D. Rotavirus stimulates IL-8 secretion from cultured epithelial cells. Virology 1996, 221, 251–259. [Google Scholar] [CrossRef]
- Zheng, K.; Chen, M.; Xiang, Y.; Ma, K.; Jin, F.; Wang, X.; Wang, X.; Wang, S.; Wang, Y. Inhibition of herpes simplex virus type 1 entry by chloride channel inhibitors tamoxifen and NPPB. Biochem. Biophys. Res. Commun. 2014, 446, 990–996. [Google Scholar] [CrossRef] [PubMed]
- Malekova, L.; Tomaskova, J.; Novakova, M.; Stefanik, P.; Kopacek, J.; Lakatos, B.; Pastorekova, S.; Krizanova, O.; Breier, A.; Ondrias, K. Inhibitory effect of DIDS, NPPB, and phloretin on intracellular chloride channels. Pflugers. Arch. 2007, 455, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Dolovcak, S.; Waldrop, S.L.; Fitz, J.G.; Kilic, G. 5-Nitro-2-(3-phenylpropylamino)benzoic acid (NPPB) stimulates cellular ATP release through exocytosis of ATP-enriched vesicles. J. Biol. Chem. 2009, 284, 33894–33903. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Bai, Z.; Juan, W.; Li, X.; Chi, B.; Chen, X. ANP32A modulates cell growth by regulating p38 and Akt activity in colorectal cancer. Oncol. Rep. 2017, 38, 1605–1612. [Google Scholar] [CrossRef]
- Carrique, L.; Fan, H.; Walker, A.P.; Keown, J.R.; Sharps, J.; Staller, E.; Barclay, W.S.; Fodor, E.; Grimes, J.M. Host ANP32A mediates the assembly of the influenza virus replicase. Nature 2020, 587, 638–643. [Google Scholar] [CrossRef]
- Hei, Z.; Wu, S.; Liu, Z.; Wang, J.; Fang, P. Retractile lysyl-tRNA synthetase-AIMP2 assembly in the human multi-aminoacyl-tRNA synthetase complex. J. Biol. Chem. 2019, 294, 4775–4783. [Google Scholar] [CrossRef]
- Gao, S.; Wu, J.; Liu, R.Y.; Li, J.; Song, L.; Teng, Y.; Sheng, C.; Liu, D.; Yao, C.; Chen, H.; et al. Interaction of NS2 with AIMP2 facilitates the switch from ubiquitination to SUMOylation of M1 in influenza A virus-infected cells. J. Virol. 2015, 89, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Levy, S. Function of the tetraspanin molecule CD81 in B and T cells. Immunol. Res. 2014, 58, 179–185. [Google Scholar] [CrossRef]
- Tardif, M.R.; Tremblay, M.J. Tetraspanin CD81 provides a costimulatory signal resulting in increased human immunodeficiency virus type 1 gene expression in primary CD4+ T lymphocytes through NF-kappaB, NFAT, and AP-1 transduction pathways. J. Virol. 2005, 79, 4316–4328. [Google Scholar] [CrossRef]
- He, J.; Sun, E.; Bujny, M.V.; Kim, D.; Davidson, M.W.; Zhuang, X. Dual function of CD81 in influenza virus uncoating and budding. PLoS Pathog. 2013, 9, e1003701. [Google Scholar] [CrossRef]
- You, F.; Sun, H.; Zhou, X.; Sun, W.; Liang, S.; Zhai, Z.; Jiang, Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 2009, 10, 1300–1308. [Google Scholar] [CrossRef]
- Rossi, M.; De Laurenzi, V.; Munarriz, E.; Green, D.R.; Liu, Y.C.; Vousden, K.H.; Cesareni, G.; Melino, G. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005, 24, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Landeras-Bueno, S.; Jorba, N.; Pérez-Cidoncha, M.; Ortín, J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 2011, 7, e1002397. [Google Scholar] [CrossRef] [PubMed]
- Shav-Tal, Y.; Zipori, D. PSF and p54nrb/NonO—Multi-functional nuclear proteins. FEBS Lett. 2002, 531, 109–114. [Google Scholar] [CrossRef] [PubMed]


| Range of Fold Change | PR8 (Protein No.) | Total Significant (Protein No.) | CLIC1 KD +PR8 (Protein No.) | Total Significant (Protein No.) |
|---|---|---|---|---|
| and F.C. > 1.00 | 76 | 218 | 213 | 352 |
| and F.C. < 1.00 | 142 | 139 | ||
| and F.C. > 1.10 | 68 | 197 | 88 | 279 |
| and F.C. < −1.10 | 129 | 191 | ||
| and F.C. > 1.20 | 43 | 144 | 31 | 183 |
| and F.C. < −1.20 | 101 | 152 | ||
| and F.C. > 1.30 | 31 | 116 | 22 | 149 |
| and F.C. < −1.30 | 85 | 127 | ||
| and F.C. > 1.50 | 20 | 68 | 18 | 94 |
| and F.C. < −1.50 | 48 | 76 | ||
| and F.C. > 1.60 | 17 | 57 | 16 | 75 |
| and F.C. < −1.60 | 40 | 59 | ||
| and F.C. > 2.00 | 11 | 33 | 6 | 31 |
| and F.C. < −2.00 | 22 | 25 | ||
| and F.C. > 2.50 | 8 | 22 | 6 | 27 |
| and F.C. < −2.50 | 14 | 21 |
| Type(s) | Symbols | Entrez Gene Name | NSC + PR8 (FC) | p-Value | CLIC1 KD + PR8 (FC) | p-Value | Location |
|---|---|---|---|---|---|---|---|
| Cytokines | CXCL8 | C-X-C motif chemokine ligand 8 | 10.27 | 2.21 × 10−5 | 5.74 | 1.97 × 10−4 | Extracellular Space |
| CCL5 | C-C motif chemokine ligand 5 | 6.39 | 3.59 × 10−3 | 9.88 | 1.05 × 10−3 | Extracellular Space | |
| IFNL1 | interferon lambda 1 | 2.87 | 3.46 × 10−2 | 4.36 | 6.17 × 10−3 | Extracellular Space | |
| LTA | lymphotoxin alpha | −1.50 | 2.96 × 10−3 | −1.53 | 1.45 × 10−2 | Extracellular Space | |
| IL17D | interleukin 17D | −1.43 | 2.76 × 10−2 | −1.52 | 1.70 × 10−3 | Extracellular Space | |
| CCL2 | C-C motif chemokine ligand 2 | −1.40 | 4.31 × 10−2 | −1.54 | 5.80 × 10−3 | Extracellular Space | |
| CCL13 | C-C motif chemokine ligand 13 | −1.42 | 9.43 × 10−2 | −1.82 | 4.77 × 10−2 | Extracellular Space | |
| Enzymes | PPIF | peptidylprolyl isomerase F | 1.73 | 2.93 × 10−2 | 1.04 | 5.15 × 10−1 | Cytoplasm |
| TOP1 | DNA topoisomerase I | 1.50 | 1.82 × 10−3 | 2.99 | 4.83 × 10−2 | Nucleus | |
| EFEMP1 | EGF containing fibulin extracellular matrix protein 1 | −1.50 | 1.75 × 10−2 | −1.71 | 5.69 × 10−3 | Extracellular Space | |
| PPID | peptidylprolyl isomerase D | −1.53 | 1.02 × 10−2 | −1.15 | 4.87 × 10−1 | Cytoplasm | |
| AKR1A1 | aldo-keto reductase family 1 member A1 | −1.72 | 3.82 × 10−2 | −1.54 | 3.50 × 10−2 | Cytoplasm | |
| HAT1 | histone acetyltransferase 1 | −2.37 | 2.10 × 10−4 | −1.34 | 1.69 × 10−1 | Nucleus | |
| CNTN1 | contactin 1 | −2.93 | 1.58 × 10−2 | −4.07 | 9.07 × 10−4 | Plasma Membrane | |
| OAS1 | 2′-5′-oligoadenylate synthetase 1 | 1.28 | 1.53 × 10−1 | 1.95 | 9.62 × 10−3 | Cytoplasm | |
| PARK7 | Parkinsonism associated deglycase | 1.10 | 4.97 × 10−1 | 1.72 | 3.38 × 10−2 | Nucleus | |
| FN1 | fibronectin 1 | −1.32 | 6.79 × 10−3 | −1.50 | 8.95 × 10−3 | Extracellular Space | |
| CHST15 | carbohydrate sulfotransferase 15 | −1.40 | 1.78 × 10−1 | −1.51 | 3.64 × 10−2 | Plasma Membrane | |
| CA7 | carbonic anhydrase 7 | −1.40 | 3.24 × 10−2 | −1.52 | 7.96 × 10−3 | Cytoplasm | |
| ENTPD5 | ectonucleoside triphosphate diphosphohydrolase 5 (inactive) | −1.49 | 4.88 × 10−3 | −1.61 | 4.17 × 10−3 | Cytoplasm | |
| GNS | glucosamine (N-acetyl)−6-sulfatase | −1.22 | 7.69 × 10−2 | −1.68 | 3.01 × 10−3 | Cytoplasm | |
| RNASEH1 | ribonuclease H1 | −1.13 | 5.80 × 10−1 | −1.74 | 4.10 × 10−2 | Nucleus | |
| TYMS | thymidylate synthetase | −1.13 | 1.21 × 10−1 | −1.82 | 2.42 × 10−3 | Nucleus | |
| GPNMB | glycoprotein nmb | −1.40 | 5.17 × 10−3 | −2.20 | 3.57 × 10−3 | Plasma Membrane | |
| Growth factors | BMP6 | bone morphogenetic protein 6 | −1.56 | 3.23 × 10−2 | −1.42 | 4.85 × 10−2 | Extracellular Space |
| NRG1 | neuregulin 1 | −1.61 | 4.73 × 10−2 | −1.63 | 8.98 × 10−3 | Plasma Membrane | |
| FGF6 | fibroblast growth factor 6 | −1.63 | 5.64 × 10−3 | −1.65 | 3.08 × 10−3 | Extracellular Space | |
| GRN | granulin precursor | −2.09 | 4.60 × 10−3 | −2.51 | 5.74 × 10−3 | Extracellular Space | |
| DKK1 | dickkopf WNT signaling pathway inhibitor 1 | −3.61 | 5.46 × 10−3 | −5.88 | 9.57 × 10−5 | Extracellular Space | |
| Kinases | EPHA2 | EPH receptor A2 | 3.00 | 4.71 × 10−3 | 1.74 | 2.53 × 10−2 | Plasma Membrane |
| STC1 | stanniocalcin 1 | 2.40 | 2.88 × 10−3 | 1.54 | 1.69 × 10−3 | Extracellular Space | |
| EPHA3 | EPH receptor A3 | 1.60 | 2.03 × 10−2 | 1.21 | 7.23 × 10−2 | Plasma Membrane | |
| NAGK | N-acetylglucosamine kinase | −1.50 | 3.50 × 10−2 | −1.30 | 7.65 × 10−2 | Cytoplasm | |
| CAMK2D | calcium/calmodulin dependent protein kinase II delta | −1.50 | 4.20 × 10−2 | −1.24 | 4.63 × 10−2 | Cytoplasm | |
| CAMK2B | calcium/calmodulin dependent protein kinase II beta | −1.55 | 5.96 × 10−3 | −1.31 | 5.47 × 10−2 | Cytoplasm | |
| PRKCG | protein kinase C gamma | −1.61 | 1.95 × 10−2 | −1.72 | 6.67 × 10−3 | Cytoplasm | |
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha | −1.63 | 4.25 × 10−2 | −1.39 | 1.87 × 10−2 | Cytoplasm | |
| PIK3R1 | phosphoinositide-3-kinase regulatory subunit 1 | −1.63 | 4.25 × 10−2 | −1.39 | 1.87 × 10−2 | Cytoplasm | |
| EFNA2 | ephrin A2 | −1.66 | 4.84 × 10−3 | −1.86 | 5.30 × 10−4 | Plasma Membrane | |
| FGFR1 | fibroblast growth factor receptor 1 | −2.07 | 3.29 × 10−3 | −3.77 | 1.83 × 10−3 | Plasma Membrane | |
| CDK2 | cyclin dependent kinase 2 | −1.26 | 2.15 × 10−1 | 1.65 | 3.12 × 10−02 | Nucleus | |
| RPS6KA3 | ribosomal protein S6 kinase A3 | −1.53 | 6.31 × 10−2 | −1.54 | 4.80 × 10−02 | Cytoplasm | |
| ERBB3 | erb-b2 receptor tyrosine kinase 3 | −1.49 | 8.60 × 10−2 | −1.55 | 4.50 × 10−03 | Plasma Membrane | |
| CHEK2 | checkpoint kinase 2 | −1.35 | 5.81 × 10−2 | −1.62 | 1.00 × 10−02 | Nucleus | |
| AKT1 | AKT serine/threonine kinase 1 | −1.37 | 1.55 × 10−1 | −1.69 | 1.96 × 10−2 | Cytoplasm | |
| AKT3 | AKT serine/threonine kinase 3 | −1.37 | 1.55 × 10−1 | −1.69 | 1.96 × 10−2 | Cytoplasm | |
| INSR | insulin receptor | −1.43 | 2.23 × 10−2 | −1.69 | 4.86 × 10−2 | Plasma Membrane | |
| EPHB2 | EPH receptor B2 | −1.08 | 2.10 × 10−1 | −1.71 | 4.86 × 10−3 | Plasma Membrane | |
| MAPKAPK3 | MAPK activated protein kinase 3 | −1.67 | 6.14 × 10−2 | −1.90 | 1.07 × 10−2 | Nucleus | |
| MET | MET proto-oncogene, receptor tyrosine kinase | −1.50 | 9.39 × 10−2 | −2.96 | 4.18 × 10−4 | Plasma Membrane | |
| Peptidase | CTSA | cathepsin A | −2.11 | 4.83 × 10−3 | −3.02 | 2.56 × 10−3 | Cytoplasm |
| C1R | complement C1r | 1.51 | 2.00 × 10−1 | 1.62 | 4.90 × 10−2 | Extracellular Space | |
| ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 | −1.37 | 9.97 × 10−3 | −1.50 | 2.30 × 10−2 | Extracellular Space | |
| PCSK9 | proprotein convertase subtilisin/kexin type 9 | −5.21 | 7.16 × 10−3 | −5.05 | 1.15 × 10−4 | Extracellular Space | |
| Phosphatase | PTPN6 | protein tyrosine phosphatase non-receptor type 6 | −1.54 | 1.17 × 10−3 | −1.55 | 6.93 × 10−3 | Cytoplasm |
| Transcription regulator | NACA | nascent polypeptide associated complex subunit alpha | 2.11 | 1.80 × 10−2 | 1.40 | 9.21 × 10−2 | Cytoplasm |
| HMGB1 | high mobility group box 1 | −1.51 | 1.38 × 10−2 | −1.09 | 6.00 × 10−1 | Nucleus | |
| AIP | aryl hydrocarbon receptor interacting protein | −1.55 | 1.13 × 10−2 | −1.29 | 1.48 × 10−1 | Nucleus | |
| SMAD2 | SMAD family member 2 | −1.62 | 3.48 × 10−2 | −1.56 | 1.27 × 10−2 | Nucleus | |
| STAT6 | signal transducer and activator of transcription 6 | −1.66 | 2.05 × 10−2 | −1.55 | 2.17 × 10−2 | Nucleus | |
| STAT3 | signal transducer and activator of transcription 3 | −2.09 | 7.86 × 10−3 | −2.78 | 3.71 × 10−3 | Nucleus | |
| EIF4EBP2 | eukaryotic translation initiation factor 4E binding protein 2 | −1.44 | 3.52 × 10−2 | −1.53 | 7.05 × 10−3 | Cytoplasm | |
| Transmembrane receptor | TNFRSF10D | TNF receptor superfamily member 10d | 3.63 | 1.02 × 10−2 | 1.82 | 3.01 × 10−4 | Plasma Membrane |
| B2M | beta-2-microglobulin | 2.49 | 1.30 × 10−4 | 1.55 | 2.11 × 10−2 | Plasma Membrane | |
| PLAUR | plasminogen activator, urokinase receptor | 1.87 | 8.83 × 10−3 | 1.60 | 2.69 × 10−3 | Plasma Membrane | |
| KIR2DL4 | killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 4 | −1.70 | 1.01 × 10−2 | −1.85 | 1.94 × 10−3 | Plasma Membrane | |
| MICB | MHC class I polypeptide-related sequence B | −1.74 | 3.03 × 10−2 | −2.85 | 8.76 × 10−4 | Plasma Membrane | |
| GFRA1 | GDNF family receptor alpha 1 | −1.95 | 1.53 × 10−2 | −1.89 | 2.41 × 10−3 | Plasma Membrane | |
| NRP1 | neuropilin 1 | −2.15 | 1.80 × 10−2 | −3.11 | 2.71 × 10−4 | Plasma Membrane | |
| TNFRSF21 | TNF receptor superfamily member 21 | −2.52 | 8.55 × 10−4 | −3.49 | 8.35 × 10−4 | Plasma Membrane | |
| RTN4R | reticulon 4 receptor | −2.98 | 1.40 × 10−2 | −2.77 | 2.31 × 10−4 | Plasma Membrane | |
| TNFRSF1A | TNF receptor superfamily member 1A | −3.92 | 5.43 × 10−3 | −3.45 | 2.82 × 10−3 | Plasma Membrane | |
| PGLYRP1 | peptidoglycan recognition protein 1 | −1.47 | 5.55 × 10−2 | −1.52 | 2.06 × 10−2 | Plasma Membrane | |
| SFRP1 | secreted frizzled related protein 1 | −1.21 | 6.59 × 10−2 | −1.61 | 9.71 × 10−3 | Plasma Membrane | |
| RELT | RELT TNF receptor | −1.49 | 2.12 × 10−2 | −1.61 | 1.26 × 10−2 | Plasma Membrane | |
| PLXNB2 | plexin B2 | −1.33 | 3.82 × 10−2 | −1.73 | 4.14 × 10−3 | Plasma Membrane | |
| IL6ST | interleukin 6 cytokine family signal transducer | −1.21 | 5.44 × 10−3 | −2.01 | 3.57 × 10−3 | Plasma Membrane | |
| Transporter | ATP5PO | ATP synthase peripheral stalk subunit OSCP | 1.59 | 2.67 × 10−02 | 1.76 | 1.59 × 10−2 | Cytoplasm |
| BPI | bactericidal permeability increasing protein | −1.61 | 4.50 × 10−03 | −1.82 | 3.75 × 10−3 | Plasma Membrane | |
| SNX4 | sorting nexin 4 | −1.61 | 2.04 × 10−02 | −1.90 | 4.02 × 10−2 | Cytoplasm | |
| LBP | lipopolysaccharide binding protein | −1.42 | 1.72 × 10−1 | −1.68 | 1.66 × 10−3 | Plasma Membrane | |
| MCL1 | MCL1 apoptosis regulator, BCL2 family member | −1.40 | 4.72 × 10−2 | −1.69 | 1.97 × 10−3 | Cytoplasm | |
| LCN2 | lipocalin 2 | −1.23 | 1.24 × 10−1 | −1.70 | 1.24 × 10−3 | Extracellular Space | |
| Other | ISG15 | ISG15 ubiquitin like modifier | 7.21 | 4.52 × 10−3 | 9.38 | 6.99 × 10−4 | Extracellular Space |
| SERPINE1 | serpin family E member 1 | 5.98 | 3.82 × 10−3 | 1.85 | 9.03 × 10−3 | Extracellular Space | |
| H1-2 | H1.2 linker histone, cluster member | 2.77 | 7.98 × 10−3 | 6.59 | 1.82 × 10−2 | Nucleus | |
| CST3 | cystatin C | 1.97 | 3.32 × 10−3 | 1.42 | 3.29 × 10−2 | Extracellular Space | |
| IGFBP6 | insulin like growth factor binding protein 6 | 1.96 | 5.31 × 10−4 | 1.24 | 1.91 × 10−2 | Extracellular Space | |
| IGFBP2 | insulin like growth factor binding protein 2 | 1.81 | 2.12 × 10−2 | 1.17 | 6.36 × 10−3 | Extracellular Space | |
| CD55 | CD55 molecule (Cromer blood group) | 1.59 | 5.56 × 10−4 | 1.17 | 2.10 × 10−1 | Plasma Membrane | |
| TGFBI | transforming growth factor beta induced | 1.51 | 1.41 × 10−2 | −1.35 | 1.08 × 10−3 | Extracellular Space | |
| GREM1 | gremlin 1, DAN family BMP antagonist | −1.54 | 1.05 × 10−2 | −1.67 | 9.10 × 10−3 | Extracellular Space | |
| SLITRK5 | SLIT and NTRK like family member 5 | −1.56 | 1.40 × 10−2 | −1.40 | 2.51 × 10−2 | Plasma Membrane | |
| RSPO2 | R-spondin 2 | −1.65 | 1.75 × 10−2 | −1.86 | 5.72 × 10−3 | Extracellular Space | |
| MICA | MHC class I polypeptide-related sequence A | −1.77 | 4.92 × 10−2 | −2.79 | 8.23 × 10−3 | Plasma Membrane | |
| CFH | complement factor H | −1.84 | 5.70 × 10−3 | −1.87 | 8.71 × 10−3 | Extracellular Space | |
| APP | amyloid beta precursor protein | −1.88 | 1.78 × 10−2 | −3.22 | 2.95 × 10−3 | Plasma Membrane | |
| MFGE8 | milk fat globule EGF and factor V/VIII domain containing | −2.01 | 4.16 × 10−2 | −2.02 | 4.08 × 10−3 | Extracellular Space | |
| H2AZ1 | H2A.Z variant histone 1 | −2.08 | 1.21 × 10−2 | −1.84 | 6.41 × 10−2 | Nucleus | |
| AMIGO2 | adhesion molecule with Ig like domain 2 | −2.62 | 1.93 × 10−2 | −2.54 | 7.72 × 10−3 | Plasma Membrane | |
| KIF23 | kinesin family member 23 | −2.63 | 8.74 × 10−3 | −1.68 | 7.04 × 10−3 | Cytoplasm | |
| SERPINE2 | serpin family E member 2 | −2.65 | 1.65 × 10−4 | −2.29 | 2.28 × 10−4 | Extracellular Space | |
| DKK4 | dickkopf WNT signaling pathway inhibitor 4 | −3.62 | 8.22 × 10−3 | −4.96 | 9.37 × 10−5 | Extracellular Space | |
| UNC5D | unc-5 netrin receptor D | −4.32 | 9.53 × 10−4 | −4.81 | 2.97 × 10−4 | Plasma Membrane | |
| LAMA1 | laminin subunit alpha 1 | −4.38 | 1.17 × 10−3 | −6.45 | 7.32 × 10−4 | Extracellular Space | |
| LAMB1 | laminin subunit beta 1 | −4.38 | 1.17 × 10−3 | −6.45 | 7.32 × 10−4 | Extracellular Space | |
| LAMC1 | laminin subunit gamma 1 | −4.38 | 1.17 × 10−3 | −6.45 | 7.32 × 10−4 | Extracellular Space | |
| CCNA2 | cyclin A2 | −1.26 | 2.15 × 10−1 | 1.65 | 3.12 × 10−2 | Nucleus | |
| GSN | gelsolin | −1.61 | 1.79 × 10−1 | −1.50 | 4.78 × 10−2 | Extracellular Space | |
| COLEC11 | collectin subfamily member 11 | −1.42 | 2.68 × 10−2 | −1.51 | 1.63 × 10−2 | Extracellular Space | |
| VSIR | V-set immunoregulatory receptor | −1.27 | 2.37 × 10−2 | −1.56 | 3.04 × 10−3 | Plasma Membrane | |
| HSPB1 | heat shock protein family B (small) member 1 | −1.05 | 5.86 × 10−1 | −1.57 | 5.91 × 10−4 | Cytoplasm | |
| BGN | biglycan | −1.39 | 1.10 × 10−2 | −1.59 | 2.28 × 10−2 | Extracellular Space | |
| LGALS8 | galectin 8 | −1.57 | 6.70 × 10−2 | −1.63 | 7.61 × 10−3 | Extracellular Space | |
| TIMP2 | TIMP metallopeptidase inhibitor 2 | −1.03 | 6.93 × 10−1 | −1.69 | 3.88 × 10−3 | Extracellular Space | |
| LRIG3 | leucine rich repeats and immunoglobulin like domains 3 | −1.19 | 5.15 × 10−2 | −1.74 | 1.62 × 10−2 | Extracellular Space | |
| NTN4 | netrin 4 | −1.47 | 8.19 × 10−3 | −1.74 | 7.80 × 10−4 | Extracellular Space |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashid, M.-u.; Coombs, K.M. Chloride Intracellular Channel Protein 1 (CLIC1) Is a Critical Host Cellular Factor for Influenza A Virus Replication. Viruses 2024, 16, 129. https://doi.org/10.3390/v16010129
Rashid M-u, Coombs KM. Chloride Intracellular Channel Protein 1 (CLIC1) Is a Critical Host Cellular Factor for Influenza A Virus Replication. Viruses. 2024; 16(1):129. https://doi.org/10.3390/v16010129
Chicago/Turabian StyleRashid, Mahamud-ur, and Kevin M. Coombs. 2024. "Chloride Intracellular Channel Protein 1 (CLIC1) Is a Critical Host Cellular Factor for Influenza A Virus Replication" Viruses 16, no. 1: 129. https://doi.org/10.3390/v16010129
APA StyleRashid, M.-u., & Coombs, K. M. (2024). Chloride Intracellular Channel Protein 1 (CLIC1) Is a Critical Host Cellular Factor for Influenza A Virus Replication. Viruses, 16(1), 129. https://doi.org/10.3390/v16010129






