Regulatory Non-Coding RNAs during Porcine Viral Infections: Potential Targets for Antiviral Therapy
Abstract
1. Introduction
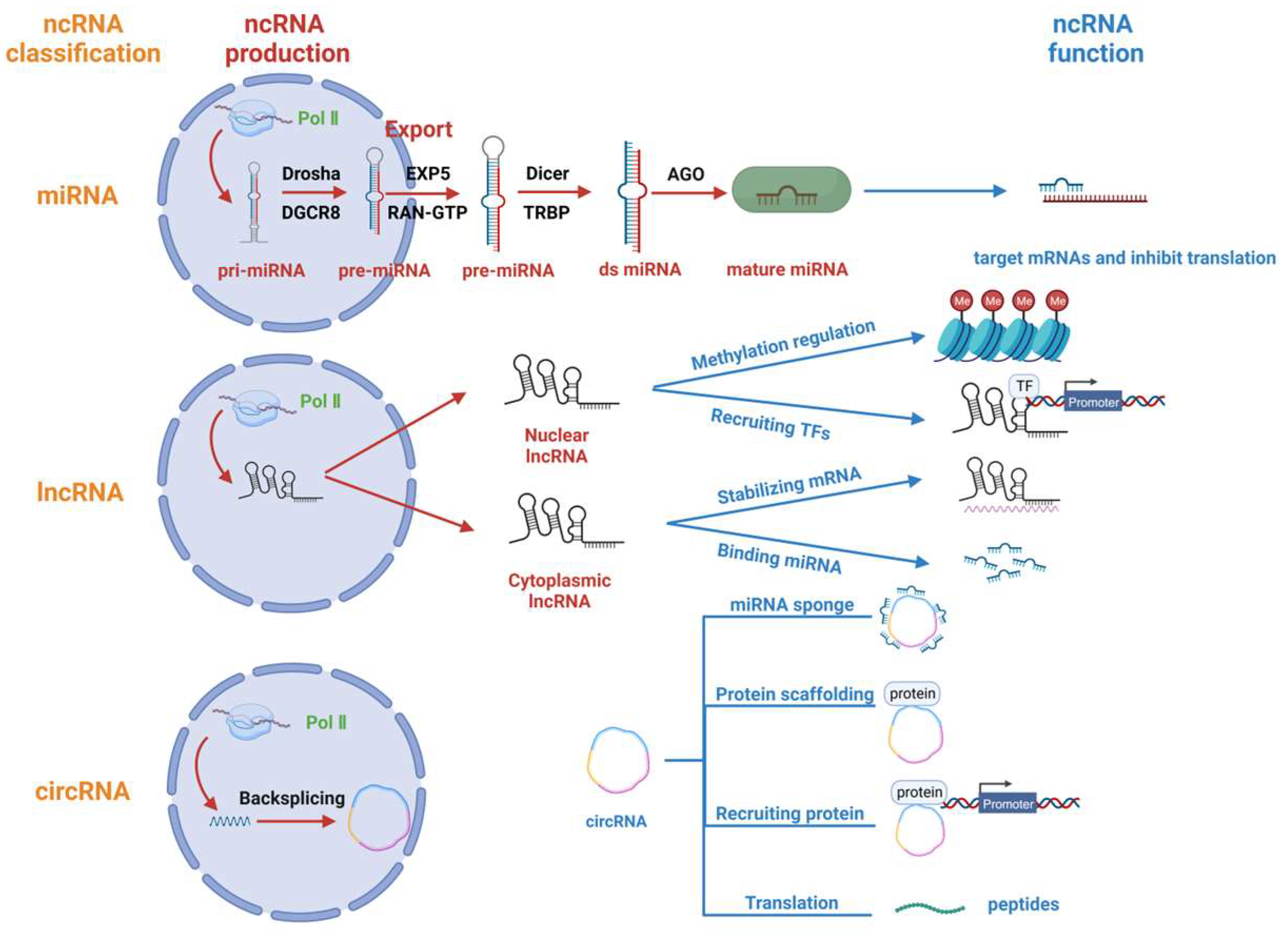
2. Regulatory ncRNA Classification, Production and Function
2.1. miRNA
2.2. lncRNA
2.3. circRNA
3. ncRNA during Porcine Virus Infection
3.1. CSFV
3.2. PEDV
3.2.1. miR-221-5p
3.2.2. miR-615
3.2.3. miRNA-328-3p
3.2.4. miR-let-7e and miR-27b
3.2.5. LncRNA446
3.3. PRRSV
3.3.1. miR-23, miR-378 and miR-505
3.3.2. miR-24-3p
3.3.3. miR-125b
3.3.4. miR-26a
3.3.5. miR-373
3.3.6. miR-22
3.3.7. miR-10a-5p
3.3.8. miR-218
3.3.9. miR-331-3p and miR-210
3.3.10. ssc-miR-27b-3p
3.3.11. miR-c89
3.3.12. ssc-miR-30d-R_1
3.3.13. miRNA let-7 Family
3.3.14. miR-142-5p
3.3.15. miR-181
3.3.16. ssc-miR-124a
3.3.17. miR-204
3.3.18. miR-382-5p
3.3.19. miR-30c
3.3.20. miR-506
3.4. PDCoV
3.5. TGEV
3.5.1. miR-30a-5p
3.5.2. miR-27b
3.5.3. miR-4331
3.6. PRV
3.6.1. LNC_000641
3.6.2. lncA02830
| Virus | ncRNA (Name) | Regulation | Targets | Effect on Virus Infection | Reference |
|---|---|---|---|---|---|
| CSFV | miR-140 | down | Rab25 | inhibit | [53] |
| PEDV | miR-221-5p | up | PEDV 3′UTR; nFKBIA; SOCS1 | inhibit | [61] |
| miR-615 | down | IRAK1 | promote | [66] | |
| miRNA-328-3p | down | ZO-3 | inhibit | [71] | |
| miR-let-7e | up | PEDV N protein | inhibit | [72] | |
| miR-27b | up | HMGB1 3′UTR | inhibit | [72] | |
| lncRNA446 | up | Alix | inhibit | [75] | |
| PRRSV | miR-23 | unchanged | PRRSV 3′UTR | inhibit | [78] |
| miR-378 | unchanged | PRRSV 3′UTR | inhibit | [78] | |
| miR-505 | unchanged | PRRSV 3′UTR | inhibit | [78] | |
| miR-24-3p | up | HO-1 3′UTR | promote | [87] | |
| miR-125b | down | κB-Ras2 | inhibit | [89] | |
| miR-26a | up | unknown | inhibit | [93,94] | |
| miR-373 | up | NFIA, NFIB, IRAK1, IRAK4, and IRF1 | promote | [98] | |
| miR-22 | up | HO-1 3′UTR | promote | [99] | |
| miR-10a-5p | up | SRP14 3′UTR | inhibit | [101] | |
| miR-218 | down | SOCS3 | inhibit | [107] | |
| miR-331-3p | up | PRRSV ORF1b; STAT1 | inhibit | [109] | |
| miR-210 | up | PRRSV ORF1b | inhibit | [109] | |
| ssc-miR-27b-3p | down | unknown | inhibit | [114] | |
| miR-c89 | up | RXRβ | inhibit | [116] | |
| ssc-miR-30d-R_1 | down | TLR4 3′UTR | inhibit | [117] | |
| miRNA let-7 family | difference | PRRSV genomic RNAs; MYH9 | inhibit | [121] | |
| miR-142-5p | up | FAM134B | inhibit | [127] | |
| miR-181 | unchanged | PRRSV RNA; CD163 3′UTR | inhibit | [129] | |
| ssc-miR-124a | down | CD163 mRNA | inhibit | [137] | |
| miR-382-5p | up | HSP60 | promote | [149] | |
| miR-30c | up | JAK; IFNAR2 | promote | [150,151] | |
| miR-506 | unknown | CD151 3′UTR | inhibit | [152] | |
| PDCoV | ssc-miR-30c-3p | up | unknown | inhibit | [156] |
| ssc-miR-374b-3p | up | unknown | inhibit | [156] | |
| TGEV | miR-30a-5p | down | SOCS1; SOCS3 | inhibit | [167] |
| miR-27b | down | SOCS6 | inhibit | [73] | |
| miR-4331 | up | CDCA7 | inhibit | [176] | |
| ssc_circ_009380 | down | miR-22 | undefined | [181] | |
| lncRNA TCONS_00058367 | down | p-p65 | undefined | [180] | |
| PRV | lnc_000641 | up | unknown | promote | [184] |
| lncA02830 | down | unknown | promote | [185] |
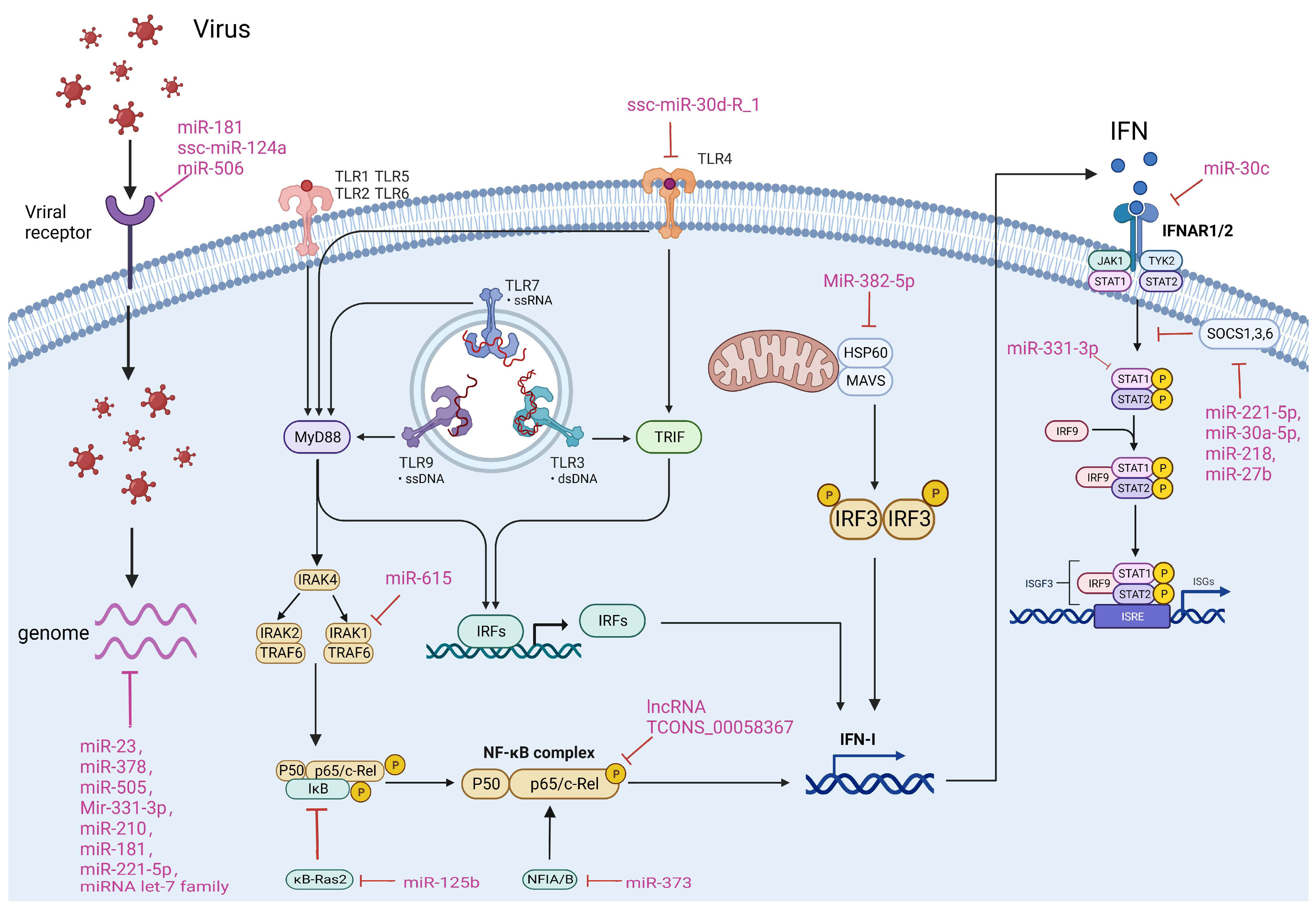
4. The Ways ncRNAs Regulate Viral Infection
4.1. Targeting Viral Genomes
4.2. Targeting Viral Receptors
4.3. Regulating TLR, NF-κB and IFN Pathways
4.4. Targeting Other Cellular Factors
5. amiRNA-Based Antiviral Therapy
6. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Eddy, S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Al-Haidari, A.A.; Syk, I.; Thorlacius, H. MiR-155-5p positively regulates CCL17-induced colon cancer cell migration by targeting RhoA. Oncotarget 2017, 8, 14887–14896. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, X.F. HIF-miR-215-KDM1B promotes glioma-initiating cell adaptation to hypoxia. Cell Cycle 2016, 15, 1939–1940. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, X.F. MiR-215 Is Induced Post-transcriptionally via HIF-Drosha Complex and Mediates Glioma-Initiating Cell Adaptation to Hypoxia by Targeting KDM1B. Cancer Cell 2016, 29, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.N.; Chen, J.; Liu, J.; Luo, Z.G.; Jiang, W.R.; Huang, J.P.; Qiu, Z.B.; Yue, W.J.; Wu, L.J. Expression of microRNA let-7a positively correlates with hepatitis B virus replication in hepatocellular carcinoma tissues. Exp. Biol. Med. 2017, 242, 939–944. [Google Scholar] [CrossRef]
- Takata, A.; Otsuka, M.; Ohno, M.; Kishikawa, T.; Yoshikawa, T. Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci. Rep. 2016, 6, 10. [Google Scholar] [CrossRef]
- Cheng, J.C.; Yeh, Y.J.; Tseng, C.P.; Hsu, S.D.; Chang, Y.L.; Sakamoto, N.; Huang, H.D. Let-7b is a novel regulator of hepatitis C virus replication. Cell. Mol. Life Sci. 2012, 69, 2621–2633. [Google Scholar] [CrossRef]
- Fan, H.X.; Tang, H. Complex interactions between microRNAs and hepatitis B/C viruses. World J. Gastroenterol. 2014, 20, 13477–13492. [Google Scholar] [CrossRef]
- Cheng, M.; Si, Y.H.; Niu, Y.Q.; Liu, X.Y.; Li, X.; Zhao, J.; Jin, Q.; Yang, W. High-Throughput Profiling of Alpha Interferon- and Interleukin-28B-Regulated MicroRNAs and Identification of let-7s with Anti-Hepatitis C Virus Activity by Targeting IGF2BP1. J. Virol. 2013, 87, 9707–9718. [Google Scholar] [CrossRef]
- Sajjad, E.A.; Radkowski, M.; Perkowska-Ptasinska, A.; Pacholczyk, M.; Durlik, M.; Fedorowicz, M.; Pietrzak, R.; Ziarkiewicz-Wróblewska, B.; Wlodarski, P.; Malejczyk, J. Negative Correlation Between Hepatitis C Virus (HCV) and Let-7 MicroRNA Family in Transplanted Livers: The Role of rs868 Single-Nucleotide Polymorphism. Ann. Transplant. 2017, 22, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, S.; Pan, Q.; Blencowe, B.J.; Claycomb, J.M.; Frappier, L. Epstein-Barr Virus EBNA1 Protein Regulates Viral Latency through Effects on let-7 MicroRNA and Dicer. J. Virol. 2014, 88, 11166–11177. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zheng, G.X.; Di, C.H.; Zhang, J.X.; Wang, X.B.; Hong, Y.; Song, Y.; Chen, R.; Yang, Y.; Yan, Y.T.; et al. Latency-associated nuclear antigen inhibits lytic replication of Kaposi’s sarcoma-associated herpesvirus by regulating let-7a/RBPJ signaling. Virology 2019, 531, 69–78. [Google Scholar] [CrossRef]
- Zhang, J.X.; Tan, X.H.; Yuan, Z.; Li, Y.H.; Qi, Y.; Nan, X.; Qi, M.J.; Gao, H.; Lian, F.Z.; Yang, L. Let-7 miRNA silencing promotes Kaposi’s sarcoma-associated herpesvirus lytic replication via activating mitogen-activated protein kinase kinase kinase kinase 4 and its downstream factors. Zhonghua Zhong Liu Za Zhi Chin. J. Oncol. 2016, 38, 485–491. [Google Scholar]
- Shishodia, G.; Verma, G.; Srivastava, Y.; Mehrotra, R.; Das, B.C.; Bharti, A.C. Deregulation of microRNAs Let-7a and miR-21 mediate aberrant STAT3 signaling during human papillomavirus-induced cervical carcinogenesis: Role of E6 oncoprotein. BMC Cancer 2014, 14, 13. [Google Scholar] [CrossRef]
- Shishodia, G.; Shukla, S.; Srivastava, Y.; Masaldan, S.; Mehta, S.; Bhambhani, S.; Sharma, S.; Mehrotra, R.; Das, B.C.; Bharti, A.C. Alterations in microRNAs miR-21 and let-7a correlate with aberrant STAT3 signaling and downstream effects during cervical carcinogenesis. Mol. Cancer 2015, 14, 13. [Google Scholar] [CrossRef]
- Kedkovid, R.; Sirisereewan, C.; Thanawongnuwech, R. Major swine viral diseases: An Asian perspective after the African swine fever introduction. Porc. Health Manag. 2020, 6, 11. [Google Scholar] [CrossRef]
- Peng, R.C.; Wu, L.A.; Wang, Q.L.; Qi, J.X.; Gao, G.F. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021, 46, 848–860. [Google Scholar] [CrossRef]
- Ma, Z.; Damania, B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016, 19, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? In Annual Review of Virology; Enquist, L., DiMaio, D., Demody, T., Eds.; Annual Reviews: Palo Alto, CA, USA, 2019; Volume 6, pp. 567–584. [Google Scholar]
- García-Sastre, A. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 2017, 22, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, Y.; Zhu, B.; Jang, K.J.; Yoo, J.S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 53, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Sun, Q.Q.; Zhang, B.; Zhao, W.; Shen, C.G. The regulation of lncRNAs and miRNAs in SARS-CoV-2 infection. Front. Cell Dev. Biol. 2023, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Pfeffer, S.; Sewer, A.; Lagos-Quintana, M.; Sheridan, R.; Sander, C.; Grässer, F.A.; van Dyk, L.F.; Ho, C.K.; Shuman, S.; Chien, M.C.; et al. Identification of microRNAs of the herpesvirus family. Nat. Methods 2005, 2, 269–276. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Görlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Knight, S.W.; Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001, 293, 2269–2271. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef]
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002, 16, 720–728. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. In Annual Review of Biochemistry; Kornberg, R.D., Ed.; Annual Reviews: Palo Alto, CA, USA, 2012; Volume 81, pp. 145–166. [Google Scholar]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Melé, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef]
- Zuckerman, B.; Ron, M.; Mikl, M.; Segal, E.; Ulitsky, I. Gene Architecture and Sequence Composition Underpin Selective Dependency of Nuclear Export of Long RNAs on NXF1 and the TREX Complex. Mol. Cell 2020, 79, 251–267. [Google Scholar] [CrossRef]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.M.J.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The Human Mitochondrial Transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Orom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.H.; et al. Long Noncoding RNAs with Enhancer-like Function in Human Cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; Laurent, G.S.; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Abe, N.; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y.; et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015, 5, 9. [Google Scholar] [CrossRef]
- Paton, D.J.; McGoldrick, A.; Greiser-Wilke, I.; Parchariyanon, S.; Song, J.Y.; Liou, P.P.; Stadejek, T.; Lowings, J.P.; Björklund, H.; Belák, S. Genetic typing of classical swine fever virus. Vet. Microbiol. 2000, 73, 137–157. [Google Scholar] [CrossRef]
- Xu, P.P.; Jia, S.K.; Wang, K.; Fan, Z.X.; Zheng, H.Q.; Lv, J.M.; Jiang, Y.F.; Hou, Y.F.; Lou, B.H.; Zhou, H.C.; et al. MiR-140 inhibits classical swine fever virus replication by targeting Rab25 in swine umbilical vein endothelial cells. Virulence 2020, 11, 10. [Google Scholar] [CrossRef]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Saif, L.J. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015, 204, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Li, B.X.; Ge, J.W.; Li, Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 2007, 365, 166–172. [Google Scholar] [CrossRef]
- Huang, J.; Lang, Q.; Li, X.; Xu, Z.; Zhu, L.; Zhou, Y. MicroRNA Expression Profiles of Porcine Kidney 15 Cell Line Infected with Porcine Epidemic Diahorrea Virus. Bing Du Xue Bao Chin. J. Virol. 2016, 32, 465–471. [Google Scholar]
- Shi, X.J.; Zhang, Q.; Wang, J.J.; Zhang, Y.T.; Yan, Y.C.; Liu, Y.; Yang, N.L.; Wang, Q.Q.; Xu, X.G. Differential expression analysis of mRNAs, lncRNAs, and miRNAs expression profiles and construction of ceRNA networks in PEDV infection. BMC Genom. 2022, 23, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Q.; Li, C.; Zhang, B.Z.; Li, Z.H.; Zeng, W.; Luo, R.; Cao, J.Y.; Cheng, G.F.; Fan, S.X.; He, Q.G. Differential expression and correlation analysis of miRNA-mRNA profiles in swine testicular cells infected with porcine epidemic diarrhea virus. Sci. Rep. 2021, 11, 14. [Google Scholar] [CrossRef]
- Chen, J.N.; Wang, H.W.; Jin, L.; Wang, L.Y.; Huang, X.; Chen, W.W.; Yan, M.M.; Liu, G.L. Profile analysis of circRNAs induced by porcine endemic diarrhea virus infection in porcine intestinal epithelial cells. Virology 2019, 527, 169–179. [Google Scholar] [CrossRef]
- Zheng, H.Q.; Xu, L.; Liu, Y.Z.; Li, C.; Zhang, L.; Wang, T.; Zhao, D.; Xu, X.G.; Zhang, Y.M. MicroRNA-221-5p Inhibits Porcine Epidemic Diarrhea Virus Replication by Targeting Genomic Viral RNA and Activating the NF-κB Pathway. Int. J. Mol. Sci. 2018, 19, 3381. [Google Scholar] [CrossRef]
- Bai, Y.; Li, J.Y.; Li, J.; Liu, Y.H.; Zhang, B. MiR-615 inhibited cell proliferation and cell cycle of human breast cancer cells by suppressing of AKT2 expression. Int. J. Clin. Exp. Med. 2015, 8, 3801–3808. [Google Scholar]
- Liu, J.T.; Jia, Y.L.; Jia, L.J.; Li, T.T.; Yang, L.; Zhang, G.W. MicroRNA 615-3p Inhibits the Tumor Growth and Metastasis of NSCLC via Inhibiting IGF2. Oncol. Res. 2019, 27, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.Y.; Ge, X.Y.; Gao, Y.; Ren, Y.D.; Ren, X.F.; Li, G.X. Porcine epidemic diarrhea virus infection induces NF-κB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 2015, 96, 1757–1767. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Karin, M. Regulation and Function of NF-κB Transcription Factors in the Immune System. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef]
- Zheng, H.Q.; Li, C.; Zhu, X.F.; Wang, W.X.; Yin, B.Y.; Zhang, W.J.; Feng, S.L.; Yin, X.H.; Huang, H.; Zhang, Y.M. miR-615 facilitates porcine epidemic diarrhea virus replication by targeting IRAK1 to inhibit type III interferon expression. Front. Microbiol. 2022, 13, 14. [Google Scholar] [CrossRef]
- Yin, L.; Shen, X.H.; Yin, D.D.; Wang, J.R.; Zhao, R.H.; Dai, Y.; Pan, X.C. Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses 2022, 14, 806. [Google Scholar] [CrossRef] [PubMed]
- Nighot, P.; Ma, T. Endocytosis of Intestinal Tight Junction Proteins: In Time and Space. Inflamm. Bowel Dis. 2021, 27, 283–290. [Google Scholar] [CrossRef]
- Zong, Q.F.; Huang, Y.J.; Wu, L.S.; Wu, Z.C.; Wu, S.L.; Bao, W.B. Effects of porcine epidemic diarrhea virus infection on tight junction protein gene expression and morphology of the intestinal mucosa in pigs. Pol. J. Vet. Sci. 2019, 22, 345–353. [Google Scholar] [PubMed]
- Luo, X.L.; Guo, L.J.; Zhang, J.; Xu, Y.F.; Gu, W.H.; Feng, L.; Wang, Y. Tight Junction Protein Occludin Is a Porcine Epidemic Diarrhea Virus Entry Factor. J. Virol. 2017, 91, 14. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, J.X.; Wang, Q.; Cui, Z.D.; Li, D.L.; Niu, J.T.; Guo, Y.B.; Zhang, Q.; Zhang, S.; Zhao, Y.L.; et al. Exosomal miRNA-328-3p targets ZO-3 and inhibits porcine epidemic diarrhea virus proliferation. Arch. Virol. 2022, 167, 901–910. [Google Scholar] [CrossRef]
- Liang, J.Q.; Xie, M.Y.; Hou, L.J.; Wang, H.L.; Luo, J.Y.; Sun, J.J.; Xi, Q.Y.; Jiang, Q.Y.; Chen, T.; Zhang, Y.L. miRNAs derived from milk small extracellular vesicles inhibit porcine epidemic diarrhea virus infection. Antivir. Res. 2023, 212, 14. [Google Scholar] [CrossRef]
- Gao, R.; Zhang, Y.; Kang, Y.; Xu, W.; Jiang, L.; Guo, T.; Huan, C. Glycyrrhizin Inhibits PEDV Infection and Proinflammatory Cytokine Secretion via the HMGB1/TLR4-MAPK p38 Pathway. Int. J. Mol. Sci. 2020, 21, 2961. [Google Scholar] [CrossRef]
- Qin, W.; Qi, X.; Xie, Y.; Wang, H.; Wu, S.; Sun, M.-A.; Bao, W. LncRNA446 Regulates Tight Junctions by Inhibiting the Ubiquitinated Degradation of Alix after Porcine Epidemic Diarrhea Virus Infection. J. Virol. 2023, 97, e0188422. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.S.; Gao, J.K.; Zhu, L.Q.; Yang, Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. 2014, 192, 34–45. [Google Scholar] [CrossRef]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.H.; Li, Y.H.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. In Annual Review of Animal Biosciences; Lewin., H.A., Roberts, R.M., Eds.; Annual Reviews: Palo Alto, CA, USA, 2016; Volume 4, pp. 129–154. [Google Scholar]
- Van Reeth, K.; Nauwynck, H.; Pensaert, M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: A clinical and virological study. Vet. Microbiol. 1996, 48, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Guo, X.K.; Gao, L.; Huang, C.; Li, N.; Jia, X.J.; Liu, W.J.; Feng, W.H. MicroRNA-23 inhibits PRRSV replication by directly targeting PRRSV RNA and possibly by upregulating type I interferons. Virology 2014, 450, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.W.; Hall, S.R.; Perrella, M.A. Role of haem oxygenase-1 in microbial host defence. Cell. Microbiol. 2009, 11, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, W.N.; Mathahs, M.M.; Zhu, Z.W. Herne and HO-1 inhibition of HCV, HBV, and HIV. Front. Pharmacol. 2012, 3, 13. [Google Scholar] [CrossRef]
- Zhu, Z.W.; Wilson, A.T.; Mathahs, M.M.; Wen, F.; Brown, K.E.; Luxon, B.A.; Schmidt, W.N. Heme Oxygenase-1 Suppresses Hepatitis C Virus Replication and Increases Resistance of Hepatocytes to Oxidant Injury. Hepatology 2008, 48, 1430–1439. [Google Scholar] [CrossRef]
- Devadas, K.; Dhawan, S. Hemin activation ameliorates HIV-1 infection via heme oxygenase-1 induction. J. Immunol. 2006, 176, 4252–4257. [Google Scholar] [CrossRef]
- Hashiba, T.; Suzuki, M.; Nagashima, Y.; Suzuki, S.; Inoue, S.; Tsubarai, T.; Matsuses, T.; Ishigatubo, Y. Adenovirus-mediated transfer of heme oxygenase-1 cDNA attenuates severe lung injury induced by the influenza virus in mice. Gene Ther. 2001, 8, 1499–1507. [Google Scholar] [CrossRef]
- Xiao, S.Q.; Jia, J.Y.; Mo, D.L.; Wang, Q.W.; Qin, L.M.; He, Z.Y.; Zhao, X.A.; Huang, Y.K.; Li, A.N.; Yu, J.W.; et al. Understanding PRRSV Infection in Porcine Lung Based on Genome-Wide Transcriptome Response Identified by Deep Sequencing. PLoS ONE 2010, 5, 16. [Google Scholar] [CrossRef]
- Wang, L.L.; Xiao, S.Q.; Gao, J.T.; Liu, M.R.; Zhang, X.Y.; Li, M.; Zhao, G.Y.; Mo, D.L.; Liu, X.H.; Chen, Y.S. Inhibition of replication of porcine reproductive and respiratory syndrome virus by hemin is highly dependent on heme oxygenase-1, but independent of iron in MARC-145 cells. Antivir. Res. 2014, 105, 39–46. [Google Scholar] [CrossRef]
- Xiao, S.Q.; Zhang, A.K.; Zhang, C.; Ni, H.B.; Gao, J.M.; Wang, C.B.; Zhao, Q.; Wang, X.P.; Wang, X.; Ma, C.; et al. Heme oxygenase-1 acts as an antiviral factor for porcine reproductive and respiratory syndrome virus infection and over-expression inhibits virus replication in vitro. Antivir. Res. 2014, 110, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.Q.; Wang, X.; Ni, H.B.; Li, N.; Zhang, A.K.; Liu, H.L.; Pu, F.X.; Xu, L.L.; Gao, J.M.; Zhao, Q.; et al. MicroRNA miR-24-3p Promotes Porcine Reproductive and Respiratory Syndrome Virus Replication through Suppression of Heme Oxygenase-1 Expression. J. Virol. 2015, 89, 4494–4503. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.K.; Wang, Z.; Li, G.D. MicroRNA-125 in immunity and cancer. Cancer Lett. 2019, 454, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Cao, L.; Xu, Z.; Fang, L.R.; Zhong, Y.; Chen, Q.G.; Luo, R.; Chen, H.C.; Li, K.; Xiao, S.B. MiR-125b Reduces Porcine Reproductive and Respiratory Syndrome Virus Replication by Negatively Regulating the NF-κB Pathway. PLoS ONE 2013, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.J.; Guyre, P.M.; Pioli, P.A. Estradiol Suppresses NF-κB Activation through Coordinated Regulation of let-7a and miR-125b in Primary Human Macrophages. J. Immunol. 2010, 184, 5029–5037. [Google Scholar] [CrossRef]
- Lee, S.M.; Kleiboeker, S.B. Porcine arterivirus activates the NF-κB pathway through IκB degradation. Virology 2005, 342, 47–59. [Google Scholar] [CrossRef]
- Luo, R.; Xiao, S.B.; Jiang, Y.B.; Jin, H.; Wang, D.; Liu, M.L.; Chen, H.C.; Fang, L.R. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-β production by interfering with the RIG-I signaling pathway. Mol. Immunol. 2008, 45, 2839–2846. [Google Scholar] [CrossRef]
- Li, L.W.; Wei, Z.Z.; Zhou, Y.J.; Gao, F.; Jiang, Y.F.; Yu, L.X.; Zheng, H.; Tong, W.; Yang, S.; Zheng, H.H.; et al. Host miR-26a suppresses replication of porcine reproductive and respiratory syndrome virus by upregulating type I interferons. Virus Res. 2015, 195, 86–94. [Google Scholar] [CrossRef]
- Jia, X.J.; Bi, Y.H.; Li, J.; Xie, Q.; Yang, H.C.; Liu, W.J. Cellular microRNA miR-26a suppresses replication of porcine reproductive and respiratory syndrome virus by activating innate antiviral immunity. Sci. Rep. 2015, 5, 14. [Google Scholar] [CrossRef]
- Wei, F.R.; Cao, C.H.; Xu, X.Q.; Wang, J.F. Diverse functions of miR-373 in cancer. J. Transl. Med. 2015, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.Y.; Liu, H.Y.; Mitchelson, K.; Rao, H.Y.; Luo, M.Y.; Xie, L.; Sun, Y.M.; Zhang, L.; Lu, Y.; Liu, R.Y.; et al. MicroRNAs-372/373 Promote the Expression of Hepatitis B Virus Through the Targeting of Nuclear Factor I/B. Hepatology 2011, 54, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Di Bisceglie, A.M.; Ray, R.B. Hepatitis C Virus-Mediated Enhancement of MicroRNA miR-373 Impairs the JAK/STAT Signaling Pathway. J. Virol. 2015, 89, 3356–3365. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shi, X.B.; Zhang, X.Z.; Wang, A.P.; Wang, L.; Yang, Y.Y.; Deng, R.G.; Zhang, G.P. MicroRNA 373 Facilitates the Replication of Porcine Reproductive and Respiratory Syndrome Virus by Its Negative Regulation of Type I Interferon Induction. J. Virol. 2017, 91, 19. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.Q.; Du, T.F.; Wang, X.; Ni, H.B.; Yan, Y.H.; Li, N.; Zhang, C.; Zhang, A.K.; Gao, J.M.; Liu, H.L.; et al. MiR-22 promotes porcine reproductive and respiratory syndrome virus replication by targeting the host factor HO-1. Vet. Microbiol. 2016, 192, 226–230. [Google Scholar] [CrossRef]
- Li, J.; Chen, Z.S.; Zhao, J.L.; Fang, L.R.; Fang, R.; Xiao, J.; Chen, X.; Zhou, A.; Zhang, Y.Y.; Ren, L.M.; et al. Difference in microRNA expression and editing profile of lung tissues from different pig breeds related to immune responses to HP-PRRSV. Sci. Rep. 2015, 5, 13. [Google Scholar] [CrossRef]
- Zhao, G.W.; Hou, J.Y.; Xu, G.X.; Xiang, A.Q.; Kang, Y.M.; Yan, Y.H.; Zhang, X.B.; Yang, G.S.; Xiao, S.Q.; Sun, S.D. Cellular microRNA miR-10a-5p inhibits replication of porcine reproductive and respiratory syndrome virus by targeting the host factor signal recognition particle 14. J. Gen. Virol. 2017, 98, 624–632. [Google Scholar] [CrossRef]
- Tie, J.; Pan, Y.L.; Zhao, L.N.; Wu, K.C.; Liu, J.; Sun, S.R.; Guo, X.G.; Wang, B.A.L.; Gang, Y.; Zhang, Y.G.; et al. MiR-218 Inhibits Invasion and Metastasis of Gastric Cancer by Targeting the Robo1 Receptor. PLoS Genet. 2010, 6, 11. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Martinez, I.; Gardiner, A.S.; Board, K.F.; Monzon, F.A.; Edwards, R.P.; Khan, S.A. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 2008, 27, 2575–2582. [Google Scholar] [CrossRef] [PubMed]
- Petrocca, F.; Visone, R.; Onelli, M.R.; Shah, M.H.; Nicoloso, M.S.; de Martino, I.; Iliopoulos, D.; Pilozzi, E.; Liu, C.G.; Negrini, M.; et al. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008, 13, 272–286. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Tanaka, T.; Yokota, J.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Res. 2006, 66, 2. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Pan, Y.; Gao, J.X.; Xu, Y.F.; Li, X.; Tian, Z.J.; Chen, H.Y.; Wang, Y. Downregulation of miR-218 by porcine reproductive and respiratory syndrome virus facilitates viral replication via inhibition of type I interferon responses. J. Biol. Chem. 2021, 296, 14. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.J.; Liu, Y.Z.; Li, C.Y.; Qu, X.F.; Zheng, G.F.; Zhang, Q.; Pan, Z.C.; Wang, Y.L.; Rong, J.J. microRNA-331-3p maintains the contractile type of vascular smooth muscle cells by regulating TNF-α and CD14 in intracranial aneurysm. Neuropharmacology 2020, 164, 11. [Google Scholar] [CrossRef]
- You, X.B.; Qu, Y.L.; Zhang, Y.; Huang, J.S.; Gao, X.X.; Huang, C.Y.; Luo, G.; Liu, Q.; Liu, M.; Xu, D.Q. Mir-331-3p Inhibits PRRSV-2 Replication and Lung Injury by Targeting PRRSV-2 ORF1b and Porcine TNF-α. Front. Immunol. 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Narita, K.; Kuwabara, Y.; Fujii, Y. Lung injury after intestinal ischemia-reperfusion may be avoided by the reduced absorption of locally produced cytokines. Surg. Today 2004, 34, 937–942. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef]
- Buck, A.H.; Perot, J.; Chisholm, M.A.; Kumar, D.S.; Tuddenham, L.; Cognat, V.; Marcinowski, L.; Dölken, L.; Pfeffer, S. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA 2010, 16, 307–315. [Google Scholar] [CrossRef]
- Yao, M.; Gao, W.H.; Yang, J.; Liang, X.Y.; Luo, J.B.; Huang, T.H. The regulation roles of miR-125b, miR-221 and miR-27b in porcine Salmonella infection signalling pathway. Biosci. Rep. 2016, 36, 11. [Google Scholar] [CrossRef]
- Wu, J.J.; Ji, Z.Y.; Qiao, M.; Peng, X.W.; Wu, H.Y.; Song, Z.X.; Zhao, H.Z.; Liu, G.S.; Li, F.G.; Mei, S.Q. MicroRNA transcriptome analysis of poly I:C-stimulated and PRRSV-infected porcine alveolar macrophages. J. Appl. Genet. 2019, 60, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kakuta, H. Retinoid X Receptor Antagonists. Int. J. Mol. Sci. 2018, 19, 2354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Feng, Y.T.; Yan, Y.H.; Zheng, Z.F.; Wang, W.J.; Zhang, Y.C.; Zhou, E.M.; Xiao, S.Q. Cellular microRNA miR-c89 inhibits replication of porcine reproductive and respiratory syndrome virus by targeting the host factor porcine retinoid X receptor β. J. Gen. Virol. 2019, 100, 1407–1416. [Google Scholar] [CrossRef]
- Wang, C.M.; Zhang, Y.Y.; Luo, J.; Ding, H.; Liu, S.L.; Amer, S.; Xie, L.; Lyv, W.; Su, W.; Li, M.; et al. Identification of miRNomes reveals ssc-miR-30d-R_1 as a potential therapeutic target for PRRS viral infection. Sci. Rep. 2016, 6, 13. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Letafati, A.; Najafi, S.; Mottahedi, M.; Karimzadeh, M.; Shahini, A.; Garousi, S.; Abbasi-Kolli, M.; Nahand, J.S.; Zadeh, S.S.T.; Hamblin, M.R.; et al. MicroRNA let-7 and viral infections: Focus on mechanisms of action. Cell. Mol. Biol. Lett. 2022, 27, 47. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An Epigenetic Switch Involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 Links Inflammation to Cell Transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef]
- You, X.B.; Liu, M.; Liu, Q.; Li, H.J.; Qu, Y.L.; Gao, X.X.; Huang, C.Y.; Luo, G.; Cao, G.; Xu, D.Q. miRNA let-7 family regulated by NEAT1 and ARID3A/NF-κB inhibits PRRSV-2 replication in vitro and in vivo. PLoS Pathog. 2022, 18, 22. [Google Scholar] [CrossRef]
- Gao, J.M.; Xiao, S.Q.; Xiao, Y.H.; Wang, X.P.; Zhang, C.; Zhao, Q.; Nan, Y.C.; Huang, B.C.; Liu, H.L.; Liu, N.N.; et al. MYH9 is an Essential Factor for Porcine Reproductive and Respiratory Syndrome Virus Infection. Sci. Rep. 2016, 6, 13. [Google Scholar] [CrossRef]
- Li, N.; Du, T.F.; Yan, Y.H.; Zhang, A.K.; Gao, J.M.; Hou, G.P.; Xiao, S.Q.; Zhou, E.M. MicroRNA let-7f-5p Inhibits Porcine Reproductive and Respiratory Syndrome Virus by Targeting MYH9. Sci. Rep. 2016, 6, 16. [Google Scholar] [CrossRef]
- Ferro-Novick, S.; Reggiori, F.; Brodsky, J.L. ER-Phagy, ER Homeostasis, and ER Quality Control: Implications for Disease. Trends Biochem. Sci. 2021, 46, 630–639. [Google Scholar] [CrossRef]
- Lennemann, N.J.; Coyne, C.B. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 2017, 13, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Chiramel, A.I.; Dougherty, J.D.; Nair, V.; Robertson, S.J.; Best, S.M. FAM134B, the Selective Autophagy Receptor for Endoplasmic Reticulum Turnover, Inhibits Replication of Ebola Virus Strains Makona and Mayinga. J. Infect. Dis. 2016, 214, S319–S325. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.; Su, Q.; Kuang, K.; Meng, X.; Zhou, X.; Liu, B. MiR-142-5p/FAM134B Axis Manipulates ER-Phagy to Control PRRSV Replication. Front. Immunol. 2022, 13, 2970. [Google Scholar] [CrossRef]
- Guo, X.K.; Zhang, Q.; Gao, L.; Li, N.; Chen, X.X.; Feng, W.H. Increasing Expression of MicroRNA 181 Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication and Has Implications for Controlling Virus Infection. J. Virol. 2013, 87, 1159–1171. [Google Scholar] [CrossRef]
- Gao, L.; Guo, X.K.; Wang, L.H.; Zhang, Q.; Li, N.; Chen, X.X.; Wang, Y.Q.; Feng, W.H. MicroRNA 181 Suppresses Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Infection by Targeting PRRSV Receptor CD163. J. Virol. 2013, 87, 8808–8812. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.X.; Liu, Y.L.; Ding, Y.Z.; Zhang, Y.G.; Zhang, J. PRRSV receptors and their roles in virus infection. Arch. Microbiol. 2015, 197, 503–512. [Google Scholar] [CrossRef]
- Delputte, P.L.; Vanderheijden, N.; Nauwynck, H.J.; Pensaert, M.B. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 2002, 76, 4312–4320. [Google Scholar] [CrossRef]
- Kim, J.K.; Fahad, A.M.; Shanmukhappa, K.; Kapil, S. Defining the cellular target(s) of porcine reproductive and respiratory syndrome virus blocking monoclonal antibody 7G10. J. Virol. 2006, 80, 689–696. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, R.; Zhang, Y.; Zhang, X.; Xia, X.; Sun, H. Establishment of a porcine CD151 transgenic PK-15 cell line susceptible to porcine reproductive and respiratory syndrome virus. Wei Sheng Wu Xue Bao Acta Microbiol. Sin. 2013, 53, 507–514. [Google Scholar]
- Welch, S.K.W.; Calvert, J.G. A brief review of CD163 and its role in PRRSV infection. Virus Res. 2010, 154, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Delputte, P.L.; Van Breedam, W.; Delrue, I.; Oetke, C.; Crocker, P.R.; Nauwynck, H.J. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. J. Virol. 2007, 81, 9546–9550. [Google Scholar] [CrossRef] [PubMed]
- Piñeyro, P.E.; Subramaniam, S.; Kenney, S.P.; Heffron, C.L.; Giménez-Lirola, L.G.; Meng, X.J. Modulation of Proinflammatory Cytokines in Monocyte-Derived Dendritic Cells by Porcine Reproductive and Respiratory Syndrome Virus Through Interaction with the Porcine Intercellular-Adhesion-Molecule-3-Grabbing Nonintegrin. Viral Immunol. 2016, 29, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Huang, K.; Chen, Y.J.; Huang, Z.Y.; Zhang, Y.Y.; Leng, C.L.; Liu, Y.K.; Shi, J.Z.; Xiao, S.Q.; Yao, L.G. MicroRNA ssc-miR-124a exhibits antiviral activity against porcine reproductive and respiratory syndrome virus via suppression of host genes CD163. Vet. Microbiol. 2021, 261, 8. [Google Scholar] [CrossRef] [PubMed]
- McCaskill, J.L.; Ressel, S.; Alber, A.; Redford, J.; Power, U.F.; Schwarze, J.; Dutia, B.M.; Buck, A.H. Broad-Spectrum Inhibition of Respiratory Virus Infection by MicroRNA Mimics Targeting p38 MAPK Signaling. Mol. Ther. Nucleic Acids 2017, 7, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.B.; Pei, Y.; Li, X.Y.; Zhao, S.H.; Zhu, M.J.; Zhao, A. miR-124 attenuates Japanese encephalitis virus replication by targeting DNM2. Virol. J. 2016, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J. Autophagy revisited. Autophagy 2008, 4, 740–743. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Yang, Y.; Klionsky, D.J. Autophagy and disease: Unanswered questions. Cell Death Differ. 2020, 27, 858–871. [Google Scholar] [CrossRef]
- Xu, C.M.; Wang, M.; Song, Z.B.; Wang, Z.J.; Liu, Q.Y.; Jiang, P.; Bai, J.; Li, Y.F.; Wang, X.W. Pseudorabies virus induces autophagy to enhance viral replication in mouse neuro-2a cells in vitro. Virus Res. 2018, 248, 44–52. [Google Scholar] [CrossRef]
- Zhou, A.; Li, S.F.; Khan, F.A.; Zhang, S.J. Autophagy postpones apoptotic cell death in PRRSV infection through Bad-Beclin1 interaction. Virulence 2016, 7, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhang, Y.S.; Pan, J.; Tong, X.Y.; Zhang, X.; Hu, X.L.; Gong, C.L. Grass Carp Reovirus triggers autophagy enhancing virus replication via the Akt/mTOR pathway. Fish Shellfish Immunol. 2022, 128, 148–156. [Google Scholar] [CrossRef]
- Sun, M.X.; Huang, L.; Wang, R.; Yu, Y.L.; Li, C.; Li, P.P.; Hu, X.C.; Hao, H.P.; Ishag, H.A.; Mao, X. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy 2012, 8, 1434–1447. [Google Scholar] [CrossRef]
- Yao, Y.; Li, S.H.; Zhu, Y.Q.; Xu, Y.Y.; Hao, S.Y.; Guo, S.Y.; Feng, W.H. miR-204 suppresses porcine reproductive and respiratory syndrome virus (PRRSV) replication via inhibiting LC3B-mediated autophagy. Virol. Sin. 2023, 38, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xie, T.; Xue, N.; Kuang, Q.; Wei, Z.; Liang, M.; Ding, X. miR-382 Contributes to Renal Tubulointerstitial Fibrosis by Downregulating HSPD1. Oxidative Med. Cell. Longev. 2017, 2017, 4708516. [Google Scholar] [CrossRef]
- Chang, X.B.; Shi, X.B.; Zhang, X.Z.; Chen, J.; Fan, X.M.; Yang, Y.H.; Wang, L.; Wang, A.P.; Deng, R.G.; Zhou, E.M.; et al. miR-382-5p promotes porcine reproductive and respiratory syndrome virus (PRRSV) replication by negatively regulating the induction of type I interferon. Faseb J. 2020, 34, 4497–4511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, C.; Yang, Q.; Gao, L.; Liu, H.C.; Tang, J.; Feng, W.H. MicroRNA-30c Modulates Type I IFN Responses To Facilitate Porcine Reproductive and Respiratory Syndrome Virus Infection by Targeting JAK1. J. Immunol. 2016, 196, 2272–2282. [Google Scholar] [CrossRef]
- Liu, F.; Wang, H.L.; Du, L.; Wei, Z.Y.; Zhang, Q.; Feng, W.H. MicroRNA-30c targets the interferon-alpha/beta receptor beta chain to promote type 2 PRRSV infection. J. Gen. Virol. 2018, 99, 1671–1680. [Google Scholar] [CrossRef]
- Wu, J.J.; Peng, X.W.; Zhou, A.; Qiao, M.; Wu, H.Y.; Xiao, H.W.; Liu, G.S.; Zheng, X.M.; Zhang, S.J.; Mei, S.Q. MiR-506 inhibits PRRSV replication in MARC-145 cells via CD151. Mol. Cell. Biochem. 2014, 394, 275–281. [Google Scholar] [CrossRef]
- Li, G.; Chen, Q.; Harmon, K.M.; Yoon, K.-J.; Schwartz, K.J.; Hoogland, M.J.; Gauger, P.C.; Main, R.G.; Zhang, J. Full-Length Genome Sequence of Porcine Deltacoronavirus Strain USA/IA/2014/8734. Genome Announ. 2014, 2, 10-1028. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Y.; Ji, C.M.; Yang, Y.L.; Liang, Q.Z.; Zhao, P.; Xu, L.D.; Lei, X.M.; Luo, W.T.; Qin, P.; et al. Porcine Deltacoronavirus Engages the Transmissible Gastroenteritis Virus Functional Receptor Porcine Aminopeptidase N for Infectious Cellular Entry. J. Virol. 2018, 92, 13. [Google Scholar] [CrossRef]
- Jung, K.; Hu, H.; Saif, L.J. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016, 226, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Chen, J.F.; Li, X.L.; Ren, W.K.; Li, F.X.; Wang, T.; Li, C.; Dong, Z.M.; Tian, X.X.; Zhang, L.; et al. Identification and integrated analysis of lncRNAs and miRNAs in IPEC-J2 cells provide novel insight into the regulation of the innate immune response by PDCoV infection. BMC Genom. 2022, 23, 17. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Z.; Cheng, A.C.; Cui, M.; Pan, Y.H.; Wang, M.S.; Huang, J.; Zhu, D.K.; Chen, S.; Liu, M.F.; Zhao, X.X.; et al. Duck Tembusu virus promotes the expression of suppressor of cytokine signaling 1 by downregulating miR-148a-5p to facilitate virus replication. Infect. Genet. Evol. 2020, 85, 10. [Google Scholar] [CrossRef]
- Alonso, S.; Izeta, A.; Sola, I.; Enjuanes, L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 2002, 76, 1293–1308. [Google Scholar] [CrossRef]
- Delmas, B.; Gelfi, J.; L’Haridon, R.; Vogel, L.K.; Sjostrom, H.; Noren, O.; Laude, H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 1992, 357, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Enjuanes, L.; Smerdou, C.; Castilla, J.; Anton, I.M.; Torres, J.M.; Sola, I.; Golvano, J.; Sanchez, J.M.; Pintado, B. Development of protection against coronavirus induced diseases. A review. Adv. Exp. Med. Biol. 1995, 380, 197–211. [Google Scholar]
- Wang, C.L.; Cai, L.C.; Liu, J.; Wang, G.; Li, H.M.; Wang, X.X.; Xu, W.H.; Ren, M.H.; Feng, L.; Liu, P.H.; et al. MicroRNA-30a-5p Inhibits the Growth of Renal Cell Carcinoma by Modulating GRP78 Expression. Cell. Physiol. Biochem. 2017, 43, 2405–2419. [Google Scholar] [CrossRef]
- Jia, Z.F.; Wang, K.; Wang, G.X.; Zhang, A.L.; Pu, P.Y. MiR-30a-5p Antisense Oligonucleotide Suppresses Glioma Cell Growth by Targeting SEPT7. PLoS ONE 2013, 8, 9. [Google Scholar] [CrossRef]
- Fu, Y.X.; Xu, W.T.; Chen, D.Y.; Feng, C.H.; Zhang, L.; Wang, X.H.; Lv, X.W.; Zheng, N.; Jin, Y.; Wu, Z.W. Enterovirus 71 induces autophagy by regulating has-miR-30a expression to promote viral replication. Antivir. Res. 2015, 124, 43–53. [Google Scholar] [CrossRef]
- Cruz, J.L.G.; Sola, I.; Becares, M.; Alberca, B.; Plana, J.; Enjuanes, L.; Zuñiga, S. Coronavirus Gene 7 Counteracts Host Defenses and Modulates Virus Virulence. PLoS Pathog. 2011, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Minakshi, R.; Padhan, K.; Rani, M.; Khan, N.; Ahmad, F.; Jameel, S. The SARS Coronavirus 3a Protein Causes Endoplasmic Reticulum Stress and Induces Ligand-Independent Downregulation of the Type 1 Interferon Receptor. PLoS ONE 2009, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.L.; Wang, C.L.; Xue, M.; Fu, F.; Zhang, X.; Li, L.; Yin, L.D.; Xu, W.H.; Feng, L.; Liu, P.H. The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response through IRE1α-Mediated Manipulation of the MicroRNA miR-30a-5p/SOCS1/3 Axis. J. Virol. 2018, 92, 21. [Google Scholar] [CrossRef] [PubMed]
- Okabayashi, T.; Kariwa, H.; Yokota, S.; Iki, S.; Indoh, T.; Yokosawa, N.; Takashima, I.; Tsutsumi, H.; Fujii, N. Cytokine regulation in SARS coronavirus infection compared to other respiratory virus infections. J. Med. Virol. 2006, 78, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.; Yokosawa, N.; Okabayashi, T.; Suzutani, T.; Miura, S.; Jimbow, K.; Fujii, N. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 contributes to inhibition of the interferon signaling pathway. J. Virol. 2004, 78, 6282–6286. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.W.; Yang, P.; Tang, Y.; Pan, Z.S.; Zhao, D.C. Respiratory Syncytial Virus Nonstructural Proteins Upregulate SOCS1 and SOCS3 in the Different Manner from Endogenous IFN Signaling. J. Immunol. Res. 2015, 2015, 11. [Google Scholar] [CrossRef]
- Xu, G.; Yang, F.; Ding, C.L.; Wang, J.; Zhao, P.; Wang, W.; Ren, H. MiR-221 accentuates IFN's anti-HCV effect by downregulating SOCS1 and SOCS3. Virology 2014, 462, 343–350. [Google Scholar] [CrossRef]
- Pauli, E.K.; Schmolke, M.; Wolff, T.; Viemann, D.; Roth, J.; Bode, J.G.; Ludwig, S. Influenza A Virus Inhibits Type I IFN Signaling via NF-κB-Dependent Induction of SOCS-3 Expression. PLoS Pathog. 2008, 4, 15. [Google Scholar] [CrossRef]
- Ding, L.; Xu, X.G.; Huang, Y.; Li, Z.C.; Zhang, K.; Chen, G.D.; Yu, G.S.; Wang, Z.S.; Li, W.; Tong, D.W. Transmissible gastroenteritis virus infection induces apoptosis through FasL- and mitochondria-mediated pathways. Vet. Microbiol. 2012, 158, 12–22. [Google Scholar] [CrossRef]
- Wu, D.; Ozaki, T.; Yoshihara, Y.; Kubo, N.; Nakagawara, A. Runt-related Transcription Factor 1 (RUNX1) Stimulates Tumor Suppressor p53 Protein in Response to DNA Damage through Complex Formation and Acetylation. J. Biol. Chem. 2013, 288, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.M.; Song, X.J.; Bai, X.Y.; Fei, N.J.; Huang, Y.; Zhao, Z.M.; Du, Q.; Zhang, H.L.; Zhang, L.; Tong, D.W. miR-27b attenuates apoptosis induced by transmissible gastroenteritis virus (TGEV) infection via targeting runt-related transcription factor 1 (RUNX1). PEERJ 2016, 4, 14. [Google Scholar] [CrossRef]
- Song, X.J.; Zhao, X.M.; Huang, Y.; Xiang, H.L.; Zhang, W.L.; Tong, D.W. Transmissible Gastroenteritis Virus (TGEV) Infection Alters the Expression of Cellular MicroRNA Species That Affect Transcription of TGEV Gene 7. Int. J. Biol. Sci. 2015, 11, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.L.; Xue, M.; Wu, P.; Wang, H.L.; Liu, Z.Q.; Wu, G.Z.; Liu, P.H.; Wang, K.L.; Xu, W.H.; Feng, L. Coronavirus transmissible gastroenteritis virus antagonizes the antiviral effect of the microRNA miR-27b via the IRE1 pathway. Sci. China-Life Sci. 2022, 65, 1413–1429. [Google Scholar] [CrossRef]
- Penzes, Z.; González, J.M.; Calvo, E.; Izeta, A.; Smerdou, C.; Méndez, A.; Sánchez, C.M.; Sola, I.; Almazán, F.; Enjuanes, L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the purdue virus cluster. Virus Genes 2001, 23, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Korotayev, K.; Ginsberg, D. Many pathways to apoptosis: E2F1 regulates splicing of apoptotic genes. Cell Death Differ. 2008, 15, 1813–1814. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, X.L.; Zhao, X.M.; Wang, K.L.; Tang, X.Y.; Guo, J.X.; Mi, M.; Qi, Y.P.; Chang, L.L.; Huang, Y.; Tong, D.W. Identification and analysis of long non-coding RNAs that are involved in inflammatory process in response to transmissible gastroenteritis virus infection. BMC Genom. 2019, 20, 13. [Google Scholar] [CrossRef]
- Ma, X.L.; Zhao, X.M.; Zhang, Z.C.; Guo, J.X.; Guan, L.J.; Li, J.J.; Mi, M.; Huang, Y.; Tong, D.W. Differentially expressed non-coding RNAs induced by transmissible gastroenteritis virus potentially regulate inflammation and NF-κB pathway in porcine intestinal epithelial cell line. BMC Genom. 2018, 19, 13. [Google Scholar] [CrossRef]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biololgy of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef]
- Tan, L.; Yao, J.; Yang, Y.D.; Luo, W.; Yuan, X.M.; Yang, L.C.; Wang, A.B. Current Status and Challenge of Pseudorabies Virus Infection in China. Virol. Sin. 2021, 36, 588–607. [Google Scholar] [CrossRef]
- Fang, L.L.; Gao, Y.N.; Liu, X.; Bai, J.; Jiang, P.; Wang, X.W. Long non-coding RNA LNC_000641 regulates pseudorabies virus replication. Vet. Res. 2021, 52, 13. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.L.; Zhang, K.L.; Huang, W.X.; Tang, W.; Li, H.M.; Dong, W.R.; Gu, J.Y.; Zhou, J.Y. Identification of functional lncRNAs in pseudorabies virus type II infected cells. Vet. Microbiol. 2020, 242, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.F.; Qian, J.H.; Shen, Z.J.; Shao, H.X.; Qian, K.; Jin, W.J.; Qin, A.J. Vector-delivered artificial miRNA effectively inhibits Porcine epidemic diarrhea virus replication. Virol. J. 2023, 20, 10. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.Q.; Wang, Q.W.; Gao, J.T.; Wang, L.L.; He, Z.Y.; Mo, D.L.; Liu, X.H.; Chen, Y.S. Inhibition of highly pathogenic PRRSV replication in MARC-145 cells by artificial microRNAs. Virol. J. 2011, 8, 11. [Google Scholar] [CrossRef]
- Zhu, L.; Song, H.Q.; Zhang, X.Y.; Xia, X.L.; Sun, H.C. Inhibition of porcine reproductive and respiratory syndrome virus infection by recombinant adenovirus- and/or exosome-delivered the artificial microRNAs targeting sialoadhesin and CD163 receptors. Virol. J. 2014, 11, 13. [Google Scholar] [CrossRef]
- Zhu, L.; Bao, L.P.; Zhang, X.Y.; Xia, X.L.; Sun, H.C. Inhibition of porcine reproductive and respiratory syndrome virus replication with exosome-transferred artificial microRNA targeting the 3′ untranslated region. J. Virol. Methods 2015, 223, 61–68. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Yu, H.; Qi, A.; Zhang, T.; Huo, Y.; Tu, Q.; Qi, C.; Wu, H.; Wang, X.; Zhou, J.; et al. Regulatory Non-Coding RNAs during Porcine Viral Infections: Potential Targets for Antiviral Therapy. Viruses 2024, 16, 118. https://doi.org/10.3390/v16010118
Li F, Yu H, Qi A, Zhang T, Huo Y, Tu Q, Qi C, Wu H, Wang X, Zhou J, et al. Regulatory Non-Coding RNAs during Porcine Viral Infections: Potential Targets for Antiviral Therapy. Viruses. 2024; 16(1):118. https://doi.org/10.3390/v16010118
Chicago/Turabian StyleLi, Feng, Hao Yu, Aosi Qi, Tianyi Zhang, Yuran Huo, Qiuse Tu, Chunyun Qi, Heyong Wu, Xi Wang, Jian Zhou, and et al. 2024. "Regulatory Non-Coding RNAs during Porcine Viral Infections: Potential Targets for Antiviral Therapy" Viruses 16, no. 1: 118. https://doi.org/10.3390/v16010118
APA StyleLi, F., Yu, H., Qi, A., Zhang, T., Huo, Y., Tu, Q., Qi, C., Wu, H., Wang, X., Zhou, J., Hu, L., Ouyang, H., Pang, D., & Xie, Z. (2024). Regulatory Non-Coding RNAs during Porcine Viral Infections: Potential Targets for Antiviral Therapy. Viruses, 16(1), 118. https://doi.org/10.3390/v16010118








