Antibody-Dependent Enhancement with a Focus on SARS-CoV-2 and Anti-Glycan Antibodies
Abstract
1. Introduction
2. Terminological Debate
3. Antibody-Dependent Enhancement (ADE): Phenomenon Outline
4. ADE in Viral Infections: Paradoxes and Key Debatable Issues
4.1. Dengue Fever Is a Classic Example of ADE
4.2. Can Virus-Specific Antibodies Intensify Infection Instead of Protecting against It?
4.3. What Is the Role of ADE in the Infection Enhancement?
4.4. What Factors Trigger the Development of ADE?
- the type of the infected cell’s Fcγ receptor to which the antibodies bind;
- G class antibody isotype;
- epitope specificity of antibodies.
- (i)
- interaction of the immunoglobulin molecule with specific epitopes of a particular virus should normally lead to virus neutralization and block its penetration into the cell, but in ADE, this process is characterized by incomplete neutralization and activation of effector cells;
- (ii)
- antibodies binding to homologous epitopes of heterologous virus genotypes do not neutralize the virus but only provide a binding effect and may be involved in antigen masking or effector reactions [72].
4.5. Which Antibodies Are Associated with ADE?
5. Infection Caused by SARS-CoV-2: Does ADE Develop?
6. Anti-Glycan Antibodies as a Possible ADE Trigger in COVID-19: Pros and Cons
6.1. Do SARS-CoV-2 Glycans Induce an Immune Response?
6.2. Anti-Glycan Antibodies as a Pool of Antibodies with Antiviral Activity
- (i)
- AGA are part of the pool of natural pre-existing antibodies and are able to recognize virus-associated glycans;
- (ii)
- Most of AGA are polyreactive antibodies, i.e., they can bind to a significant number of epitopes due to the conformational lability of epitopes and paratopes of the antibodies [134]. The broad epitope specificity of AGA determines their binding not only to carbohydrate antigens but also to glycolipid and glycopeptide antigens [135];
- (iii)
- A significant number of AGA have low affinity compared to protein–protein interactions. Their affinity for the monovalent hapten (in KD terms) is in the range of 10–4–10–6 M. AGA content of some specificities (this refers to “top” antibodies) in healthy persons reaches the level of ~0.5% of the total amount of immunoglobulin M class [20], which indicates their rather high concentration in blood.
7. Anti-Glycan Antibodies as ADE Trigger
7.1. Involvement of Anti-Glycan Antibodies in ADE Development in Bacterial Infections
7.2. Anti-Glycan Antibodies in Coronavirus Infection: Data from Previous Studies
7.2.1. Autoantibodies Induction in Coronavirus Infection
7.2.2. AGA Virus-Neutralizing Activity
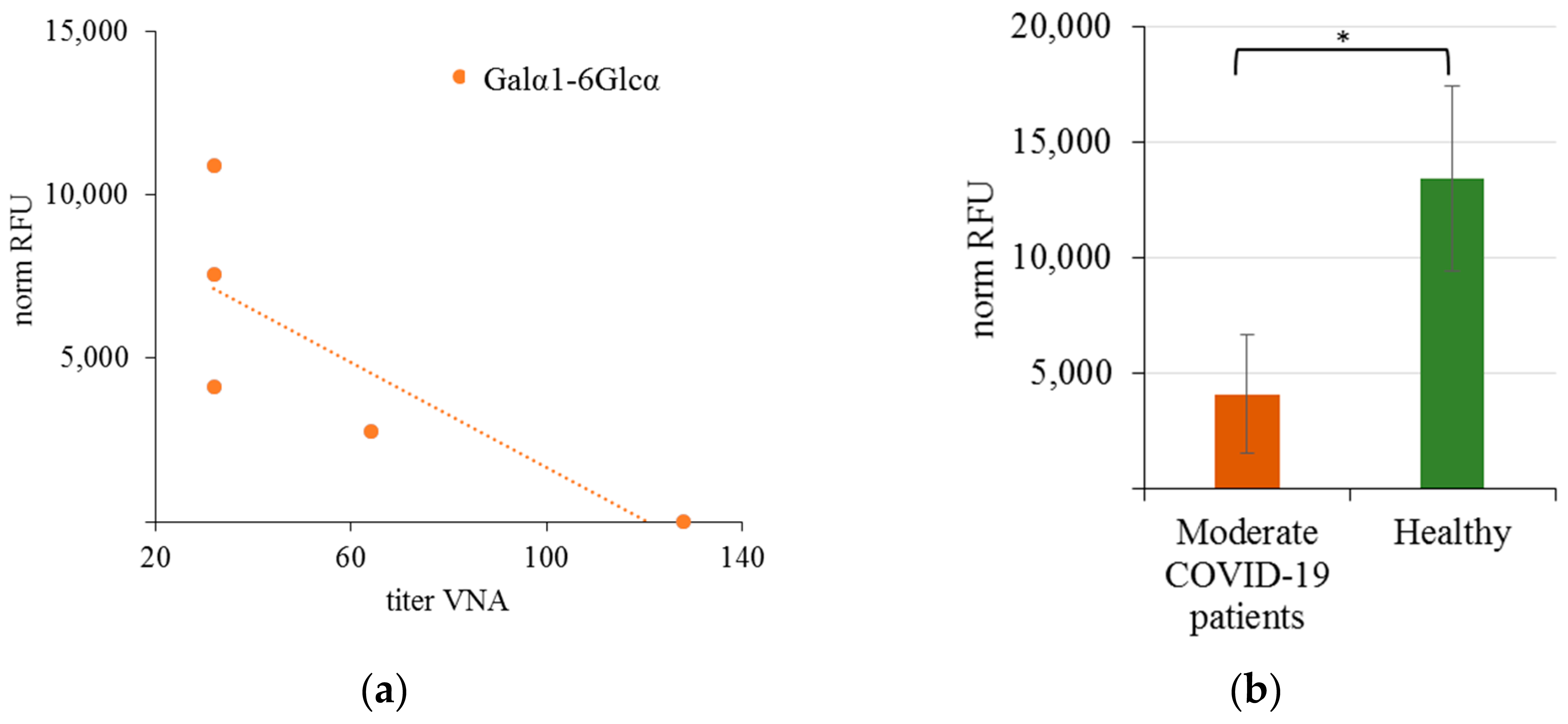
7.3. Anti-Glycan Antibodies in COVID-19: Own Data
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Angiotensin-converting enzyme 2 | ACE2 |
| Antibody-dependent enhancement | ADE |
| Anti-glycan antibodies | AGA |
| already existing antibodies | AEAbs |
| Complement receptors | CRs |
| Fc-receptors | FcRs |
| Receptor-binding domain | RBD |
| Pathogen-associated molecular patterns | PAMPs |
| Printed glycan array | PGA |
| Virus-neutralizing activity | VNA |
| Carbohydrate abbreviations | |
| Galβ1-4GlcNAcβ | LacNAc |
| GalNAcβ1-4GlcNAcβ | LDN |
| Galβ1-3(Fucα1-4)GlcNAcβ | Lewis X, LeX |
| Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc | LNnO |
| Fucα1-2Galβ1-3GlcNAcβ | H type 1 |
| GalNAcα1-3GalNAcβ1-3Galα1-4Galβ1-4Glcβ | Forssman antigen, Fs |
| GalNAcα | Tn |
| (GlcNAcβ1-4)n | chito-oligosaccharides |
References
- Wen, J.; Cheng, Y.; Ling, R.; Dai, Y.; Huang, B.; Huang, W.; Zhang, S.; Jiang, Y. Antibody-dependent enhancement of coronavirus. Int. J. Infect. Dis. 2020, 100, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Zuno, G.A.; Matuz-Flores, M.G.; González-Estevez, G.; Nicoletti, F.; Turrubiates-Hernández, F.J.; Mangano, K.; Muñoz-Valle, J.F. A review: Antibody-dependent enhancement in COVID-19: The not so friendly side of antibodies. Int. J. Immunopathol. Pharmacol. 2021, 35, 205873842110501. [Google Scholar] [CrossRef] [PubMed]
- Gartlan, C.; Tipton, T.; Salguero, F.J.; Sattentau, Q.; Gorringe, A.; Carroll, M.W. Vaccine-Associated Enhanced Disease and Pathogenic Human Coronaviruses. Front. Immunol. 2022, 13, 882972. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020, 5, 1185–1191. [Google Scholar] [CrossRef]
- Ramakrishnan, B.; Viswanathan, K.; Tharakaraman, K.; Dančík, V.; Raman, R.; Babcock, G.J.; Shriver, Z.; Sasisekharan, R. A Structural and Mathematical Modeling Analysis of the Likelihood of Antibody-Dependent Enhancement in Influenza. Trends Microbiol. 2016, 24, 933–943. [Google Scholar] [CrossRef]
- Takada, A.; Kawaoka, Y. Antibody-dependent enhancement of viral infection: Molecular mechanisms andin vivo implications. Rev. Med. Virol. 2003, 13, 387–398. [Google Scholar] [CrossRef]
- Rockstroh, A.; Wolf, J.; Fertey, J.; Kalbitz, S.; Schroth, S.; Lübbert, C.; Ulbert, S.; Borte, S. Correlation of humoral immune responses to different SARS-CoV-2 antigens with virus neutralizing antibodies and symptomatic severity in a German COVID-19 cohort. Emerg. Microbes Infect. 2021, 10, 774–781. [Google Scholar] [CrossRef]
- Iyer, A.S.; Jones, F.K.; Nodoushani, A.; Kelly, M.; Becker, M.; Slater, D.; Mills, R.; Teng, E.; Kamruzzaman, M.; Garcia-Beltran, W.F.; et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020, 5, eabe0367. [Google Scholar] [CrossRef]
- Dolscheid-Pommerich, R.; Bartok, E.; Renn, M.; Kümmerer, B.M.; Schulte, B.; Schmithausen, R.M.; Stoffel-Wagner, B.; Streeck, H.; Saschenbrecker, S.; Steinhagen, K.; et al. Correlation between a quantitative anti-SARS-CoV-2 IgG ELISA and neutralization activity. J. Med. Virol. 2022, 94, 388–392. [Google Scholar] [CrossRef]
- Ryzhov, I.M.; Tuzikov, A.B.; Nizovtsev, A.V.; Baidakova, L.K.; Galanina, O.E.; Shilova, N.V.; Ziganshina, M.M.; Dolgushina, N.V.; Bayramova, G.R.; Sukhikh, G.T.; et al. SARS-CoV-2 Peptide Bioconjugates Designed for Antibody Diagnostics. Bioconjug. Chem. 2021, 32, 1606–1616. [Google Scholar] [CrossRef]
- Nagappan, R.; Flegel, W.A.; Srivastava, K.; Williams, E.C.; Ryzhov, I.; Tuzikov, A.; Galanina, O.; Shilova, N.; Sukhikh, G.; Perry, H.; et al. COVID -19 antibody screening with SARS-CoV-2 red cell kodecytes using routine serologic diagnostic platforms. Transfusion 2021, 61, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Sanda, M.; Morrison, L.; Goldman, R. N- and O-Glycosylation of the SARS-CoV-2 Spike Protein. Anal. Chem. 2021, 93, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Brun, J.; Vasiljevic, S.; Gangadharan, B.; Hensen, M.; Chandran, A.V.; Hill, M.L.; Kiappes, J.; Dwek, R.A.; Alonzi, D.S.; Struwe, W.B.; et al. Assessing Antigen Structural Integrity through Glycosylation Analysis of the SARS-CoV-2 Viral Spike. ACS Central Sci. 2021, 7, 586–593. [Google Scholar] [CrossRef]
- Watanabe, Y.; Berndsen, Z.T.; Raghwani, J.; Seabright, G.E.; Allen, J.D.; Pybus, O.G.; McLellan, J.S.; Wilson, I.A.; Bowden, T.A.; Ward, A.B.; et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020, 11, 2688. [Google Scholar] [CrossRef] [PubMed]
- Butler, D.L.; Gildersleeve, J.C. Abnormal antibodies to self-carbohydrates in SARS-CoV-2 infected patients. PNAS Nexus 2020, 1, pgac062. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, S.; Lidbury, B.A. Antibody-dependent enhancement of infection: Bacteria do it too. Trends Immunol. 2003, 24, 465–467. [Google Scholar] [CrossRef]
- Skurnik, D.; Kropec, A.; Roux, D.; Theilacker, C.; Huebner, J.; Pier, G.B. Natural Antibodies in Normal Human Serum Inhibit Staphylococcus aureus Capsular Polysaccharide Vaccine Efficacy. Clin. Infect. Dis. 2012, 55, 1188–1197. [Google Scholar] [CrossRef]
- DBello-Gil, D.; Manez, R. Exploiting natural anti-carbohydrate antibodies for therapeutic purposes. Biochemistry 2015, 80, 836–845. [Google Scholar] [CrossRef]
- Breiman, A.; Ruvoën-Clouet, N.; Deleers, M.; Beauvais, T.; Jouand, N.; Rocher, J.; Bovin, N.; Labarrière, N.; El Kenz, H.; Le Pendu, J. Low Levels of Natural Anti-α-N-Acetylgalactosamine (Tn) Antibodies Are Associated With COVID-19. Front. Microbiol. 2021, 12, 641460. [Google Scholar] [CrossRef]
- Bovin, N.V. Natural antibodies to glycans. Biochemistry 2013, 78, 786–797. [Google Scholar] [CrossRef]
- Khandia, R.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Malik, Y.S.; Singh, R.K.; Chaicumpa, W. Modulation of Dengue/Zika Virus Pathogenicity by Antibody-Dependent Enhancement and Strategies to Protect against Enhancement in Zika Virus Infection. Front. Immunol. 2018, 9, 597. [Google Scholar] [CrossRef]
- Sarker, A.; Dhama, N.; Gupta, R.D. Dengue virus neutralizing antibody: A review of targets, cross-reactivity, and antibody-dependent enhancement. Front. Immunol. 2023, 14, 1200195. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R. Antibody-Dependent Enhancement of Viral Infections. In Dynamics of Immune Activation in Viral Diseases; Springer: Singapore, 2020; pp. 9–41. [Google Scholar] [CrossRef]
- Klasse, P.J. Neutralization of Virus Infectivity by Antibodies: Old Problems in New Perspectives. Adv. Biol. 2014, 2014, 157895. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Yang, Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 339–341. [Google Scholar] [CrossRef] [PubMed]
- Ricke, D.; Malone, R.W. Medical Countermeasures Analysis of 2019-nCoV and Vaccine Risks for Antibody-Dependent Enhancement (ADE). SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Karthik, K.; Senthilkumar, T.M.A.; Udhayavel, S.; Raj, G.D. Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum. Vaccines Immunother. 2020, 16, 3055–3060. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Sun, S.; Tai, W.; Chen, J.; Geng, Q.; He, L.; Chen, Y.; Wu, J.; Shi, Z.; et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J. Virol. 2020, 94, e02015–e02019. [Google Scholar] [CrossRef]
- Langerak, T.; Mumtaz, N.; Tolk, V.I.; van Gorp, E.C.M.; Martina, B.E.; Rockx, B.; Koopmans, M.P.G. The possible role of cross-reactive dengue virus antibodies in Zika virus pathogenesis. PLoS Pathog. 2019, 15, e1007640. [Google Scholar] [CrossRef]
- Gan, E.S.; Ting, D.H.R.; Chan, K.R. The mechanistic role of antibodies to dengue virus in protection and disease pathogenesis. Expert Rev. Anti-Infect. Ther. 2017, 15, 111–119. [Google Scholar] [CrossRef]
- Dustin, M.L. Complement Receptors in Myeloid Cell Adhesion and Phagocytosis. Microbiol. Spectr. 2016, 4, 429–445. [Google Scholar] [CrossRef]
- Dunkelberger, J.R.; Song, W.-C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef] [PubMed]
- French, M.A. Antibody-mediated control of HIV-1 infection through an alternative pathway. Aids 2019, 33, 1961–1966. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nie, J.; Wang, H.; Zhao, Q.; Xiong, Y.; Deng, L.; Song, S.; Ma, Z.; Mo, P.; Zhang, Y. Characteristics of Peripheral Lymphocyte Subset Alteration in COVID-19 Pneumonia. J. Infect. Dis. 2020, 221, 1762–1769. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Su, X.; Pan, P.; Zhang, L.; Hu, Y.; Tan, H.; Wu, D.; Liu, B.; Li, H.; Li, H.; et al. Neutrophil extracellular traps are indirectly triggered by lipopolysaccharide and contribute to acute lung injury. Sci. Rep. 2016, 6, 37252. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.-Y.; Yuen, K.-S.; Ye, Z.-W.; Chan, C.-P.; Jin, D.-Y. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: Lessons from other pathogenic viruses. Emerg. Microbes Infect. 2020, 9, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Korteweg, C. Pathology and Pathogenesis of Severe Acute Respiratory Syndrome. Am. J. Pathol. 2007, 170, 1136–1147. [Google Scholar] [CrossRef]
- Pang, X.; Zhang, R.; Cheng, G. Progress towards understanding the pathogenesis of dengue hemorrhagic fever. Virol. Sin. 2017, 32, 16–22. [Google Scholar] [CrossRef]
- Rothman, A.L. Immunity to dengue virus: A tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 2011, 11, 532–543. [Google Scholar] [CrossRef]
- Campos, G.S.; Pinho, A.C.O.; Brandão, C.J.D.F.; Bandeira, A.C.; Sardi, S.I. Dengue Virus 4 (DENV-4) Re-Emerges after 30 Years in Brazil: Cocirculation of DENV-2, DENV-3, and DENV-4 in Bahia. Jpn. J. Infect. Dis. 2015, 68, 45–49. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Ooi, E.-E.; Horstick, O.; Wills, B. Dengue. Lancet 2019, 393, 350–363. [Google Scholar] [CrossRef]
- Roy, S.K.; Bhattacharjee, S. Dengue virus: Epidemiology, biology, and disease aetiology. Can. J. Microbiol. 2021, 67, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Tsai, Y.-T.; Wang, S.-F.; Wang, W.-H.; Chen, Y.-H. Dengue vaccine: An update. Expert Rev. Anti-Infect. Ther. 2021, 19, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Nunez, N.V.; Hoornweg, T.E.; van de Pol, D.P.; Sjollema, K.A.; Flipse, J.; van der Schaar, H.M.; Smit, J.M. How antibodies alter the cell entry pathway of dengue virus particles in macrophages. Sci. Rep. 2016, 6, 28768. [Google Scholar] [CrossRef] [PubMed]
- Nanaware, N.; Banerjee, A.; Bagchi, S.M.; Bagchi, P.; Mukherjee, A. Dengue Virus Infection: A Tale of Viral Exploitations and Host Responses. Viruses 2021, 13, 1967. [Google Scholar] [CrossRef]
- Narayan, R.; Tripathi, S. Intrinsic ADE: The Dark Side of Antibody Dependent Enhancement During Dengue Infection. Front. Cell. Infect. Microbiol. 2020, 10, 580096. [Google Scholar] [CrossRef]
- Feng, T.; Zhang, J.; Chen, Z.; Pan, W.; Chen, Z.; Yan, Y.; Dai, J. Glycosylation of viral proteins: Implication in virus–host interaction and virulence. Virulence 2022, 13, 670–683. [Google Scholar] [CrossRef]
- Lei, Y.; Yu, H.; Dong, Y.; Yang, J.; Ye, W.; Wang, Y.; Chen, W.; Jia, Z.; Xu, Z.; Li, Z.; et al. Characterization of N-Glycan Structures on the Surface of Mature Dengue 2 Virus Derived from Insect Cells. PLoS ONE 2015, 10, e0132122. [Google Scholar] [CrossRef]
- Yap, S.S.L.; Nguyen-Khuong, T.; Rudd, P.M.; Alonso, S. Dengue Virus Glycosylation: What Do We Know? Front. Microbiol. 2017, 8, 1415. [Google Scholar] [CrossRef]
- Idris, F.; Muharram, S.H.; Diah, S. Glycosylation of dengue virus glycoproteins and their interactions with carbohydrate receptors: Possible targets for antiviral therapy. Arch. Virol. 2016, 161, 1751–1760. [Google Scholar] [CrossRef]
- Chan, K.R.; Ong, E.Z.; Tan, H.C.; Zhang, S.L.-X.; Zhang, Q.; Tang, K.F.; Kaliaperumal, N.; Lim, A.P.C.; Hibberd, M.L.; Chan, S.H.; et al. Leukocyte immunoglobulin-like receptor B1 is critical for antibody-dependent dengue. Proc. Natl. Acad. Sci. USA 2014, 111, 2722–2727. [Google Scholar] [CrossRef]
- Flipse, J.; Diosa-Toro, M.A.; Hoornweg, T.E.; van de Pol, D.P.I.; Urcuqui-Inchima, S.; Smit, J.M. Antibody-Dependent Enhancement of Dengue Virus Infection in Primary Human Macrophages; Balancing Higher Fusion against Antiviral Responses. Sci. Rep. 2016, 6, 29201. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.-T.; Chuang, Y.-J.; Lin, Y.-S.; Wan, S.-W.; Chen, C.-L.; Lin, C.-F. An emerging role for the anti-inflammatory cytokine interleukin-10 in dengue virus infection. J. Biomed. Sci. 2013, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chi, J.; Lv, W.; Wang, Y. Obesity and diabetes as high-risk factors for severe coronavirus disease 2019 (COVID-19). Diabetes/Metabolism Res. Rev. 2021, 37, e3377. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; on behalf of the HLH across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Pang, T.; Cardosa, M.J.; Guzman, M.G. Of cascades and perfect storms: The immunopathogenesis of dengue haemorrhagic fever-dengue shock syndrome (DHF/DSS). Immunol. Cell Biol. 2007, 85, 43–45. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Gresh, L.; Halloran, M.E.; Mercado, J.C.; Kuan, G.; Gordon, A.; Balmaseda, A.; Harris, E. Antibody-dependent enhancement of severe dengue disease in humans. Science 2017, 358, 929–932. [Google Scholar] [CrossRef]
- Serrano-Collazo, C.; Pérez-Guzmán, E.X.; Pantoja, P.; Hassert, M.A.; Rodríguez, I.V.; Giavedoni, L.; Hodara, V.; Parodi, L.; Cruz, L.; Arana, T.; et al. Effective control of early Zika virus replication by Dengue immunity is associated to the length of time between the 2 infections but not mediated by antibodies. PLoS Neglected Trop. Dis. 2020, 14, e0008285. [Google Scholar] [CrossRef]
- Jaume, M.; Yip, M.S.; Cheung, C.Y.; Leung, H.L.; Li, P.H.; Kien, F.; Dutry, I.; Callendret, B.; Escriou, N.; Altmeyer, R.; et al. Anti-Severe Acute Respiratory Syndrome Coronavirus Spike Antibodies Trigger Infection of Human Immune Cells via a pH- and Cysteine Protease-Independent FcγR Pathway. J. Virol. 2011, 85, 10582–10597. [Google Scholar] [CrossRef]
- Liu, L.; Wei, Q.; Lin, Q.; Fang, J.; Wang, H.; Kwok, H.; Tang, H.; Nishiura, K.; Peng, J.; Tan, Z.; et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. J. Clin. Investig. 2019, 4, e123158. [Google Scholar] [CrossRef]
- Eroshenko, N.; Gill, T.; Keaveney, M.K.; Church, G.M.; Trevejo, J.M.; Rajaniemi, H. Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat. Biotechnol. 2020, 38, 789–791. [Google Scholar] [CrossRef]
- Ricke, D.O. Two Different Antibody-Dependent Enhancement (ADE) Risks for SARS-CoV-2 Antibodies. Front. Immunol. 2021, 12, 640093. [Google Scholar] [CrossRef] [PubMed]
- Ajmeriya, S.; Kumar, A.; Karmakar, S.; Rana, S.; Singh, H. Neutralizing Antibodies and Antibody-Dependent Enhancement in COVID-19: A Perspective. J. Indian Inst. Sci. 2022, 102, 671–687. [Google Scholar] [CrossRef]
- Willis, E.; Hensley, S.E. Characterization of Zika virus binding and enhancement potential of a large panel of flavivirus murine monoclonal antibodies. Virology 2017, 508, 1–6. [Google Scholar] [CrossRef]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef]
- Pinto, D.; Park, Y.-J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.H.; Morrison, S.L. Studies of Aglycosylated Chimeric Mouse-Human IgG. Role of Carbohydrate in the Structure and Effector Functions Mediated by the Human IgG Constant Region. J. Immunol. 1989, 143, 2595–2601. Available online: http://www.ncbi.nlm.nih.gov/pubmed/2507634 (accessed on 14 July 2023). [CrossRef] [PubMed]
- Chappel, M.S.; Isenman, D.E.; Everett, M.; Xu, Y.Y.; Dorrington, K.J.; Klein, M.H. Identification of the Fc gamma receptor class I binding site in human IgG through the use of recombinant IgG1/IgG2 hybrid and point-mutated antibodies. Proc. Natl. Acad. Sci. USA 1991, 88, 9036–9040. [Google Scholar] [CrossRef]
- Lo, M.; Kim, H.S.; Tong, R.K.; Bainbridge, T.W.; Vernes, J.-M.; Zhang, Y.; Lin, Y.L.; Chung, S.; Dennis, M.S.; Zuchero, Y.J.Y.; et al. Effector-attenuating Substitutions That Maintain Antibody Stability and Reduce Toxicity in Mice. J. Biol. Chem. 2017, 292, 3900–3908. [Google Scholar] [CrossRef]
- Yuan, F.F.; Tanner, J.; Chan, P.K.S.; Biffin, S.; Dyer, W.B.; Geczy, A.F.; Tang, J.W.; Hui, D.S.C.; Sung, J.J.Y.; Sullivan, J.S. Influence of FcgammaRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens 2005, 66, 291–296. [Google Scholar] [CrossRef]
- Krapp, S.; Mimura, Y.; Jefferis, R.; Huber, R.; Sondermann, P. Structural Analysis of Human IgG-Fc Glycoforms Reveals a Correlation Between Glycosylation and Structural Integrity. J. Mol. Biol. 2003, 325, 979–989. [Google Scholar] [CrossRef]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef]
- Della-Torre, E.; Lanzillotta, M.; Strollo, M.; Ramirez, G.A.; Dagna, L.; Tresoldi, M. Serum IgG4 level predicts COVID-19 related mortality. Eur. J. Intern. Med. 2021, 93, 107–109. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, R.J. Viruses and Glycosylation. In Glycovirology Protocols. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2007; Volume 379. [Google Scholar]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Sumbria, D.; Berber, E.; Mathayan, M.; Rouse, B.T. Virus Infections and Host Metabolism—Can We Manage the Interactions? Front. Immunol. 2021, 11, 594963. [Google Scholar] [CrossRef]
- Mazumder, N.; Lyn, R.K.; Singaravelu, R.; Ridsdale, A.; Moffatt, D.J.; Hu, C.-W.; Tsai, H.-R.; McLauchlan, J.; Stolow, A.; Kao, F.-J.; et al. Fluorescence Lifetime Imaging of Alterations to Cellular Metabolism by Domain 2 of the Hepatitis C Virus Core Protein. PLoS ONE 2013, 8, e66738. [Google Scholar] [CrossRef] [PubMed]
- Dowd, K.A.; Jost, C.A.; Durbin, A.P.; Whitehead, S.S.; Pierson, T.C. A Dynamic Landscape for Antibody Binding Modulates Antibody-Mediated Neutralization of West Nile Virus. PLoS Pathog. 2011, 7, e1002111. [Google Scholar] [CrossRef]
- Pierson, T.C.; Fremont, D.H.; Kuhn, R.J.; Diamond, M.S. Structural Insights into the Mechanisms of Antibody-Mediated Neutralization of Flavivirus Infection: Implications for Vaccine Development. Cell Host Microbe 2008, 4, 229–238. [Google Scholar] [CrossRef]
- Newton, A.H.; Cardani, A.; Braciale, T.J. The host immune response in respiratory virus infection: Balancing virus clearance and immunopathology. Semin. Immunopathol. 2016, 38, 471–482. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Z.; Li, S.; Xu, W.; Zhang, Q.; Silva, I.T.; Li, C.; Wu, Y.; Jiang, Q.; Liu, Z.; et al. Enhancement versus neutralization by SARS-CoV-2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep. 2021, 34, 108699. [Google Scholar] [CrossRef]
- Wang, S.-F.; Tseng, S.-P.; Yen, C.-H.; Yang, J.-Y.; Tsao, C.-H.; Shen, C.-W.; Chen, K.-H.; Liu, F.-T.; Liu, W.-T.; Chen, Y.-M.A.; et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014, 451, 208–214. [Google Scholar] [CrossRef]
- Klasse, P.J.; Sattentau, Q.J. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 2002, 83, 2091–2108. [Google Scholar] [CrossRef] [PubMed]
- VanBlargan, L.A.; Goo, L.; Pierson, T.C. Deconstructing the Antiviral Neutralizing-Antibody Response: Implications for Vaccine Development and Immunity. Microbiol. Mol. Biol. Rev. 2016, 80, 989–1010. [Google Scholar] [CrossRef] [PubMed]
- Arvin, A.M.; Fink, K.; Schmid, M.A.; Cathcart, A.; Spreafico, R.; Havenar-Daughton, C.; Lanzavecchia, A.; Corti, D.; Virgin, H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 2020, 584, 353–363. [Google Scholar] [CrossRef]
- Kliks, S.C.; Halstead, S.B. An explanation for enhanced virus plaque formation in chick embryo cells. Nature 1980, 285, 504–505. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, L.; Kuwahara, K.; Li, L.; Liu, Z.; Li, T.; Zhu, H.; Liu, J.; Xu, Y.; Xie, J.; et al. Immunodominant SARS Coronavirus Epitopes in Humans Elicited both Enhancing and Neutralizing Effects on Infection in Non-human Primates. ACS Infect. Dis. 2016, 2, 361–376. [Google Scholar] [CrossRef]
- Ngono, A.E.; Shresta, S. Immune Response to Dengue and Zika. Annu. Rev. Immunol. 2018, 36, 279–308. [Google Scholar] [CrossRef]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Taylor, A.; Foo, S.-S.; Bruzzone, R.; Dinh, L.V.; King, N.J.C.; Mahalingam, S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015, 268, 340–364. [Google Scholar] [CrossRef]
- Mok, D.Z.L.; Chan, K.R. The Effects of Pre-Existing Antibodies on Live-Attenuated Viral Vaccines. Viruses 2020, 12, 520. [Google Scholar] [CrossRef]
- Pang, N.Y.-L.; Pang, A.S.-R.; Chow, V.T.; Wang, D.-Y. Understanding neutralising antibodies against SARS-CoV-2 and their implications in clinical practice. Mil. Med. Res. 2021, 8, 47. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Yamada, S.; Doki, T.; Hohdatsu, T. Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: Antibody-dependent enhancement infection of cats with type I FIPV via the oral route. J. Veter. Med. Sci. 2019, 81, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.-S.; Chen, W.-J.; Chen, H.-Y.; Lin, S.-F.; Wang, M.-C.; Di, J.; Lu, Y.-T.; Liu, C.-L.; Chang, S.-C.; Chao, C.-L.; et al. Neutralizing Antibody Response and SARS Severity. Emerg. Infect. Dis. 2005, 11, 1730–1737. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Chan, P.; Ip, M.; Wong, E.; Ho, J.; Ho, C.; Cockram, C.; Hui, D.S. Anti-SARS-CoV IgG response in relation to disease severity of severe acute respiratory syndrome. J. Clin. Virol. 2006, 35, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, F.; Yu, W.; He, T.; Yu, J.; Yi, C.E.; Ba, L.; Li, W.; Farzan, M.; Chen, Z.; et al. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J. Med. Virol. 2006, 78, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yip, M.S.; Leung, H.L.; Li, P.H.; Cheung, C.Y.; Dutry, I.; Li, D.; Daëron, M.; Bruzzone, R.; Peiris, J.S.M.; Jaume, M. Antibody-Dependent Enhancement of SARS Coronavirus Infection and Its Role in the Pathogenesis of SARS. Hong Kong Med. J. 2016, 22, 25–31. Available online: http://www.ncbi.nlm.nih.gov/pubmed/27390007 (accessed on 14 July 2023). [CrossRef] [PubMed]
- Du, L.; Zhao, G.; Yang, Y.; Qiu, H.; Wang, L.; Kou, Z.; Tao, X.; Yu, H.; Sun, S.; Tseng, C.-T.K.; et al. A Conformation-Dependent Neutralizing Monoclonal Antibody Specifically Targeting Receptor-Binding Domain in Middle East Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 2014, 88, 7045–7053. [Google Scholar] [CrossRef]
- Ou, J.; Zhou, Z.; Dai, R.; Zhang, J.; Zhao, S.; Wu, X.; Lan, W.; Ren, Y.; Cui, L.; Lan, Q.; et al. V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity. J. Virol. 2021, 95, JVI0061721. [Google Scholar] [CrossRef]
- Jin, X.; Xu, K.; Jiang, P.; Lian, J.; Hao, S.; Yao, H.; Jia, H.; Zhang, Y.; Zheng, L.; Zheng, N.; et al. Virus strain from a mild COVID-19 patient in Hangzhou represents a new trend in SARS-CoV-2 evolution potentially related to Furin cleavage site. Emerg. Microbes Infect. 2020, 9, 1474–1488. [Google Scholar] [CrossRef]
- Kissler, S.M.; Tedijanto, C.; Goldstein, E.; Grad, Y.H.; Lipsitch, M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 2020, 368, 860–868. [Google Scholar] [CrossRef]
- Cegolon, L.; Pichierri, J.; Mastrangelo, G.; Cinquetti, S.; Sotgiu, G.; Bellizzi, S.; Pichierri, G. Hypothesis to explain the severe form of COVID-19 in Northern Italy. BMJ Glob. Health 2020, 5, e002564. [Google Scholar] [CrossRef]
- Tetro, J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020, 22, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Zaichuk, T.A.; Nechipurenko, Y.D.; Adzhubey, A.A.; Onikienko, S.B.; Chereshnev, V.A.; Zainutdinov, S.S.; Kochneva, G.V.; Netesov, S.V.; Matveeva, O.V. The Challenges of Vaccine Development against Betacoronaviruses: Antibody Dependent Enhancement and Sendai Virus as a Possible Vaccine Vector. Mol. Biol. 2020, 54, 812–826. [Google Scholar] [CrossRef]
- Ng, K.W.; Faulkner, N.; Cornish, G.H.; Rosa, A.; Harvey, R.; Hussain, S.; Ulferts, R.; Earl, C.; Wrobel, A.G.; Benton, D.J.; et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020, 370, 1339–1343. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Li, P.; Ji, Y.; Ikram, A.; Pan, Q. Cross-reactivity towards SARS-CoV-2: The potential role of low-pathogenic human coronaviruses. Lancet Microbe 2020, 1, e151. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Wu, N.C.; Tsang, O.T.-Y.; Yuan, M.; Perera, R.A.P.M.; Leung, W.S.; So, R.T.Y.; Chan, J.M.C.; Yip, G.K.; Chik, T.S.H.; et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020, 31, 107725. [Google Scholar] [CrossRef]
- Okba, N.M.A.; Müller, M.A.; Li, W.; Wang, C.; GeurtsvanKessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; De Bruin, E.; Chandler, F.D.; et al. Severe Acute Respiratory Syndrome Coronavirus 2−Specific Antibody Responses in Coronavirus Disease Patients. Emerg. Infect. Dis. 2020, 26, 478–1488. [Google Scholar] [CrossRef]
- Joyner, M.J.; Bruno, K.A.; Klassen, S.A.; Kunze, K.L.; Johnson, P.W.; Lesser, E.R.; Wiggins, C.C.; Senefeld, J.W.; Klompas, A.M.; Hodge, D.O.; et al. Safety Update: COVID-19 Convalescent Plasma in 20,000 Hospitalized Patients. Mayo Clin. Proc. 2020, 95, 1888–1897. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Helle, F.; Duverlie, G.; Dubuisson, J. The Hepatitis C Virus Glycan Shield and Evasion of the Humoral Immune Response. Viruses 2011, 3, 1909–1932. [Google Scholar] [CrossRef] [PubMed]
- Tate, M.D.; Job, E.R.; Deng, Y.-M.; Gunalan, V.; Maurer-Stroh, S.; Reading, P.C. Playing Hide and Seek: How Glycosylation of the Influenza Virus Hemagglutinin Can Modulate the Immune Response to Infection. Viruses 2014, 6, 1294–1316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, W.; Mao, Y.; Chen, Y.; Zheng, S.; Cao, W.; Zhu, J.; Hu, L.; Gong, M.; Cheng, J.; et al. O-Glycosylation Landscapes of SARS-CoV-2 Spike Proteins. Front. Chem. 2021, 9, 689521. [Google Scholar] [CrossRef] [PubMed]
- Lenza, M.P.; Oyenarte, I.; Diercks, T.; Quintana, J.I.; Gimeno, A.; Coelho, H.; Diniz, A.; Peccati, F.; Delgado, S.; Bosch, A.; et al. Structural Characterization of N-Linked Glycans in the Receptor Binding Domain of the SARS-CoV-2 Spike Protein and their Interactions with Human Lectins. Angew. Chem. Int. Ed. 2020, 59, 23763–23771. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.D. The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 2009, 5, 1087–1104. [Google Scholar] [CrossRef]
- Wang, D.; Lu, J. Glycan arrays lead to the discovery of autoimmunogenic activity of SARS-CoV. Physiol. Genom. 2004, 18, 245–248. [Google Scholar] [CrossRef]
- Wielgat, P.; Rogowski, K.; Godlewska, K.; Car, H. Coronaviruses: Is Sialic Acid a Gate to the Eye of Cytokine Storm? From the Entry to the Effects. Cells 2020, 9, 1963. [Google Scholar] [CrossRef]
- Remoortere, A.V.; Hokke, C.H.; Dam, G.J.; Die, I.V.; M.Deelder, A.; Eijnden, D.H.D. Various stages of Schistosoma express Lewisx, LacdiNAc, GalNAc 1-4 (Fuc 1-3)GlcNAc and GalNAc 1-4(Fuc 1-2Fuc 1-3)GlcNAc carbohydrate epitopes: Detection with monoclonal antibodies that are characterized by enzymatically synthesized neoglycoproteins. Glycobiology 2000, 10, 601–609. [Google Scholar] [CrossRef]
- Luyai, A.E.; Heimburg-Molinaro, J.; Prasanphanich, N.S.; Mickum, M.L.; Lasanajak, Y.; Song, X.; Nyame, A.K.; Wilkins, P.; Rivera-Marrero, C.A.; Smith, D.F.; et al. Differential expression of anti-glycan antibodies in schistosome-infected humans, rhesus monkeys and mice. Glycobiology 2014, 24, 602–618. [Google Scholar] [CrossRef]
- Mickum, M.L.; Prasanphanich, N.S.; Heimburg-Molinaro, J.; Leon, K.E.; Cummings, R.D. Deciphering the glycogenome of schistosomes. Front. Genet. 2014, 5, 262. [Google Scholar] [CrossRef]
- Crispin, M.; Doores, K.J. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr. Opin. Virol. 2015, 11, 63–69. [Google Scholar] [CrossRef]
- Shajahan, A.; Pepi, L.E.; Rouhani, D.S.; Heiss, C.; Azadi, P. Glycosylation of SARS-CoV-2: Structural and functional insights. Anal. Bioanal. Chem. 2021, 413, 7179–7193. [Google Scholar] [CrossRef] [PubMed]
- Temme, J.S.; Butler, D.L.; Gildersleeve, J.C. Anti-glycan antibodies: Roles in human disease. Biochem. J. 2021, 478, 1485–1509. [Google Scholar] [CrossRef] [PubMed]
- Shilova, N.; Huflejt, M.E.; Vuskovic, M.; Obukhova, P.; Navakouski, M.; Khasbiullina, N.; Pazynina, G.; Galanina, O.; Bazhenov, A.; Bovin, N. Natural Antibodies Against Sialoglycans. Biosynthesis 2015, 366, 169–181. [Google Scholar] [CrossRef]
- Huflejt, M.E.; Vuskovic, M.; Vasiliu, D.; Xu, H.; Obukhova, P.; Shilova, N.; Tuzikov, A.; Galanina, O.; Arun, B.; Lu, K.; et al. Anti-carbohydrate antibodies of normal sera: Findings, surprises and challenges. Mol. Immunol. 2009, 46, 3037–3049. [Google Scholar] [CrossRef] [PubMed]
- Lobo, P.I. Role of Natural Autoantibodies and Natural IgM Anti-Leucocyte Autoantibodies in Health and Disease. Front. Immunol. 2016, 7, 198. [Google Scholar] [CrossRef]
- Silverman, G.J. Regulatory natural autoantibodies to apoptotic cells: Pallbearers and protectors. Arthritis Rheum. 2011, 63, 597–602. [Google Scholar] [CrossRef]
- Ziganshina, M.M.; Bovin, N.V.; Sukhikh, G.T. Natural antibodies as a key element of the mechanism supporting homeostasis in immune system. Immunoloriya 2013, 34, 277–281. [Google Scholar]
- Vollmers, H.P.; Brändlein, S. Natural IgM antibodies: The orphaned molecules in immune surveillance. Adv. Drug Deliv. Rev. 2006, 58, 755–765. [Google Scholar] [CrossRef]
- Maddur, M.S.; Lacroix-Desmazes, S.; Dimitrov, J.D.; Kazatchkine, M.D.; Bayry, J.; Kaveri, S.V. Natural Antibodies: From First-Line Defense Against Pathogens to Perpetual Immune Homeostasis. Clin. Rev. Allergy Immunol. 2020, 58, 213–228. [Google Scholar] [CrossRef]
- Zhou, Z.-H.; Zhang, Y.; Hu, Y.-F.; Wahl, L.M.; Cisar, J.O.; Notkins, A.L. The Broad Antibacterial Activity of the Natural Antibody Repertoire Is Due to Polyreactive Antibodies. Cell Host Microbe 2007, 1, 51–61. [Google Scholar] [CrossRef]
- Kaveri, S.V.; Silverman, G.J.; Bayry, J. Natural IgM in Immune Equilibrium and Harnessing Their Therapeutic Potential. J. Immunol. 2012, 188, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Bovin, N.V.; Obukhova, P.S.; Galanina, O.E.; Dobrochaeva, K.L.; Khasbiullina, N.R.; Shilova, N.V.; Antipova, N.V. Is it time to switch over to glyco molecular patterns? In Glycome: The Hidden Code in Biology; Banerjee, D., Ed.; Nova Science Publishers: New York, NY, USA, 2021; pp. 377–395. [Google Scholar]
- Dobrochaeva, K.; Khasbiullina, N.; Shilova, N.; Antipova, N.; Obukhova, P.; Ovchinnikova, T.; Galanina, O.; Blixt, O.; Kunz, H.; Filatov, A.; et al. Specificity of human natural antibodies referred to as anti-Tn. Mol. Immunol. 2020, 120, 74–82. [Google Scholar] [CrossRef]
- Hamadeh, R.M.; Jarvis, G.A.; Galili, U.; Mandrell, R.E.; Zhou, P.; Griffiss, J.M. Human natural anti-Gal IgG regulates alternative complement pathway activation on bacterial surfaces. J. Clin. Investig. 1992, 89, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Hamadeh, R.M.; Estabrook, M.M.; Zhou, P.; Jarvis, G.A.; Griffiss, J.M. Anti-Gal binds to pili of Neisseria meningitidis: The immunoglobulin A isotype blocks complement-mediated killing. Infect. Immun. 1995, 63, 4900–4906. [Google Scholar] [CrossRef] [PubMed]
- Manez, R.; Perez-Cruz, M.; Bello, D.; Dominguez, M.; Costa, C. 0728. Removal of natural anti-galactose α1,3 galactose antibodies with GAS914 enhances humoral immunity and prevents sepsis mortality in mice. Intensiv. Care Med. Exp. 2014, 2, P50. [Google Scholar] [CrossRef]
- Matyushkina, D.; Shokina, V.; Tikhonova, P.; Manuvera, V.; Shirokov, D.; Kharlampieva, D.; Lazarev, V.; Varizhuk, A.; Vedekhina, T.; Pavlenko, A.; et al. Autoimmune Effect of Antibodies against the SARS-CoV-2 Nucleoprotein. Viruses 2022, 14, 1141. [Google Scholar] [CrossRef] [PubMed]
- Boukhari, R.; Breiman, A.; Jazat, J.; Ruvoën-Clouet, N.; Martinez, S.; Damais-Cepitelli, A.; Le Niger, C.; Devie-Hubert, I.; Penasse, F.; Mauriere, D.; et al. ABO Blood Group Incompatibility Protects against SARS-CoV-2 Transmission. Front. Microbiol. 2022, 12, 799519. [Google Scholar] [CrossRef]
- Guillon, P.; Clément, M.; Sébille, V.; Rivain, J.-G.; Chou, C.-F.; Ruvoën-Clouet, N.; Le Pendu, J. Inhibition of the interaction between the SARS-CoV Spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology 2008, 18, 1085–1093. [Google Scholar] [CrossRef]
- Cheng, Y.; Cheng, G.; Chui, C.H.; Lau, F.Y.; Chan, P.K.S.; Ng, M.H.L.; Sung, J.J.Y.; Wong, R.S.M. ABO Blood Group and Susceptibility to Severe Acute Respiratory Syndrome. JAMA 2005, 293, 1447–1451. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, Y.; Huang, H.; Li, D.; Gu, D.; Lu, X.; Zhang, Z.; Liu, L.; Liu, T.; Liu, Y.; et al. Relationship between the ABO Blood Group and the Coronavirus Disease 2019 (COVID-19) Susceptibility. Clin. Infect. Dis. 2021, 73, 328–331. [Google Scholar] [CrossRef]
- Breiman, A.; Ruvën-Clouet, N.; Le Pendu, J. Harnessing the natural anti-glycan immune response to limit the transmission of enveloped viruses such as SARS-CoV-2. PLoS Pathog. 2020, 16, e1008556. [Google Scholar] [CrossRef] [PubMed]
- Deleers, M.; Breiman, A.; Daubie, V.; Maggetto, C.; Barreau, I.; Besse, T.; Clémenceau, B.; Ruvoën-Clouet, N.; Fils, J.-F.; Maillart, E.; et al. Covid-19 and blood groups: ABO antibody levels may also matter. Int. J. Infect. Dis. 2021, 104, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Smorodin, E.P.; Kurtenkov, O.A.; Sergeyev, B.L.; Kodar, K.E.; Chuzmarov, V.I.; Afanasyev, V.P. Postoperative change of anti-Thomsen-Friedenreich and Tn IgG level: The follow-up study of gastrointestinal cancer patients. World J. Gastroenterol. 2008, 14, 4352–4358. [Google Scholar] [CrossRef]
- Zhang, T.; de Waard, A.A.; Wuhrer, M.; Spaapen, R.M. The Role of Glycosphingolipids in Immune Cell Functions. Front. Immunol. 2019, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Obukhova, P.; Piskarev, V.; Severov, V.; Pazynina, G.; Tuzikov, A.; Navakouski, M.; Shilova, N.; Bovin, N. Profiling of serum antibodies with printed glycan array: Room for data misinterpretation. Glycoconj. J. 2011, 28, 501–505. [Google Scholar] [CrossRef]
- Perepelov, A.V.; Liu, B.; Senchenkova, S.N.; Guo, D.; Shevelev, S.D.; Feng, L.; Shashkov, A.S.; Wang, L.; Knirel, Y.A. O-antigen structure and gene clusters of Escherichia coli O51 and Salmonellaenterica O57; another instance of identical O-antigens in the two species. Carbohydr. Res. 2011, 346, 828–832. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziganshina, M.M.; Shilova, N.V.; Khalturina, E.O.; Dolgushina, N.V.; V. Borisevich, S.; Yarotskaya, E.L.; Bovin, N.V.; Sukhikh, G.T. Antibody-Dependent Enhancement with a Focus on SARS-CoV-2 and Anti-Glycan Antibodies. Viruses 2023, 15, 1584. https://doi.org/10.3390/v15071584
Ziganshina MM, Shilova NV, Khalturina EO, Dolgushina NV, V. Borisevich S, Yarotskaya EL, Bovin NV, Sukhikh GT. Antibody-Dependent Enhancement with a Focus on SARS-CoV-2 and Anti-Glycan Antibodies. Viruses. 2023; 15(7):1584. https://doi.org/10.3390/v15071584
Chicago/Turabian StyleZiganshina, Marina M., Nadezhda V. Shilova, Eugenia O. Khalturina, Natalya V. Dolgushina, Sergey V. Borisevich, Ekaterina L. Yarotskaya, Nicolai V. Bovin, and Gennady T. Sukhikh. 2023. "Antibody-Dependent Enhancement with a Focus on SARS-CoV-2 and Anti-Glycan Antibodies" Viruses 15, no. 7: 1584. https://doi.org/10.3390/v15071584
APA StyleZiganshina, M. M., Shilova, N. V., Khalturina, E. O., Dolgushina, N. V., V. Borisevich, S., Yarotskaya, E. L., Bovin, N. V., & Sukhikh, G. T. (2023). Antibody-Dependent Enhancement with a Focus on SARS-CoV-2 and Anti-Glycan Antibodies. Viruses, 15(7), 1584. https://doi.org/10.3390/v15071584






