Altered Proteomic Profile of Exosomes Secreted from Vero Cells Infected with Porcine Epidemic Diarrhea Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and PEDV Strain
2.2. Exosome Isolation and Purification
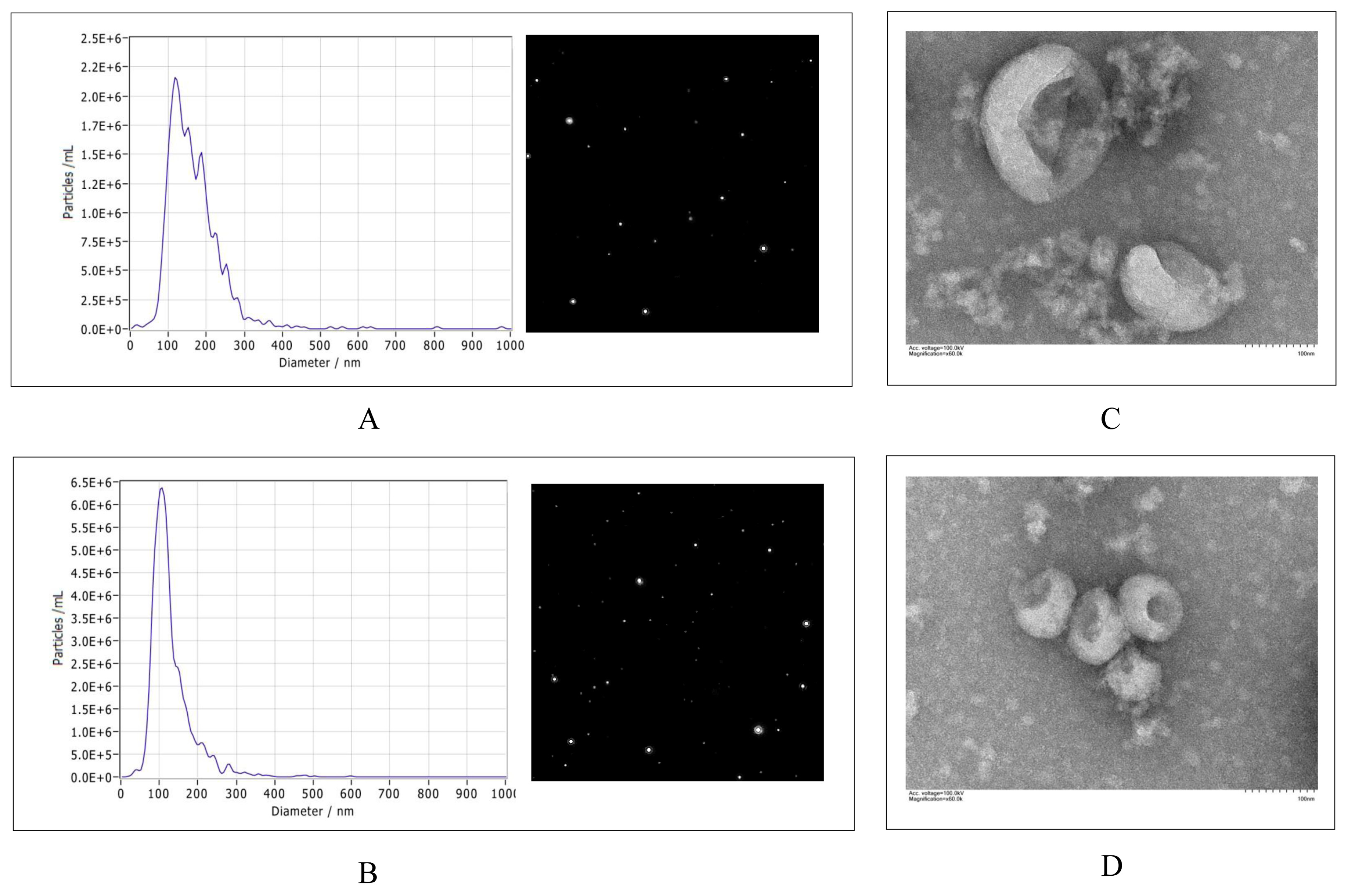
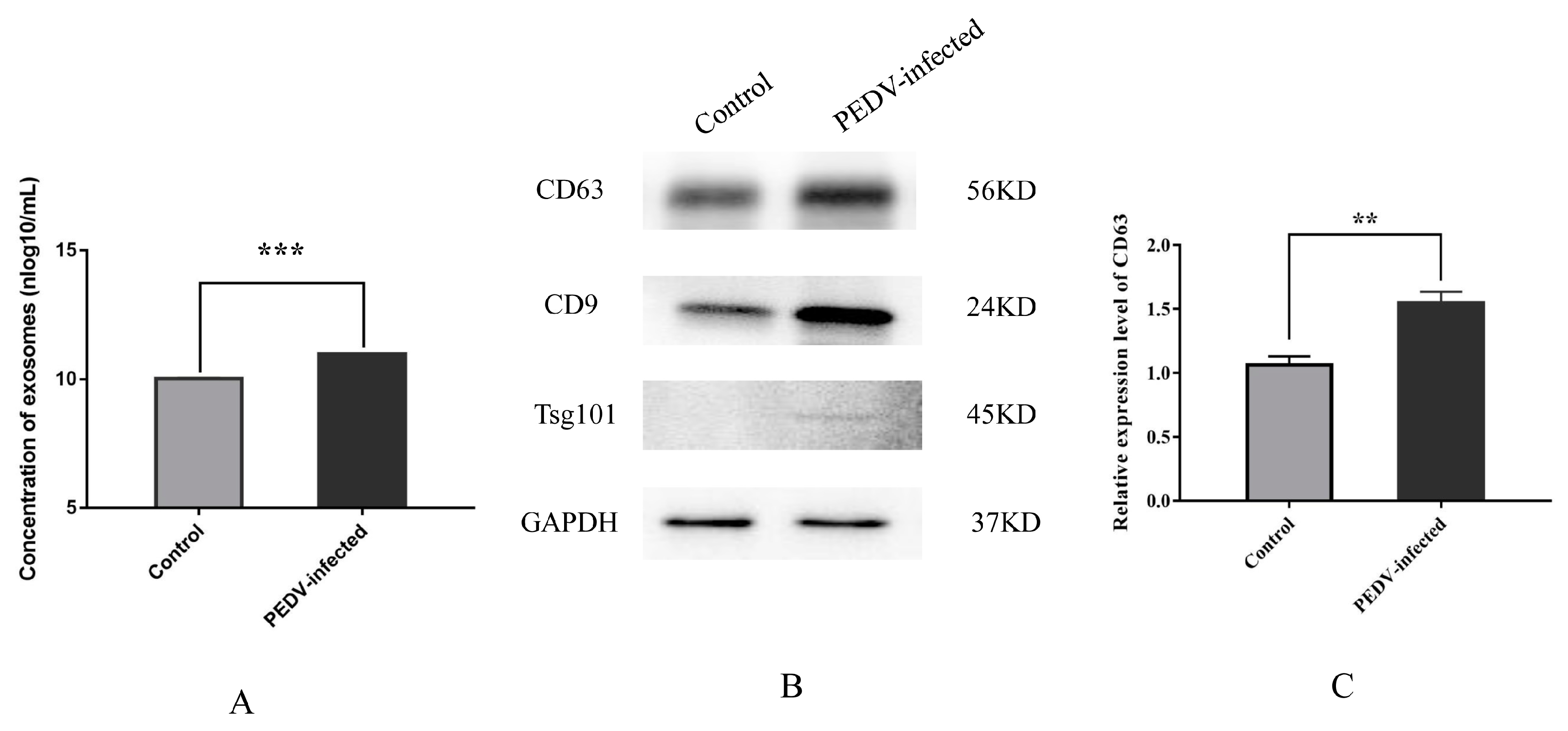
2.3. Nanoparticle Tracking Analysis (NTA)
2.4. Transmission Electron Microscopy (TEM)
2.5. Western Blot Analysis of Exosomal Proteins
2.6. Exosomal Protein Preparation
2.7. Proteome Sequencing
2.8. Database Search
2.9. Statistical Analysis
3. Results
3.1. Characterization and Secretion of Exosomes from Control and PEDV–Infected Vero Cells
3.2. Identification of Exosomal Proteins from Vero Cells
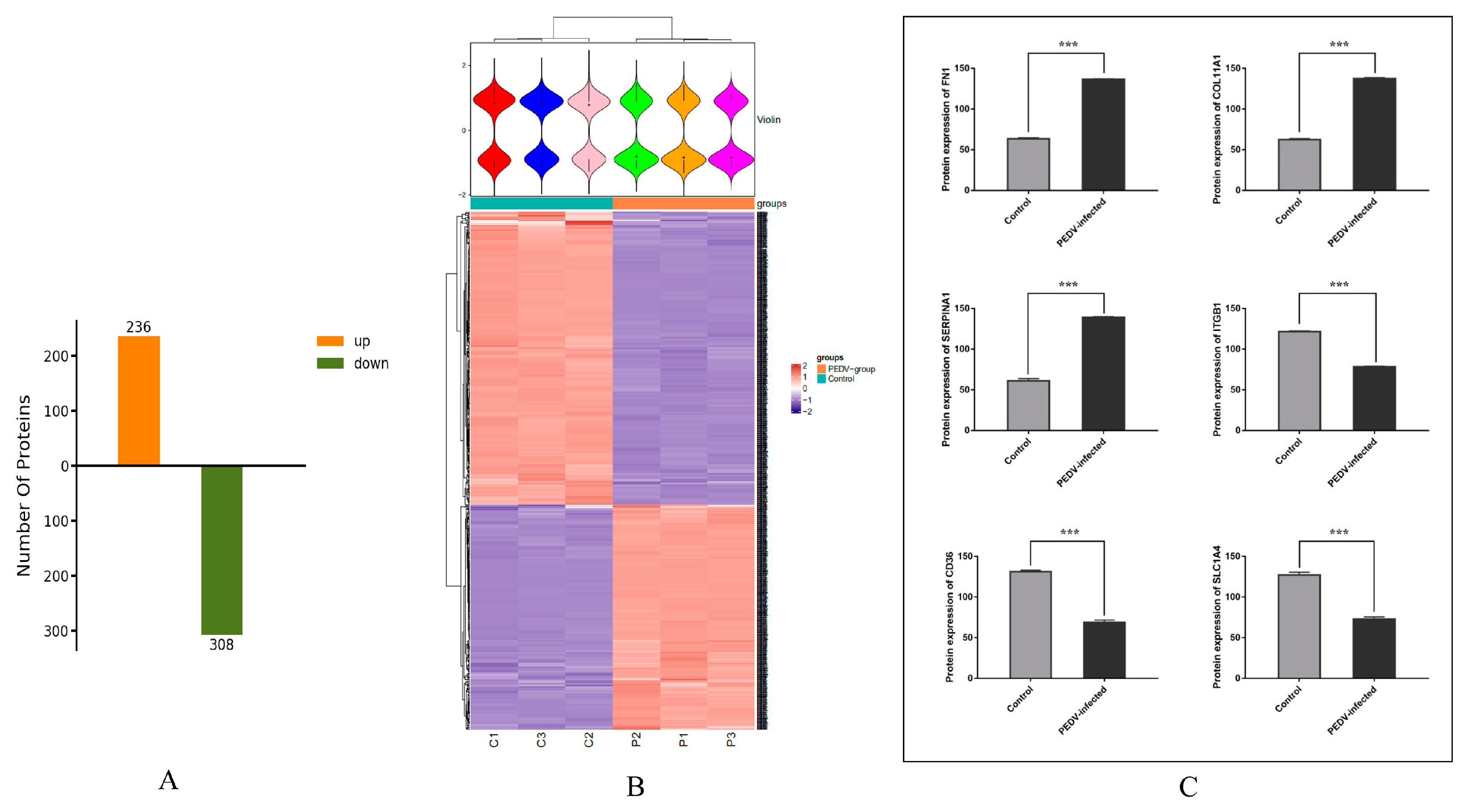
3.3. Analysis of Differentially Expressed Proteins in Exosomes from Control and PEDV–Infected Groups
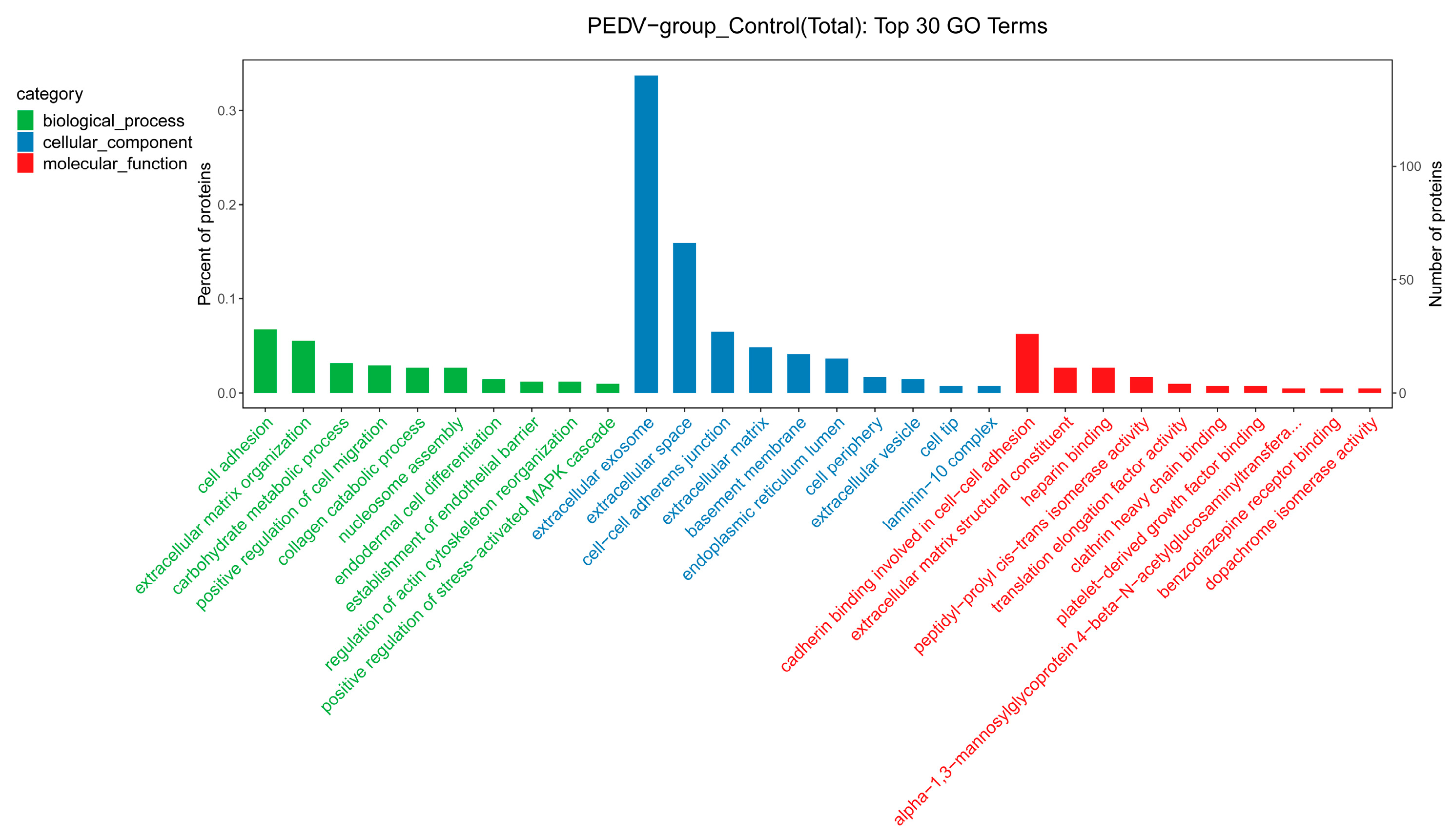
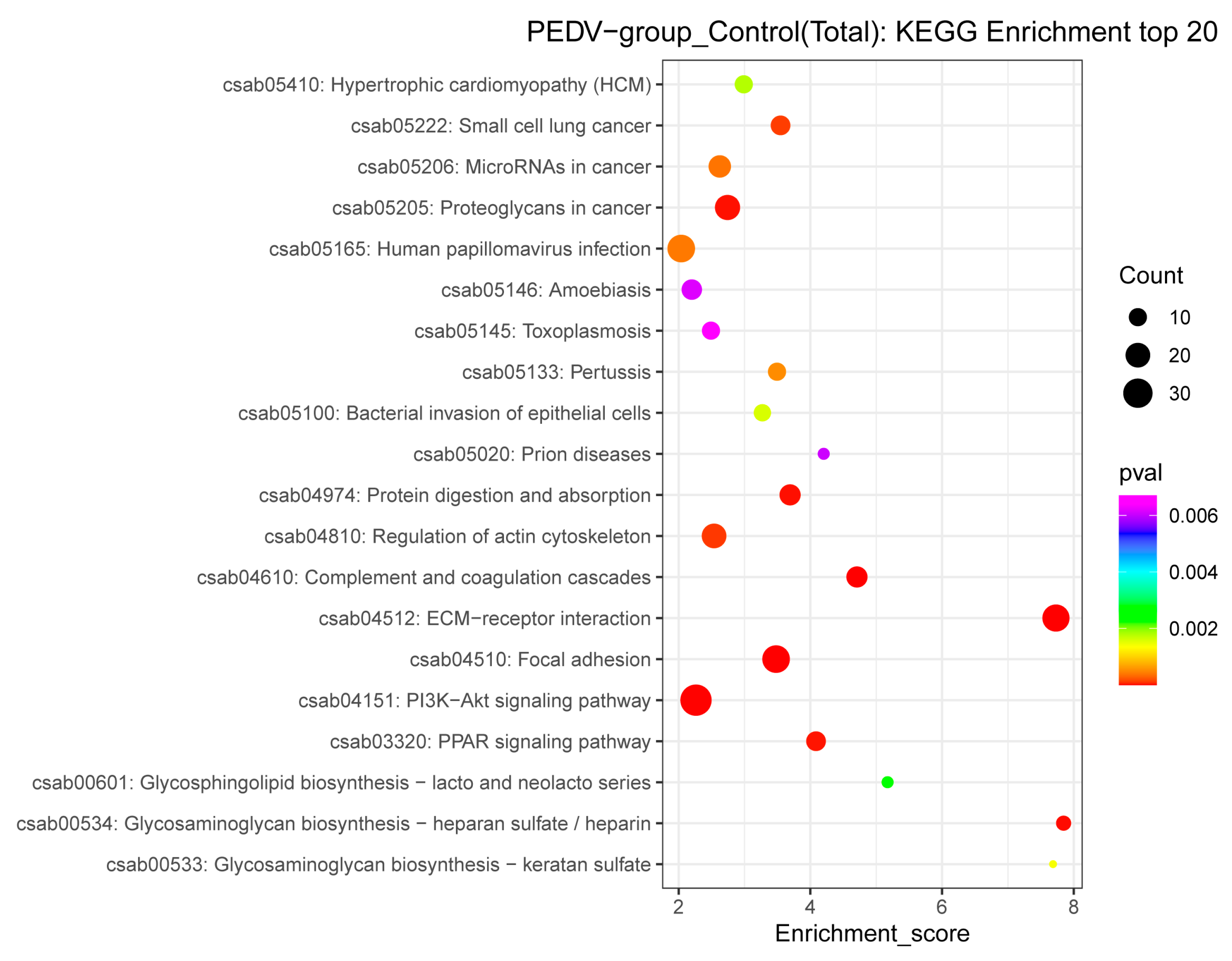
3.4. GO and KEGG Pathway Annotations of the DEPs
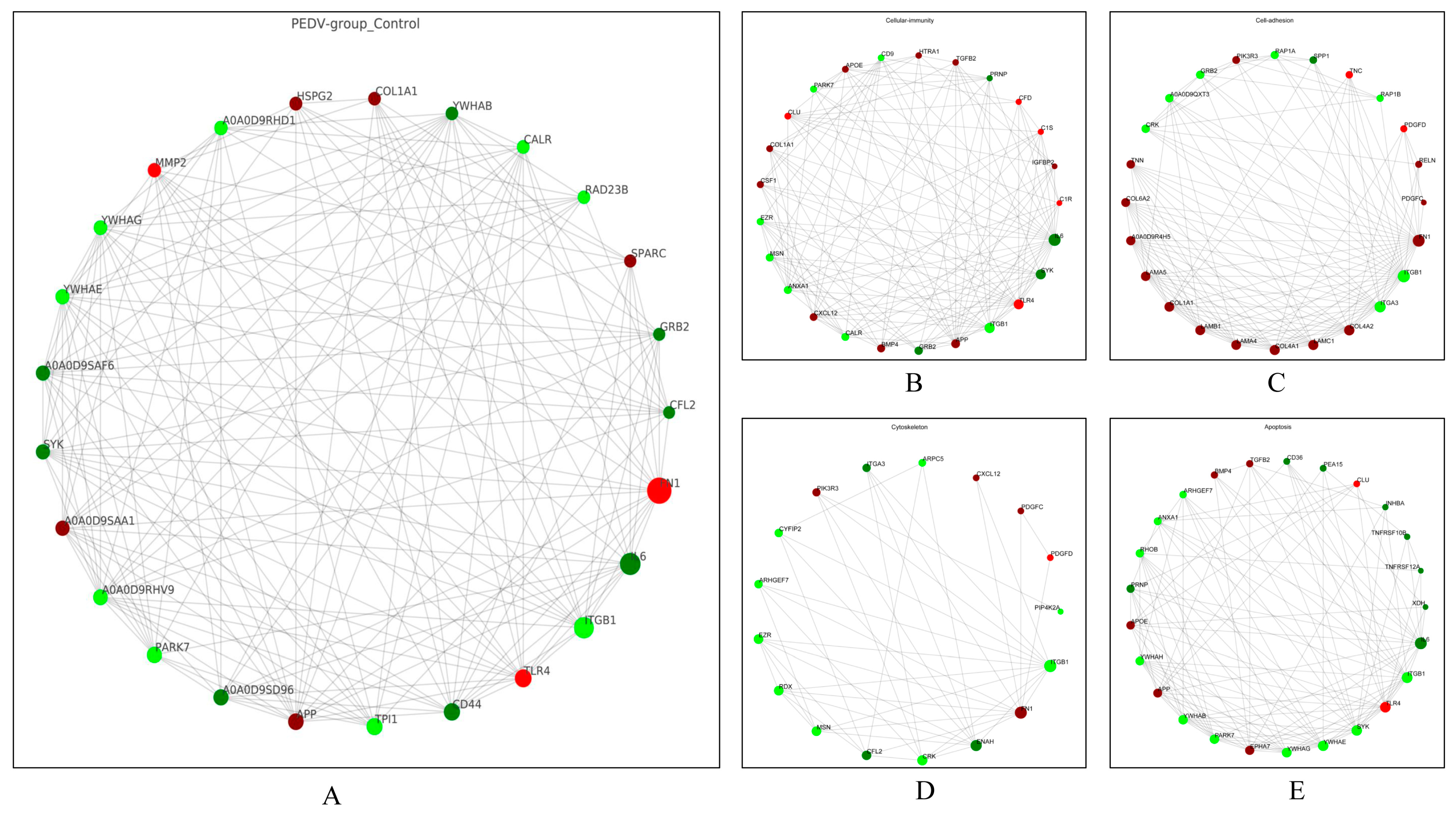
3.5. PPI Network Analysis of the DEPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xie, H.; Ai, Q.; Tong, T.; Liao, M.; Fan, H. PEDV infection affects the expression of polyamine-related genes inhibiting viral proliferation. Virus Res. 2022, 312, 198708. [Google Scholar] [CrossRef]
- Jung, K.; Annamalai, T.; Lu, Z.; Saif, L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 2015, 178, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Zhang, H.; Li, L.; Yang, Y.; Zou, X.; Chen, J.; Tang, X. PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition. Viruses 2022, 14, 1744. [Google Scholar] [CrossRef]
- Zhang, H.; Zou, C.; Peng, O.; Ashraf, U.; Xu, Q.; Gong, L.; Fan, B.; Zhang, Y.; Xu, Z.; Xue, C.; et al. Global Dynamics of Porcine Enteric Coronavirus PEDV Epidemiology, Evolution, and Transmission. Mol. Biol. Evol. 2023, 40, msad052. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Raab-Traub, N.; Dittmer, D.P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 2017, 15, 559–572. [Google Scholar] [CrossRef]
- Schwab, A.; Meyering, S.S.; Lepene, B.; Iordanskiy, S.; van Hoek, M.L.; Hakami, R.M.; Kashanchi, F. Extracellular vesicles from infected cells: Potential for direct pathogenesis. Front. Microbiol. 2015, 6, 1132. [Google Scholar] [CrossRef]
- Yin, Y.; Zhao, Y.; Chen, Q.; Chen, Y.; Mao, L. Dual roles and potential applications of exosomes in HCV infections. Front. Microbiol. 2022, 13, 1044832. [Google Scholar] [CrossRef] [PubMed]
- Bedford, J.G.; Infusini, G.; Dagley, L.F.; Villalon-Letelier, F.; Zheng, M.Z.M.; Bennett-Wood, V.; Reading, P.C.; Wakim, L.M. Airway Exosomes Released During Influenza Virus Infection Serve as a Key Component of the Antiviral Innate Immune Response. Front. Immunol. 2020, 11, 887. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, S.; Shi, X.; Xu, G.; Shen, C.; Liu, X.; Zheng, H. Exosomes-mediated transmission of foot-and-mouth disease virus in vivo and in vitro. Vet. Microbiol. 2019, 233, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Pan, J.; Zhu, M.; Liang, Z.; Shen, Z.; Dai, K.; Yan, B.; Dai, Y.; Xue, R.; et al. Proteomic analysis of the exosomes secreted from Ctenopharyngodon idellus kidney cells infected with grass carp reovirus reveals their involvement in the cellular responses to viral infection. Fish. Physiol. Biochem. 2021, 47, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jin, L.; Yan, M.; Yang, Z.; Wang, H.; Geng, S.; Gong, Z.; Liu, G. Serum Exosomes from Newborn Piglets Restrict Porcine Epidemic Diarrhea Virus Infection. J. Proteome Res. 2019, 18, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Shen, X.; Yin, D.; Wang, J.; Zhao, R.; Dai, Y.; Pan, X. Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses 2022, 14, 806. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, J.; Wang, Q.; Cui, Z.; Li, D.; Niu, J.; Guo, Y.; Zhang, Q.; Zhang, S.; Zhao, Y.; et al. Exosomal miRNA-328-3p targets ZO-3 and inhibits porcine epidemic diarrhea virus proliferation. Arch. Virol. 2022, 167, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Zhou, J.; Wang, X.; Ma, L.; Li, J.; Yang, L.; Yuan, H.; Pang, D.; Ouyang, H. Porcine Epidemic Diarrhea Virus: An Updated Overview of Virus Epidemiology, Virulence Variation Patterns and Virus-Host Interactions. Viruses 2022, 14, 2434. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015, 16, 24–43. [Google Scholar] [CrossRef]
- Wang, J.; Yao, Y.; Chen, X.; Wu, J.; Gu, T.; Tang, X. Host derived exosomes-pathogens interactions: Potential functions of exosomes in pathogen infection. Biomed. Pharmacother. 2018, 108, 1451–1459. [Google Scholar] [CrossRef]
- Sun, D.; Shi, H.; Guo, D.; Chen, J.; Shi, D.; Zhu, Q.; Zhang, X.; Feng, L. Analysis of protein expression changes of the Vero E6 cells infected with classic PEDV strain CV777 by using quantitative proteomic technique. J. Virol. Methods 2015, 218, 27–39. [Google Scholar] [CrossRef]
- Sun, P.; Wu, H.; Huang, J.; Xu, Y.; Yang, F.; Zhang, Q.; Xu, X. Porcine epidemic diarrhea virus through p53-dependent pathway causes cell cycle arrest in the G0/G1 phase. Virus Res. 2018, 253, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Shi, D.; Xing, X.; Qi, S.; Yang, D.; Zhang, J.; Han, Y.; Zhu, Q.; Sun, H.; Wang, X.; et al. Coronavirus Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Interacts with p53 To Induce Cell Cycle Arrest in S-Phase and Promotes Viral Replication. J. Virol. 2021, 95, e0018721. [Google Scholar] [CrossRef]
- Cong, Y.; Li, X.; Bai, Y.; Lv, X.; Herrler, G.; Enjuanes, L.; Zhou, X.; Qu, B.; Meng, F.; Cong, C.; et al. Porcine aminopeptidase N mediated polarized infection by porcine epidemic diarrhea virus in target cells. Virology 2015, 478, 1–8. [Google Scholar] [CrossRef]
- Li, C.; Su, M.; Yin, B.; Guo, D.; Wei, S.; Kong, F.; Feng, L.; Wu, R.; Sun, D. Integrin αvβ3 enhances replication of porcine epidemic diarrhea virus on Vero E6 and porcine intestinal epithelial cells. Vet. Microbiol. 2019, 237, 108400. [Google Scholar] [CrossRef] [PubMed]
- Madison, M.N.; Okeoma, C.M. Exosomes: Implications in HIV-1 Pathogenesis. Viruses 2015, 7, 4093–4118. [Google Scholar] [CrossRef]
- Longatti, A.; Boyd, B.; Chisari, F.V. Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J. Virol. 2015, 89, 2956–2961. [Google Scholar] [CrossRef] [PubMed]
- Madison, M.N.; Jones, P.H.; Okeoma, C.M. Exosomes in human semen restrict HIV-1 transmission by vaginal cells and block intravaginal replication of LP-BM5 murine AIDS virus complex. Virology 2015, 482, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.H.; Badierah, R.; Redwan, E.M.; El-Fakharany, E.M. A Comprehensive Insight into the Role of Exosomes in Viral Infection: Dual Faces Bearing Different Functions. Pharmaceutics 2021, 13, 1405. [Google Scholar] [CrossRef] [PubMed]
- Alenquer, M.; Amorim, M.J. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses 2015, 7, 5066–5083. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.R.; Kashanchi, F.; Jacobson, S. Exosomes in Viral Disease. Neurotherapeutics 2016, 13, 535–546. [Google Scholar] [CrossRef]
- Gould, S.J.; Booth, A.M.; Hildreth, J.E. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA 2003, 100, 10592–10597. [Google Scholar] [CrossRef] [PubMed]
- Hildreth, J.E.K. HIV As Trojan Exosome: Immunological Paradox Explained? Front. Immunol. 2017, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Pawliczek, T.; Crump, C.M. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 2009, 83, 11254–11264. [Google Scholar] [CrossRef] [PubMed]
- Votteler, J.; Sundquist, W.I. Virus budding and the ESCRT pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef]
- Gerber, P.P.; Cabrini, M.; Jancic, C.; Paoletti, L.; Banchio, C.; von Bilderling, C.; Sigaut, L.; Pietrasanta, L.I.; Duette, G.; Freed, E.O.; et al. Rab27a controls HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 2015, 209, 435–452. [Google Scholar] [CrossRef]
- Bello-Morales, R.; Crespillo, A.J.; Fraile-Ramos, A.; Tabarés, E.; Alcina, A.; López-Guerrero, J.A. Role of the small GTPase Rab27a during herpes simplex virus infection of oligodendrocytic cells. BMC Microbiol. 2012, 12, 265. [Google Scholar] [CrossRef]
- Wang, J.; Wu, F.; Liu, C.; Dai, W.; Teng, Y.; Su, W.; Kong, W.; Gao, F.; Cai, L.; Hou, A.; et al. Exosomes Released from Rabies Virus-Infected Cells May be Involved in the Infection Process. Virol. Sin. 2019, 34, 59–65. [Google Scholar] [CrossRef]
- Xie, J.; Guo, T.; Zhong, Z.; Wang, N.; Liang, Y.; Zeng, W.; Liu, S.; Chen, Q.; Tang, X.; Wu, H.; et al. ITGB1 Drives Hepatocellular Carcinoma Progression by Modulating Cell Cycle Process Through PXN/YWHAZ/AKT Pathways. Front. Cell Dev. Biol. 2021, 9, 711149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shang, L.; Zhang, J.; Liu, Y.; Jin, C.; Zhao, Y.; Lei, X.; Wang, W.; Xiao, X.; Zhang, X.; et al. An antibody-based proximity labeling map reveals mechanisms of SARS-CoV-2 inhibition of antiviral immunity. Cell Chem. Biol. 2022, 29, 5–18.e6. [Google Scholar] [CrossRef]
- Zeng, C.; Waheed, A.A.; Li, T.; Yu, J.; Zheng, Y.M.; Yount, J.S.; Wen, H.; Freed, E.O.; Liu, S.L. SERINC proteins potentiate antiviral type I IFN production and proinflammatory signaling pathways. Sci. Signal 2021, 14, eabc7611. [Google Scholar] [CrossRef] [PubMed]
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021, 33, 1744–1762. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Lewin, S.; Hunt, S.; Lambert, D.W. Extracellular vesicles and the extracellular matrix: A new paradigm or old news? Biochem. Soc. Trans. 2020, 48, 2335–2345. [Google Scholar] [CrossRef] [PubMed]
- Basile, M.S.; Cavalli, E.; McCubrey, J.; Hernández-Bello, J.; Muñoz-Valle, J.F.; Fagone, P.; Nicoletti, F. The PI3K/Akt/mTOR pathway: A potential pharmacological target in COVID-19. Drug Discov. Today 2022, 27, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Wilczek, M.P.; Armstrong, F.J.; Mayberry, C.L.; King, B.L.; Maginnis, M.S. PI3K/AKT/mTOR Signaling Pathway Is Required for JCPyV Infection in Primary Astrocytes. Cells 2021, 10, 3218. [Google Scholar] [CrossRef]
- Shen, X.; Yin, L.; Pan, X.; Zhao, R.; Zhang, D. Porcine epidemic diarrhea virus infection blocks cell cycle and induces apoptosis in pig intestinal epithelial cells. Microb. Pathog. 2020, 147, 104378. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Li, B.; Chen, L.; Ma, Z.; He, K.; Fan, H. Differential Protein Analysis of IPEC-J2 Cells Infected with Porcine Epidemic Diarrhea Virus Pandemic and Classical Strains Elucidates the Pathogenesis of Infection. J. Proteome Res. 2017, 16, 2113–2120. [Google Scholar] [CrossRef]
- Lin, H.; Li, B.; Liu, M.; Zhou, H.; He, K.; Fan, H. Nonstructural protein 6 of porcine epidemic diarrhea virus induces autophagy to promote viral replication via the PI3K/Akt/mTOR axis. Vet. Microbiol. 2020, 244, 108684. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Knaut, H. Focal adhesion-mediated cell anchoring and migration: From in vitro to in vivo. Development 2022, 149, dev200647. [Google Scholar] [CrossRef]
- Mishra, Y.G.; Manavathi, B. Focal adhesion dynamics in cellular function and disease. Cell Signal 2021, 85, 110046. [Google Scholar] [CrossRef]
- Avraham, H.K.; Jiang, S.; Lee, T.H.; Prakash, O.; Avraham, S. HIV-1 Tat-mediated effects on focal adhesion assembly and permeability in brain microvascular endothelial cells. J. Immunol. 2004, 173, 6228–6233. [Google Scholar] [CrossRef]
- Albecka, A.; Owen, D.J.; Ivanova, L.; Brun, J.; Liman, R.; Davies, L.; Ahmed, M.F.; Colaco, S.; Hollinshead, M.; Graham, S.C.; et al. Dual Function of the pUL7-pUL51 Tegument Protein Complex in Herpes Simplex Virus 1 Infection. J. Virol. 2017, 91, e02196-16. [Google Scholar] [CrossRef] [PubMed]
- Ramsauer, A.S.; Kubacki, J.; Favrot, C.; Ackermann, M.; Fraefel, C.; Tobler, K. RNA-seq analysis in equine papillomavirus type 2-positive carcinomas identifies affected pathways and potential cancer markers as well as viral gene expression and splicing events. J. Gen. Virol. 2019, 100, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Wang, X.; Wu, Q.; Yang, Q. Role of intestinal extracellular matrix-related signaling in porcine epidemic diarrhea virus infection. Virulence 2021, 12, 2352–2365. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Goldman, R.D. Overview of the Cytoskeleton from an Evolutionary Perspective. Cold Spring Harb. Perspect. Biol. 2018, 10, a030288. [Google Scholar] [CrossRef]
- Li, C.C.; Chi, X.J.; Wang, J.; Potter, A.L.; Wang, X.J.; Yang, C.J. Small molecule RAF265 as an antiviral therapy acts against HSV-1 by regulating cytoskeleton rearrangement and cellular translation machinery. J. Med. Virol. 2023, 95, e28226. [Google Scholar] [CrossRef]
- Wen, Z.; Zhang, Y.; Lin, Z.; Shi, K.; Jiu, Y. Cytoskeleton-a crucial key in host cell for coronavirus infection. J. Mol. Cell Biol. 2020, 12, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Gao, J.; Zhu, L.; Yang, Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. 2014, 192, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Kang, W.; Li, Y.; Shan, Y.; Wang, S.; Liu, F. Dynamic Dissection of Dynein and Kinesin-1 Cooperatively Mediated Intercellular Transport of Porcine Epidemic Diarrhea Coronavirus along Microtubule Using Single Virus Tracking. Virulence 2021, 12, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Sheng, X.; Murray-Nerger, L.A.; Cristea, I.M. Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat. Commun. 2020, 11, 806. [Google Scholar] [CrossRef] [PubMed]
- Ramovs, V.; Te Molder, L.; Sonnenberg, A. The opposing roles of laminin-binding integrins in cancer. Matrix Biol. 2017, 57–58, 213–243. [Google Scholar] [CrossRef]
- Kruize, Z.; Cobos Jiménez, V.; Martinez, F.O.; Di Vincenzo, R.; van Dort, K.A.; van Nuenen, A.C.; Booiman, T.; Kootstra, N.A. CD9 and ITGA3 are regulated during HIV-1 infection in macrophages to support viral replication. Virology 2021, 562, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Zhao, Y.; Peng, O.; Xia, Y.; Xu, Q.; Li, H.; Xue, C.; Cao, Y.; Zhang, H. Swine acute diarrhea syndrome coronavirus induces autophagy to promote its replication via the Akt/mTOR pathway. iScience 2022, 25, 105394. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Yin, L.; Xu, S.; Wang, J.; Yin, D.; Zhao, R.; Pan, X.; Dai, Y.; Hou, H.; Zhou, X.; et al. Altered Proteomic Profile of Exosomes Secreted from Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses 2023, 15, 1640. https://doi.org/10.3390/v15081640
Shen X, Yin L, Xu S, Wang J, Yin D, Zhao R, Pan X, Dai Y, Hou H, Zhou X, et al. Altered Proteomic Profile of Exosomes Secreted from Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses. 2023; 15(8):1640. https://doi.org/10.3390/v15081640
Chicago/Turabian StyleShen, Xuehuai, Lei Yin, Shuangshuang Xu, Jieru Wang, Dongdong Yin, Ruihong Zhao, Xiaocheng Pan, Yin Dai, Hongyan Hou, Xueli Zhou, and et al. 2023. "Altered Proteomic Profile of Exosomes Secreted from Vero Cells Infected with Porcine Epidemic Diarrhea Virus" Viruses 15, no. 8: 1640. https://doi.org/10.3390/v15081640
APA StyleShen, X., Yin, L., Xu, S., Wang, J., Yin, D., Zhao, R., Pan, X., Dai, Y., Hou, H., Zhou, X., & Hu, X. (2023). Altered Proteomic Profile of Exosomes Secreted from Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses, 15(8), 1640. https://doi.org/10.3390/v15081640






