Genetic Characteristics of Wuxiang Virus in Shanxi Province, China
Abstract
1. Introduction
2. Materials and Methods
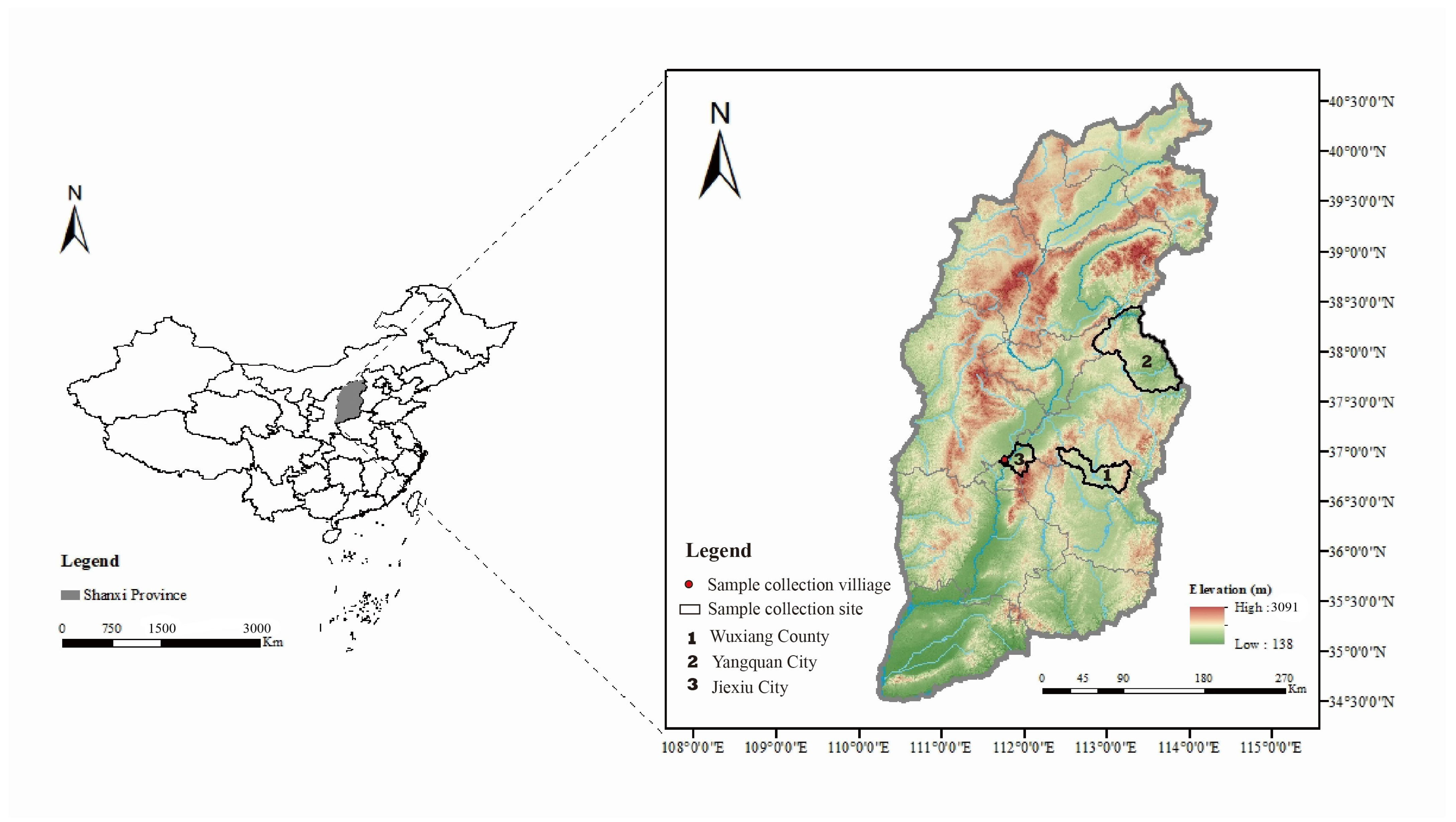
2.1. Sample Collection
2.2. Cells and Viruses
2.3. Virus Isolation and Identification
2.4. Viral Genome Sequencing
2.5. Gene Analysis
2.6. Morphological and Genetic Identification of Sandfly Species
3. Results
3.1. Sandfly Collection
3.2. Isolation and Identification of Isolated WUXV Strains
3.3. Viral Sequencing Results
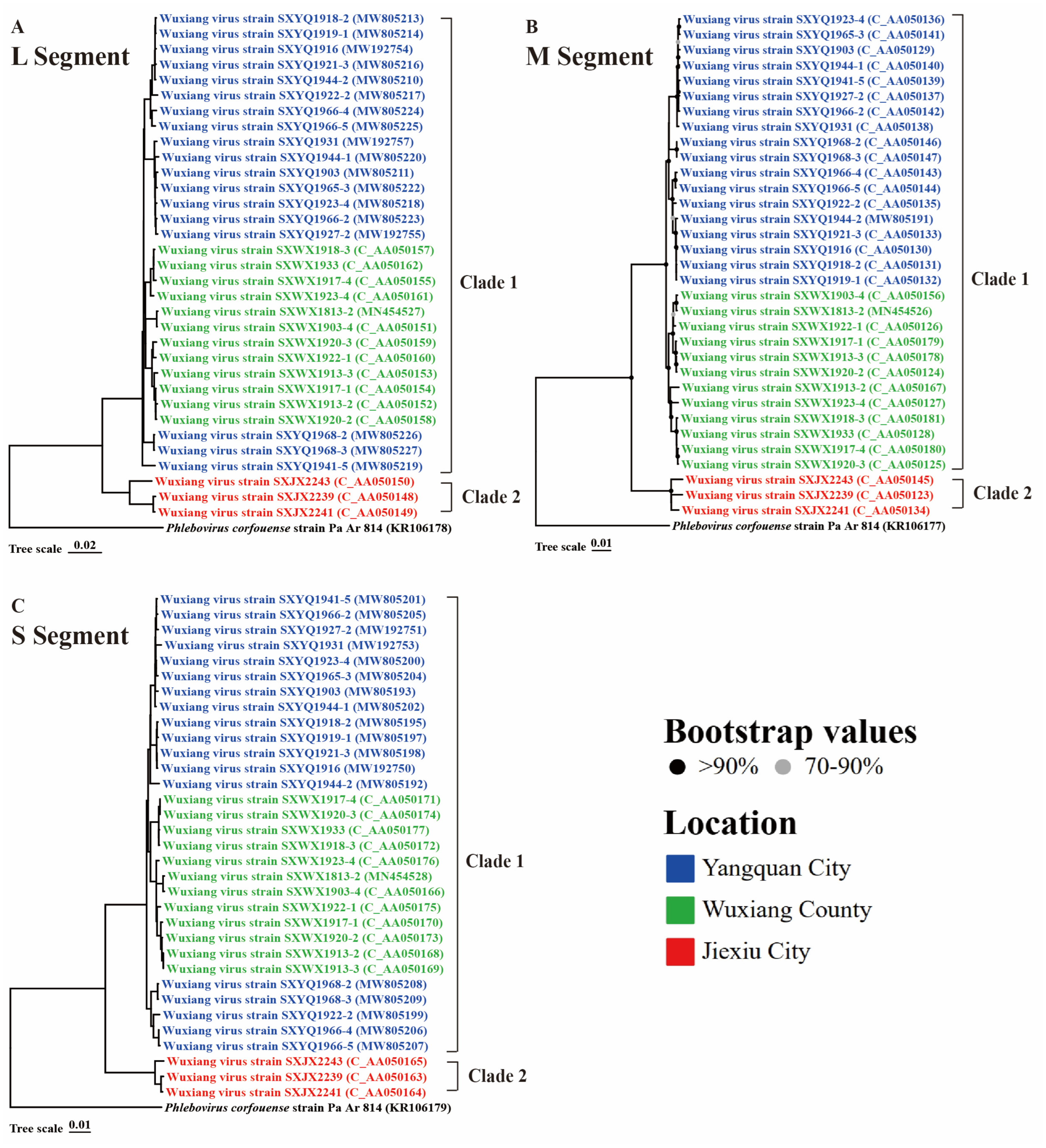
3.4. Phylogenetic Analysis and Genetic Distance
3.5. Reassortment/Recombination Analysis
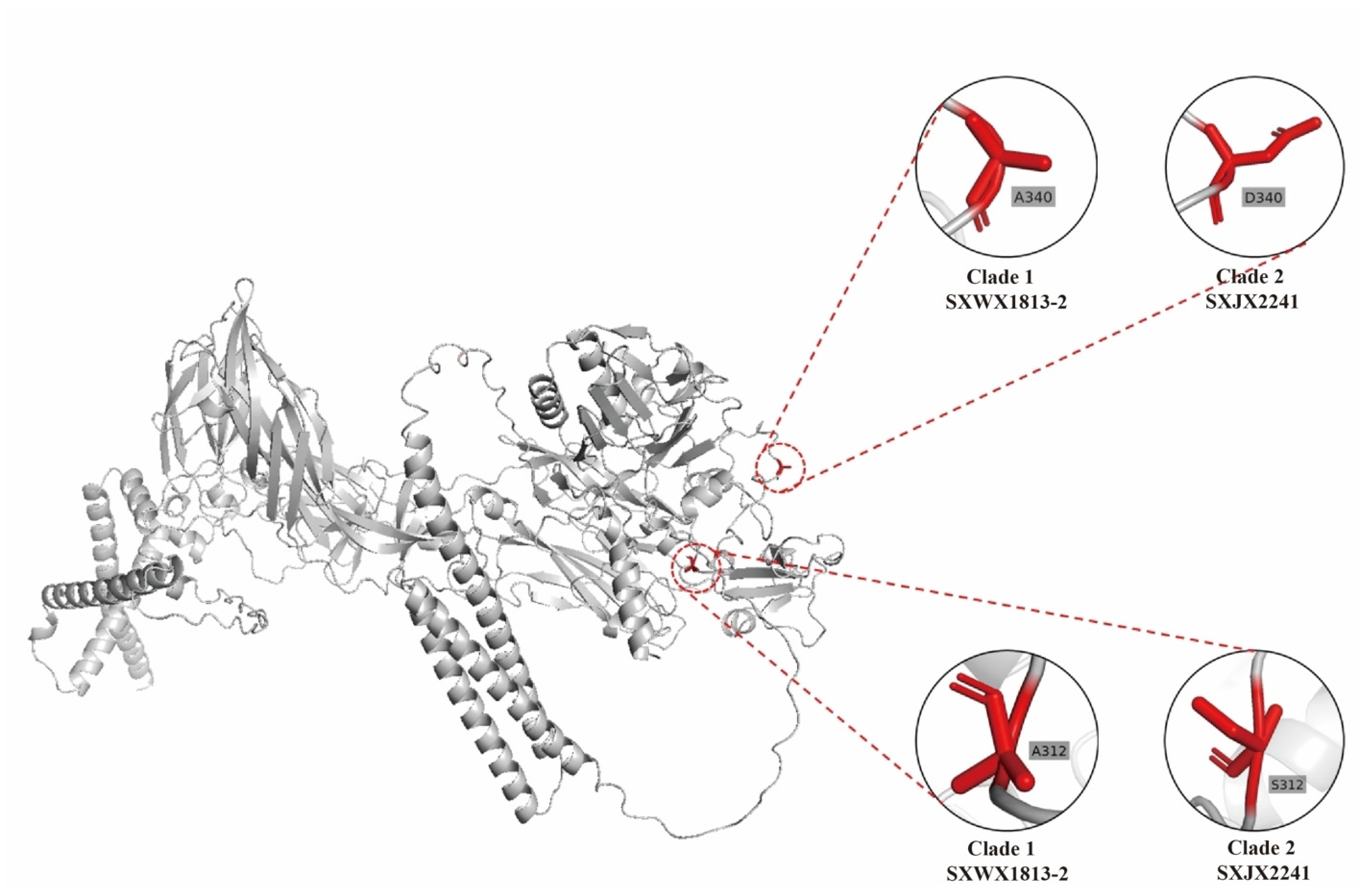
3.6. The Amino Acid Mutation and the Influence of Protein Tertiary Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Calisher, C.H.; Calzolari, M. Taxonomy of Phleboviruses, Emphasizing Those That Are Sandfly-Borne. Viruses 2021, 13, 918. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef] [PubMed]
- Alkan, C.; Bichaud, L.; de Lamballerie, X.; Alten, B.; Gould, E.A.; Charrel, R.N. Sandfly-borne phleboviruses of Eurasia and Africa: Epidemiology, genetic diversity, geographic range, control measures. Antivir. Res. 2013, 100, 54–74. [Google Scholar] [CrossRef] [PubMed]
- Ayhan, N.; Alten, B.; Ivovic, V.; Dvořák, V.; Martinkovic, F.; Omeragic, J.; Stefanovska, J.; Petric, D.; Vaselek, S.; Baymak, D.; et al. Direct evidence for an expanded circulation area of the recently identified Balkan virus (Sandfly fever Naples virus species) in several countries of the Balkan archipelago. Parasite Vector 2017, 10, 402. [Google Scholar] [CrossRef] [PubMed]
- Ergunay, K.; Ayhan, N.; Charrel, R.N. Novel and emergent sandfly-borne phleboviruses in Asia Minor: A systematic review. Rev. Med. Virol. 2017, 27, e1898. [Google Scholar] [CrossRef] [PubMed]
- Shahhosseini, N.; Paquette, S.J.; Kayedi, M.H.; Abaei, M.R.; Sedaghat, M.M. Genetic Characterization of Sandfly-Borne Viruses in Phlebotomine Sandflies in Iran. Microorganisms 2023, 11, 2754. [Google Scholar] [CrossRef]
- Lambert, A.J.; Hughes, H.R. Clinically Important Phleboviruses and Their Detection in Human Samples. Viruses 2021, 13, 1500. [Google Scholar] [CrossRef]
- Vilibic-Cavlek, T.; Zidovec-Lepej, S.; Ledina, D.; Knezevic, S.; Savic, V.; Tabain, I.; Ivic, I.; Slavuljica, I.; Bogdanic, M.; Grgic, I.; et al. Clinical, Virological, and Immunological Findings in Patients with Toscana Neuroinvasive Disease in Croatia: Report of Three Cases. Trop. Med. Infect. Dis. 2020, 5, 144. [Google Scholar] [CrossRef]
- Gashaw, A.E.; Meseret, B.A. One Health Approach for the Control of Zoonotic Diseases. Zoonoses 2022, 2, e963. [Google Scholar] [CrossRef]
- Wang, J.; Fu, S.H.; Xu, Z.Q.; Cheng, J.X.; Shi, M.; Fan, N.; Song, J.D.; Tian, X.D.; Cheng, J.S.; Ni, S.Q.; et al. Emerging Sand Fly-Borne Phlebovirus in China. Emerg. Infect. Dis. 2020, 26, 2435–2438. [Google Scholar] [CrossRef]
- Teng, A.Y.; Che, T.L.; Zhang, A.R.; Zhang, Y.Y.; Xu, Q.; Wang, T.; Sun, Y.Q.; Jiang, B.G.; Lv, C.L.; Chen, J.J.; et al. Mapping the viruses belonging to the order Bunyavirales in China. Infect. Dis. Poverty 2022, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Duan, X.M.; Wang, B.; Zhu, L.Y.; Zhang, Y.F.; Zhang, J.Y.; Wang, J.; Luo, T.; Kou, C.; Liu, D.; et al. A novel tick-borne Phlebovirus, closely related to severe fever with thrombocytopenia syndrome virus and Heartland virus, is a potential pathogen. Emerg. Microbes. Infec. 2018, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fan, N.; Fu, S.H.; Cheng, J.X.; Wu, B.; Xu, Z.Q.; Song, J.D.; Tian, X.D.; Li, Y.; He, Y.; et al. Isolation and Characterization of Wuxiang Virus from Sandflies Collected in Yangquan County, Shanxi Province, China. Vector-Borne Zoonot. 2021, 21, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; Cheng, J.X.; Fu, S.H.; Wang, Q.Y.; Wang, J.; Lu, X.Q.; Tian, X.D.; Cheng, J.S.; Ni, S.Q.; He, Y.; et al. Wuxiang Virus Is a Virus Circulated Naturally in Wuxiang County, China. Vector-Borne Zoonot. 2021, 21, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Y.; Yin, Q.K.; Fu, S.H.; Cheng, J.X.; Xu, X.Y.; Wang, J.; Wu, B.; Tian, X.D.; Li, Y.; Lu, J.; et al. Isolation and Identification of Sandfly-Borne Viruses from Sandflies Collected from June to August, 2019, in Yangquan County, China. Viruses 2022, 14, 2692. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Y.; Fu, S.H.; Cheng, J.X.; Xu, X.Y.; Wang, J.; Wu, B.; Tian, X.D.; Li, Y.; He, Y.; Li, F.; et al. Re-isolation of Wuxiang Virus from Wild Sandflies Collected from Yangquan County, China. Virol. Sin. 2021, 36, 1177–1186. [Google Scholar] [CrossRef]
- Yao, X.H.; Yin, Q.K.; Tian, X.D.; Zheng, Y.K.; Li, H.Y.; Fu, S.H.; Lian, Z.M.; Zhang, W.J.; He, Y.; Wang, R.C.; et al. Human and animal exposure to newly discovered sand fly viruses, China. Front. Cell Infect. MI 2024, 10, 402. [Google Scholar] [CrossRef]
- Yao, X.H.; Hu, D.H.; Fu, S.H.; Li, F.; He, Y.; Yin, J.Y.; Yin, Q.K.; Xu, S.T.; Liang, G.D.; Li, X.D.; et al. A Reverse-Transcription Recombinase-Aided Amplification Assay for the Rapid Detection of the Wuxiang Virus. Biomed. Environ. Sci. 2022, 35, 746–749. [Google Scholar] [CrossRef]
- Hu, D.H.; Yao, X.H.; Fu, S.H.; Li, F.; Gu, T.M.; Cheng, J.J.; He, Y.; Yin, J.Y.; Xu, S.T.; Yin, Q.K.; et al. Establishment of TaqMan RT-PCR assay for Wuxiang virus. Chin. J. Exp. Clin. Virol. 2022, 4, 460–464. [Google Scholar]
- Yao, X.H.; Yin, Q.K.; Hu, D.H.; Fu, S.H.; Zhang, W.J.; Nie, K.; Li, F.; Xu, S.T.; He, Y.; Liang, G.D.; et al. In Vitro Infection Dynamics of Wuxiang Virus in Different Cell Lines. Viruses 2022, 14, 2383. [Google Scholar] [CrossRef]
- Amroun, A.; Priet, S.; de Lamballerie, X.; Quérat, G. Bunyaviridae RdRps: Structure, motifs, and RNA synthesis machinery. Crit. Rev. Microbiol. 2017, 43, 753–778. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhu, Y.H.; Gao, F.; Jiao, Y.J.; Oladejo, B.O.; Chai, Y.; Bi, Y.H.; Lu, S.; Dong, M.Q.; Zhang, C.; et al. Structures of Phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope. Proc. Natl. Acad. Sci. USA 2017, 114, E7564–E7573. [Google Scholar] [CrossRef]
- Guardado-Calvo, P.; Rey, F.A. The Envelope Proteins of the Bunyavirales. Adv. Virus Res. 2017, 98, 83–118. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, M.; Plegge, T.; Pöhlmann, S. The Role of Phlebovirus Glycoproteins in Viral Entry, Assembly and Release. Viruses 2016, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Ly, H.J.; Ikegami, T. Rift Valley fever virus NSs protein functions and the similarity to other bunyavirus NSs proteins. Virol. J. 2016, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Leventhal, S.S.; Wilson, D.; Feldmann, H.; Hawman, D.W. A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins. Viruses 2021, 13, 314. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.H.; Song, S.; Liu, H.; Li, Y.Y.; Li, X.L.; Gao, X.Y.; Xu, Z.Q.; Liu, G.P.; Wang, D.M.; Tian, Z.Z.; et al. ZIKA virus isolated from mosquitoes: A field and laboratory investigation in China, 2016. Sci. China Life Sci. 2017, 60, 1364–1371. [Google Scholar] [CrossRef]
- Xie, J.M.; Chen, Y.R.; Cai, G.J.; Cai, R.L.; Hu, Z.; Wang, H. Tree Visualization by One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Amaro, F.; Zé-Zé, L.; Lourenço, J.; Giovanetti, M.; Becker, S.C.; Alves, M.J. Phylogenetic Analysis of Massilia phlebovirus in Portugal. Viruses 2021, 13, 1412. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky, P.S. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Kosakovsky, P.S.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky, P.S.; Scheffler, K. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Dong, H.W.; Yuan, H.; Shan, W.Q.; Zhou, Q.M.; Li, X.Y.; Peng, H.; Ma, Y.J. Mitochondrial COI and Cytb gene as valid molecular identification marker of sandfly species (Diptera: Psychodidae) in China. Acta Trop. 2023, 238, 106798. [Google Scholar] [CrossRef]
- Fu, S.H.; Li, M.H.; Meng, W.S.; Cao, Y.X.; Cheng, J.X.; Zhao, J.Y.; Kong, X.S.; Dai, P.F.; Liang, G.D. Isolation and identification of Arbovirus in southern part of Shanxi province, 2006. Dis. Surveill. 2010, 4, 95–98. [Google Scholar]
- Li, M.H.; Meng, W.S.; Fu, S.H.; Cheng, J.X.; Zhao, J.Y.; Kong, X.S.; Dai, P.F.; Liang, G.D. Arbovirus investigation in some regions of Shanxi province in 2007. Chin. J. Exp. Clin. Virol. 2009, 3, 32–34. [Google Scholar]
- Zheng, Y.Y.; Cao, Y.X.; Fu, S.H.; Cheng, J.X.; Zhao, J.Y.; Dai, P.F.; Kong, X.S.; Liang, G.D. Isolation and identification of mosquito-borne arboviruses in Yuncheng city, Shanxi province, 2012. Chin. J. Epidemiol. 2015, 36, 368–373. [Google Scholar]
- Liu, S.H.; Tian, X.D.; Zhang, W.J.; Zheng, H.M.; Zhao, J.Y.; Xu, C.X.; Zhang, Y.L.; Fu, S.H.; Nie, K.; Li, F.; et al. Nam Dinh virus was detected and isolated in arbovirus investigation in Shanxi, China. Chin. J. Exp. Clin. Virol. 2023, 37, 25–29. [Google Scholar] [CrossRef]
- Xu, Z.Q.; Fan, N.; Hou, X.; Wang, J.; Fu, S.H.; Song, J.D.; Shi, M.; Liang, G.D. Isolation and Identification of a Novel Phlebovirus, Hedi Virus, from Sandflies Collected in China. Viruses 2021, 13, 772. [Google Scholar] [CrossRef]
- Bente, D.A.; Forrester, N.L.; Watts, D.M.; McAuley, A.J.; Whitehouse, C.A.; Bray, M. Crimean-Congo hemorrhagic fever: History, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antivir. Res. 2013, 100, 159–189. [Google Scholar] [CrossRef]
- Dai, Z.N.; Peng, X.F.; Li, J.C.; Zhao, J.; Wu, Y.X.; Yang, X.; Yang, T.; Zhang, S.F.; Dai, K.; Guan, X.G.; et al. Effect of genomic variations in severe fever with thrombocytopenia syndrome virus on the disease lethality. Emerg. Microbes. Infec. 2022, 11, 1672–1682. [Google Scholar] [CrossRef]
- Liang, G.D. Research progress of natural sandfly-borne viruses in China. Chin. J. Exp. Clin. Virol. 2022, 36, 469–474. [Google Scholar] [CrossRef]
- Klempa, B. Reassortment events in the evolution of hantaviruses. Virus Genes 2018, 54, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Briese, T.; Calisher, C.H.; Higgs, S. Viruses of the family Bunyaviridae: Are all available isolates reassortants? Virology 2013, 446, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Zhang, H.J.; Jiang, Z.Y.; Zhou, M.; Min, Y.Q.; Deng, F.; Li, P.Q.; Wang, H.L.; Ning, Y.J. SFTS bunyavirus NSs protein sequestrates mTOR into inclusion bodies and deregulates mTOR-ULK1 signaling, provoking pro-viral autophagy. J. Med. Virol. 2023, 95, e28371. [Google Scholar] [CrossRef]
- Chen, Q.; Xu, X.H.; Tan, M.; Yang, L.; Wang, D.Y.; Shu, Y.L.; Zhu, W.F. NAN342K Mutation Enhances the Pathogenicity of Influenza B Virus in Mice. Zoonoses 2023, 3, e956. [Google Scholar] [CrossRef]
- Monteiro, G.; Jansen, V.V.P.; Wichgers, S.P.; Odendaal, L.; Clift, S.J.; Kortekaas, J.; Paweska, J.T. Mutation of adjacent cysteine residues in the NSs protein of Rift Valley fever virus results in loss of virulence in mice. Virus. Res. 2018, 249, 31–44. [Google Scholar] [CrossRef]

| Recombinant Sequence | Minor Parental Sequence 1 | Major Parental Sequence 2 | Begin of Recombination (nt) | End of Recombination (nt) | Detection Methods | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RDP | GENECONV | Bootscan | Maxchi | Chim- aera | SiScan | 3Seq | |||||
| SXWX1920-3 | SXWX1922-1 | SXWX1917-4 | 6261 | 10,365 | 2.96 × 10−14 | 2.96 × 10−12 | 3.72 × 10−10 | 1.03 × 10−19 | 2.51 × 10−18 | 2.28 × 10−15 | 1.68 × 10−34 |
| SXWX1913-2 | SXWX1920-2 | SXWX1813-2 | 6258 | 11,973 | 1.15 × 10−14 | 2.14 × 10−15 | 4.44 × 10−9 | 4.72 × 10−15 | 7.83 × 10−11 | 4.72 × 10−20 | 1.22 × 10−22 |
| SXYQ1941-5 | SXWX1923-4 | SXYQ1927-2 | 6247 | 10,380 | 3.24 × 10−19 | 1.20 × 10−14 | 1.83 × 10−12 | 2.50 × 10−17 | 5.10 × 10−9 | 5.57 × 10−32 | 8.58 × 10−5 |
| SXYQ1966-4 | SXYQ1965-3 | SXYQ1966-5 | 8943 | 9892 | 2.19 × 10−16 | 3.12 × 10−11 | 2.18 × 10−16 | 2.70 × 10−7 | 1.78 × 10−5 | 8.50 × 10−4 | 3.30 × 10−12 |
| Segment | Codon | Method | |||||||
|---|---|---|---|---|---|---|---|---|---|
| FEL | FUBAR | MEME | SLAC | ||||||
| dN-dS | p-Value | dN-dS | Pos.pro * | ω+ | p-value | dN-dS | p-Value | ||
| L | 237 | / | / | / | / | 112.475 | 0.076 | / | / |
| 744 | 7.760 | 0.037 | 15.411 | 0.977 | 43.582 | 0.012 | / | / | |
| M | 126 | / | / | / | / | 91.747 | 0.097 | / | / |
| 149 | / | / | 3.610 | 0.907 | / | / | / | / | |
| 182 | / | / | / | / | 132.870 | 0.081 | / | / | |
| 212 | / | / | / | / | 20.188 | 0.056 | / | / | |
| 298 | / | / | / | / | 45.696 | 0.040 | / | / | |
| 312 | 2.003 | 0.090 | / | / | / | / | / | / | |
| 340 | / | / | / | / | 426.456 | 0.098 | / | / | |
| 437 | / | / | / | / | 76.178 | 0.098 | / | / | |
| 566 | / | / | / | / | 13.571 | 0.078 | / | / | |
| 817 | / | / | / | / | 318.453 | 0.065 | / | / | |
| 1234 | / | / | / | / | 536.459 | 0.035 | / | / | |
| 1289 | 3.343 | 0.047 | / | / | 3.315 | 0.065 | / | / | |
| 1358 | / | / | / | / | 125.278 | 0.071 | / | / | |
| NS | 30 | / | / | 10.461 | 0.962 | / | / | / | / |
| N | 107 | / | / | 27.425 | 0.998 | 157.352 | 0.071 | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Tian, X.; Wang, R.; Yao, X.; Zhang, W.; Yin, Q.; Li, F.; Nie, K.; Cui, Q.; Xu, S.; et al. Genetic Characteristics of Wuxiang Virus in Shanxi Province, China. Viruses 2024, 16, 103. https://doi.org/10.3390/v16010103
Zheng Y, Tian X, Wang R, Yao X, Zhang W, Yin Q, Li F, Nie K, Cui Q, Xu S, et al. Genetic Characteristics of Wuxiang Virus in Shanxi Province, China. Viruses. 2024; 16(1):103. https://doi.org/10.3390/v16010103
Chicago/Turabian StyleZheng, Yuke, Xiaodong Tian, Ruichen Wang, Xiaohui Yao, Weijia Zhang, Qikai Yin, Fan Li, Kai Nie, Qianqian Cui, Songtao Xu, and et al. 2024. "Genetic Characteristics of Wuxiang Virus in Shanxi Province, China" Viruses 16, no. 1: 103. https://doi.org/10.3390/v16010103
APA StyleZheng, Y., Tian, X., Wang, R., Yao, X., Zhang, W., Yin, Q., Li, F., Nie, K., Cui, Q., Xu, S., Fu, S., Li, H., Cheng, J., & Wang, H. (2024). Genetic Characteristics of Wuxiang Virus in Shanxi Province, China. Viruses, 16(1), 103. https://doi.org/10.3390/v16010103






