Identification of Markers Associated with Wheat Dwarf Virus (WDV) Tolerance/Resistance in Barley (Hordeum vulgare ssp. vulgare) Using Genome-Wide Association Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
Phenotyping
2.2. Statistical Analysis
2.3. Genotyping
2.3.1. Population Structure
2.3.2. Genome -Wide Association Study (GWAS)
3. Results
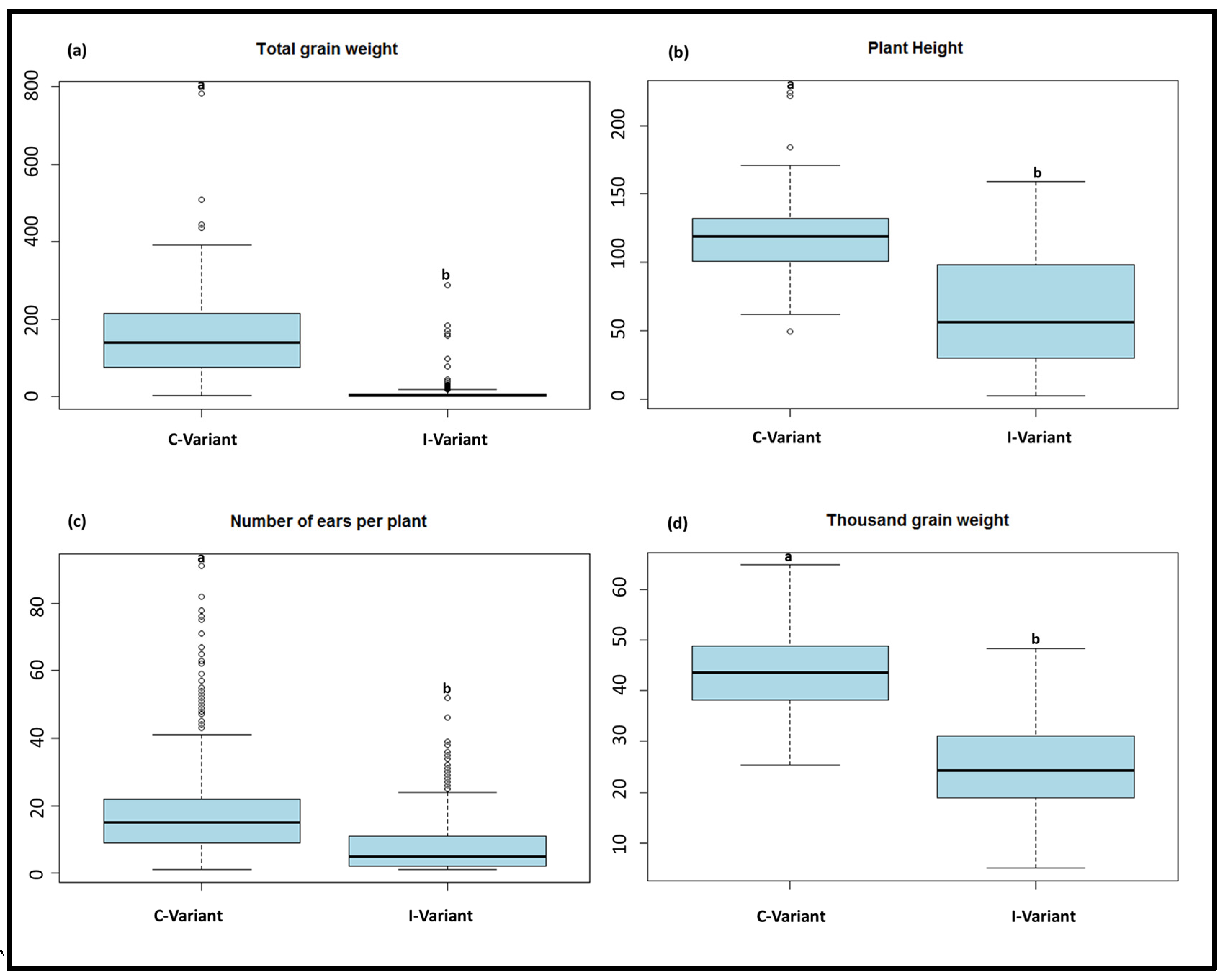
3.1. Phenotypic Data
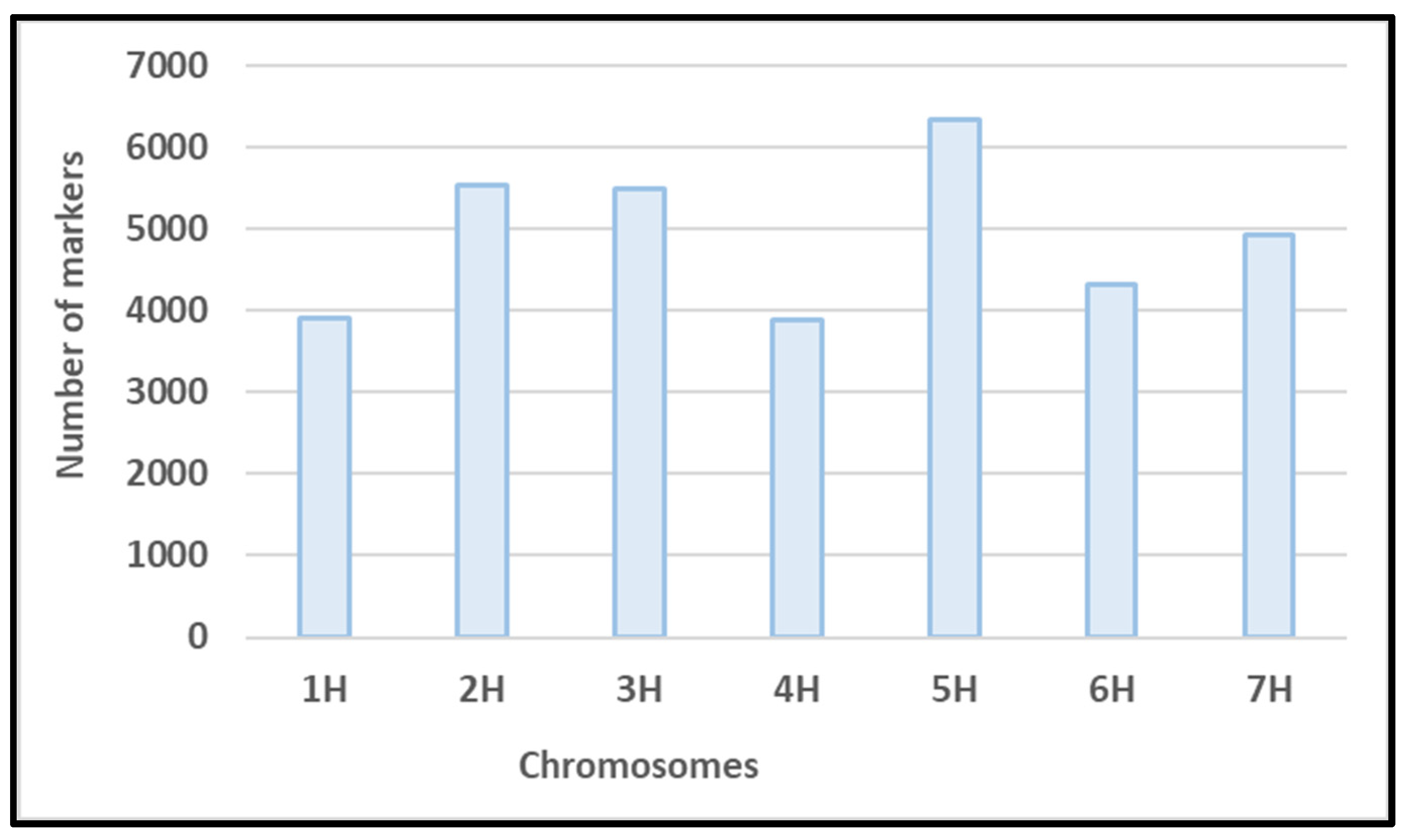
3.2. Genotyping
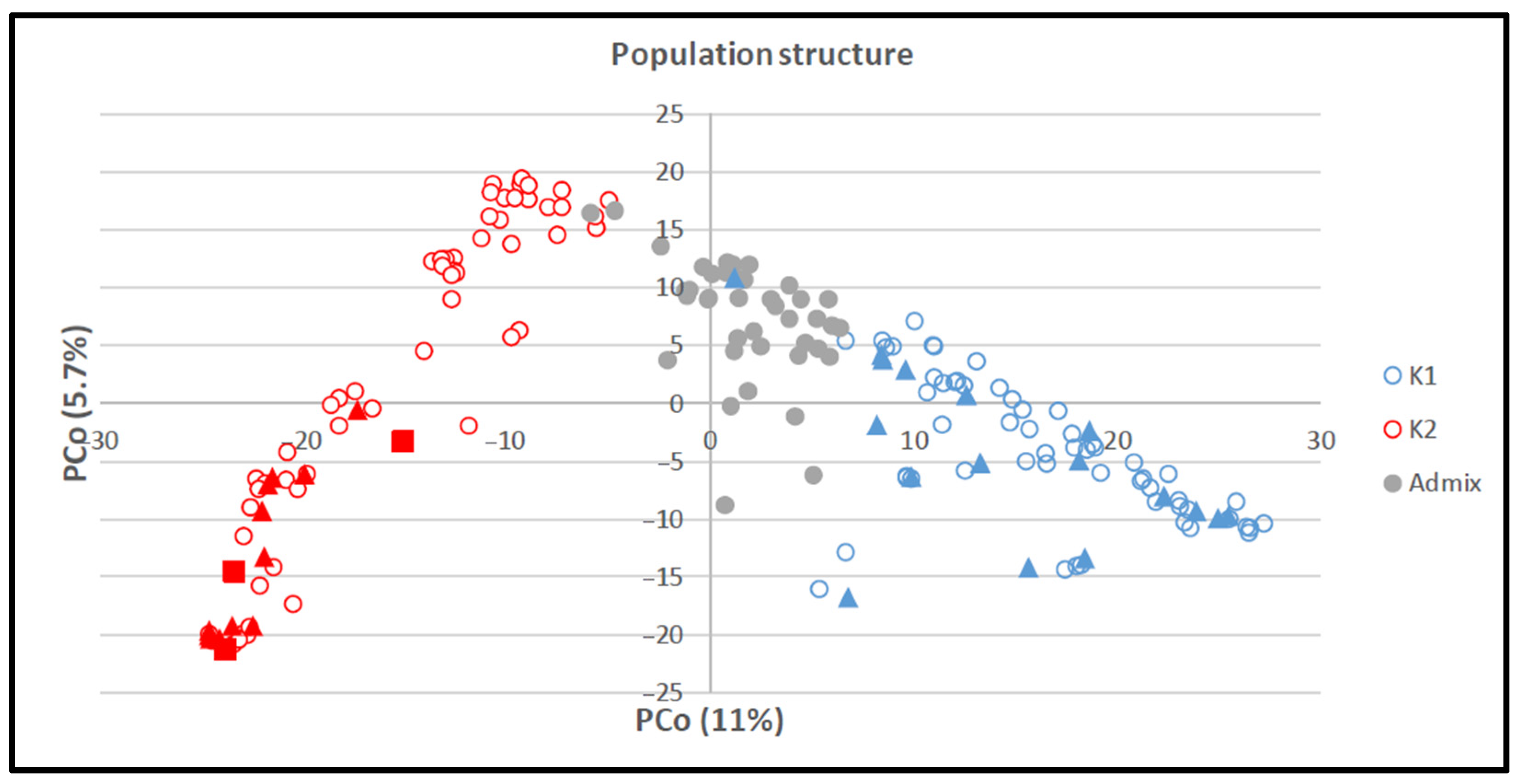
3.2.1. Population structure
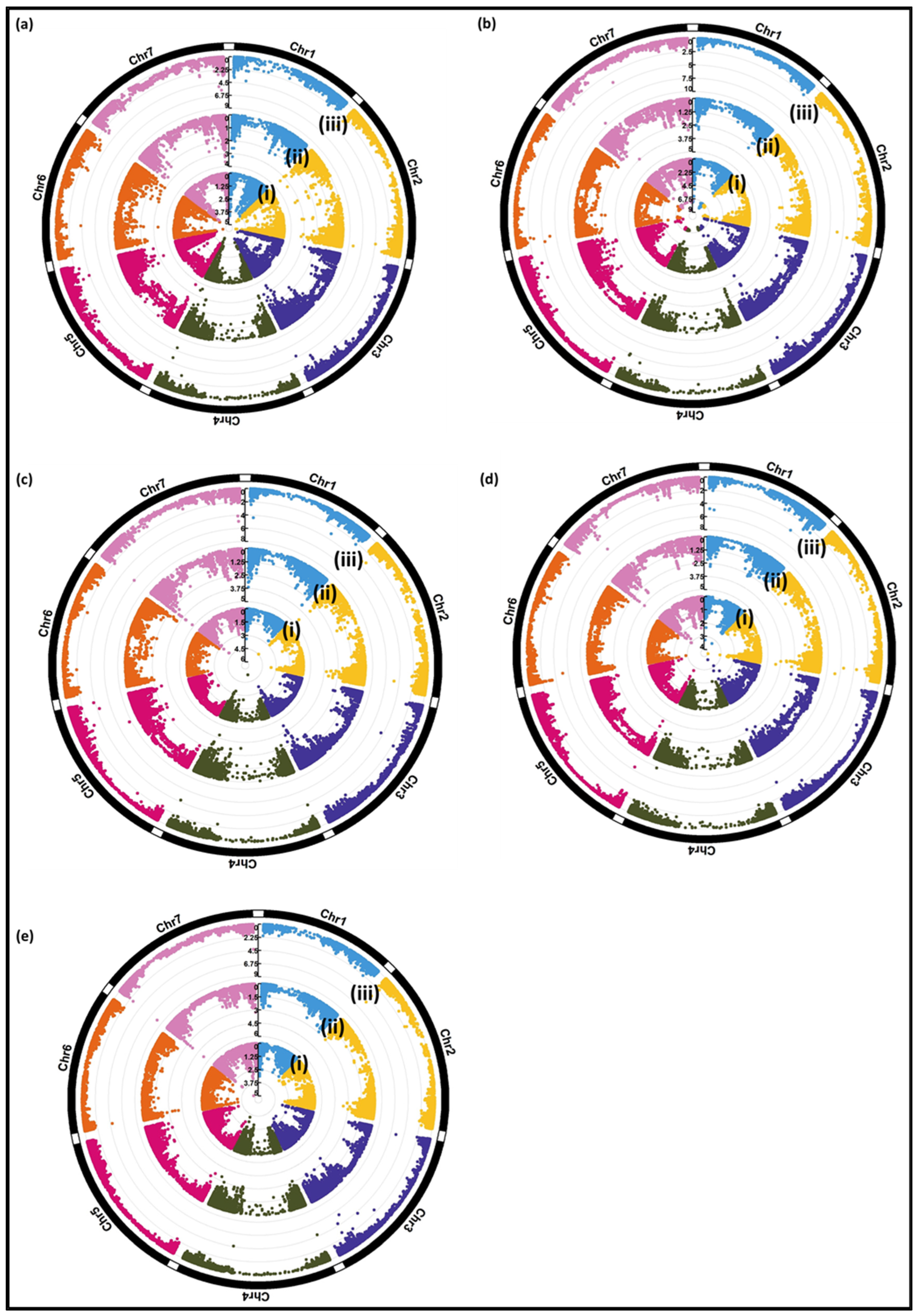
3.2.2. Genome-Wide Association Study (GWAS)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mayer, K.F.; Waugh, R.; Langridge, P.; Close, T.J.; Wise, R.P.; Graner, A.; Matsumoto, T.; Sato, K.; Schulman, A.; Muehlbauer, G.J.; et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–716. [Google Scholar] [PubMed]
- Habekuß, A.; Riedel, C.; Schliephake, E.; Ordon, F. Reeding for resistance to insect-transmitted viruses in barley-an emerging challenge due to global warming. J. Kult. 2009, 6, 53–61. [Google Scholar]
- Nygren, J.; Shad, N.; Kvarnheden, A.; Westerbergh, A. Variation in Susceptibility to Wheat dwarf virus among Wild and Domesticated Wheat. PLoS ONE 2015, 10, e0121580. [Google Scholar] [CrossRef]
- Cejnar, P.; Ohnoutková, L.; Ripl, J.; Vlčko, T.; Kundu, J.K. Two mutations in the truncated Rep gene RBR domain delayed the Wheat dwarf virus infection in transgenic barley plants. J. Integr. Agric. 2018, 17, 2492–2500. [Google Scholar] [CrossRef]
- Vacke, J. Host plants range and symptoms of wheat dwarf virus. Ěvěd Ust. Rostl Výroby Praha-Ruzyn 1972, 17, 151–162. [Google Scholar]
- Vacke, J.; Cibulka, R. Reactions of registered winter barley varieties to wheat dwarf virus infection. Czech J. Genet. Plant Breed.-UZPI 2001, 37, 50–52. [Google Scholar]
- Vacke, J. Wheat dwarf virus. Biol. Plant. 1961, 3, 228–233. [Google Scholar] [CrossRef]
- Koklu, G.; Ramsell, J.N.; Kvarnheden, A. The complete genome sequence for a Turkish isolate of Wheat dwarf virus (WDV) from barley confirms the presence of two distinct WDV strains. Virus Genes 2007, 34, 359–366. [Google Scholar] [CrossRef]
- Kanzi, A.M.; San, J.E.; Chimukangara, B.; Wilkinson, E.; Fish, M.; Ramsuran, V.; De Oliveira, T. Next Generation Sequencing and Bioinformatics Analysis of Family Genetic Inheritance. Front. Genet. 2020, 11, 544162. [Google Scholar] [CrossRef]
- Wang, S.C.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS ONE 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.Y.; Janss, L.L.; Andersen, J.R.; Orabi, J.; Jensen, J.D.; Jahoor, A.; Jensen, J. Genomic prediction and GWAS of yield, quality and disease-related traits in spring barley and winter wheat. Sci. Rep. 2020, 10, 3747. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, S.; Chao, S.; Vaish, S.S.; Singh, S.P.; Rehman, S.; Vishwakarma, S.R.; Verma, R.P.S. Genome wide association studies (GWAS) of spot blotch resistance at the seedling and the adult plant stages in a collection of spring barley. Mol. Breed. 2018, 38, 62. [Google Scholar] [CrossRef]
- Novakazi, F.; Afanasenko, O.; Lashina, N.; Platz, G.J.; Snowdon, R.; Loskutov, I.; Ordon, F. Genome-wide association studies in a barley (Hordeum vulgare) diversity set reveal a limited number of loci for resistance to spot blotch (Bipolaris sorokiniana). Plant Breed. 2020, 139, 521–535. [Google Scholar] [CrossRef]
- Wehner, G.; Kopahnke, D.; Richter, K.; Kecke, S.; Schikora, A.; Ordon, F. Priming is a suitable strategy to enhance resistance towards leaf rust in barley. Phytobiomes J. 2019, 3, 46–51. [Google Scholar] [CrossRef]
- Jabbari, M.; Fakheri, B.A.; Aghnoum, R.; Mahdi Nezhad, N.; Ataei, R. GWAS analysis in spring barley (Hordeum vulgare L.) for morphological traits exposed to drought. PLoS ONE 2018, 13, e0204952. [Google Scholar] [CrossRef]
- Rode, J.; Ahlemeyer, J.; Friedt, W.; Ordon, F. Identification of marker-trait associations in the German winter barley breeding gene pool (Hordeum vulgare L.). Mol. Breed. 2012, 30, 831–843. [Google Scholar] [CrossRef]
- Pfrieme, A.K.; Ruckwied, B.; Habekuß, A.; Will, T.; Stahl, A.; Pillen, K.; Ordon, F. Identification and validation of Quantitative Trait Loci for Wheat dwarf virus resistance in wheat (Triticum spp.). Front. Plant Sci. 2022, 13, 828639. [Google Scholar] [CrossRef]
- de Ronde, D.; Butterbach, P.; Kormelink, R. Dominant resistance against plant viruses. Front. Plant Sci. 2014, 5, 307. [Google Scholar] [CrossRef] [PubMed]
- Paudel, D.B.; Sanfacon, H. Exploring the Diversity of Mechanisms Associated With Plant Tolerance to Virus Infection. Front. Plant Sci. 2018, 9, 1575. [Google Scholar] [CrossRef]
- Schubert, J.; Habekuß, A.; Wu, B.; Thieme, T.; Wang, X. Analysis of complete genomes of isolates of the Wheat dwarf virus from new geographical locations and descriptions of their defective forms. Virus Genes 2014, 48, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W.; Waugh, R. Development and Evaluation of a Barley 50k iSelect SNP Array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar]
- Clark, M.F.; Adams, A.N. Characteristics of microplate method of enzyme-linked immunosorbent assay for detection of plant viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef]
- Doyle, J.F.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Mascher, M. Pseudomolecules and Annotation of the Second Version of the Reference Genome Sequence Assembly of Barley cv. Morex [Morex V2]; IPK Gatersleben: Seeland OT Gatersleben, Germany, 2019. [Google Scholar]
- Browning, B.L.; Browning, S.R. A Unified Approach to Genotype Imputation and Haplotype-Phase Inference for Large Data Sets of Trios and Unrelated Individuals. Am. J. Hum. Genet. 2009, 84, 210–223. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef]
- Reif, J.C.; Melchinger, A.E.; Frisch, M. Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Sci. 2005, 45, 1–7. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software: Dissimilarity Analysis and Representation for Windows. 2006. Available online: http://darwin.cirad.fr (accessed on 26 April 2019).
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. 2021. Available online: http://www.R-project.org/ (accessed on 15 December 2021).
- Shin, J.H.; Blay, S.; McNeney, B.; Graham, J. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 2006, 16, 1–9. [Google Scholar] [CrossRef]
- Warnes, G.; Gorjanc, G.; Leisch, F.; Man, M. Genetics: Population Genetics. R Package Version 1.3.8.1. 2013. Available online: http://CRAN.R-project.org/package=genetics (accessed on 1 March 2021).
- Voss-Fels, K.; Frisch, M.; Qian, L.; Kontowski, S.; Friedt, W.; Gottwald, S.; Snowdon, R.J. Subgenomic Diversity Patterns Caused by Directional Selection in Bread Wheat Gene Pools. Plant Genome 2015, 8, plantgenome2015.03.0013. [Google Scholar] [CrossRef]
- Sannemann, W.; Huang, B.E.; Mathew, B.; Léon, J. Multi-parent advanced generation inter-cross in barley: High-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 2015, 35, 86. [Google Scholar] [CrossRef]
- Lehnert, H.; Serfling, A.; Friedt, W.; Ordon, F. Genome-Wide Association Studies Reveal Genomic Regions Associated With the Response of Wheat (Triticum aestivum L.) to Mycorrhizae Under Drought Stress Conditions. Front. Plant Sci. 2018, 9, 1728. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Buerstmayr, H. Two major quantitative trait loci control wheat dwarf virus resistance in four related winter wheat populations. Theor. Appl. Genet. 2023, 136, 103. [Google Scholar] [CrossRef]
- Rousseau, E.; Bonneault, M.; Fabre, F.; Moury, B.; Mailleret, L.; Grognard, F. Virus epidemics, plant-controlled population bottlenecks and the durability of plant resistance. Philos. Trans. R. Soc. B-Biol. Sci. 2019, 374, 20180263. [Google Scholar] [CrossRef]
- Kaler, A.S.; Gillman, J.D.; Beissinger, T.; Purcell, L.C. Comparing different statistical models and multiple testing corrections for association mapping in soybean and maize. Front. Plant Sci. 2020, 10, 1794. [Google Scholar] [CrossRef]
- He, T.; Angessa, T.T.; Li, C. Pleiotropy structures plant height and seed weight scaling in barley despite long history of domestication and breeding selection. Plant Phenomics 2020, 5, 0015. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.N.; Venkatachalam, S.R. Inhibition of dihydrofolate reductase and cell growth activity by the phenanthroindolizidine alkaloids pergularinine and tylophorinidine: The in vitro cytotoxicity of these plant alkaloids and their potential as antimicrobial and anticancer agents. Toxicol. Vitr. 2000, 14, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Gorelova, V.; De Lepeleire, J.; Van Daele, J.; Pluim, D.; Meï, C.; Cuypers, A.; Leroux, O.; Rébeillé, F.; Schellens, J.H.; Blancquaert, D.; et al. Dihydrofolate Reductase/Thymidylate Synthase Fine-Tunes the Folate Status and Controls Redox Homeostasis in Plants. Plant Cell 2017, 29, 2831–2853. [Google Scholar] [CrossRef] [PubMed]
- Maule, A.J.; Caranta, C.; Boulton, M.I. Sources of natural resistance to plant viruses: Status and prospects. Mol. Plant Pathol. 2007, 8, 223–231. [Google Scholar] [CrossRef]
- Kristiansen, K.N.; Rohde, W. Structure of the Hordeum vulgare gene encoding dihydroflavonol-4-reductase and molecular analysis of ant 18 mutants blocked in flavonoid synthesis. Mol. Gen. Genet. 1991, 230, 49–59. [Google Scholar] [CrossRef]
- Badshah, S.L.; Faisal, S.; Muhammad, A.; Poulson, B.G.; Emwas, A.H.; Jaremko, M. Antiviral activities of flavonoids. Biomed. Pharmacother. 2021, 140, 111596. [Google Scholar] [CrossRef]
- Masoud, S.A.; Johnson, L.B.; White, F.F.; Reeck, G.R. Expression of a Cysteine Proteinase-Inhibitor (Oryzacystatin-I) in Transgenic Tobacco Plants. Plant Mol. Biol. 1993, 21, 655–663. [Google Scholar] [CrossRef]
- Gutierrez-Campos, R.; Torres-Acosta, J.A.; Saucedo-Arias, L.J.; Gomez-Lim, M.A. The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 1999, 17, 1223–1226. [Google Scholar] [CrossRef]
- Bouchard, E.; Michaud, D.; Cloutier, C. Molecular interactions between an insect predator and its herbivore prey on transgenic potato expressing a cysteine proteinase inhibitor from rice. Mol. Ecol. 2003, 12, 2429–2437. [Google Scholar] [CrossRef]
- Carrillo, L.; Martinez, M.; Alvarez-Alfageme, F.; Castanera, P.; Smagghe, G.; Diaz, I.; Ortego, F. A barley cysteine-proteinase inhibitor reduces the performance of two aphid species in artificial diets and transgenic Arabidopsis plants. Transgenic Res. 2011, 20, 305–319. [Google Scholar] [CrossRef]
- Soleimani, B.; Lehnert, H.; Trebing, S.; Habekuß, A.; Ordon, F.; Stahl, A.; Will, T. Dataset: Identification of markers associated with Wheat dwarf virus (WDV) tolerance/resistance in barley (Hordeum vulgare ssp. vulgare) using Genome wide association studies. OpenAgrar Repos. 2023. [Google Scholar] [CrossRef]

| Trait | Abbreviation | Method of Measurement | Unit |
|---|---|---|---|
| Relative virus titer | ELISA-60 | Extinction value of a double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) | |
| Total grain weight | ToGW | Weight all harvested seeds per plant | g |
| Plant height | HEI | Measure plant length from basis to top of the head | cm |
| Number of ears per plant | NEP | Count number of ears after harvesting | |
| Thousand grain weight | TGW | Weigh 1000 grains after threshing | g |
| Trait | Treatment a | N b | Mean c | Minimum d | Maximum d | Sd e | CV f |
|---|---|---|---|---|---|---|---|
| Relative virus titer | I-variant | 1866 | 0.3 | −0.03 | 1.84 | 0.6 | 179.7 |
| Total grain weight | I-variant | 209 | 11.3 | 0 | 287.4 | 31.7 | 280.7 |
| C-variant | 229 | 156.3 | 2.2 | 781.7 | 102.6 | 65.7 | |
| Plant height | I-variant | 520 | 62.9 | 1 | 159 | 40.4 | 64.2 |
| C-variant | 1163 | 117.5 | 49 | 224 | 20.3 | 17.2 | |
| Number of ears per plant | I-variant | 524 | 6.9 | 0 | 52 | 8.2 | 118.3 |
| C-variant | 1160 | 18.4 | 1 | 117 | 12.1 | 65.8 | |
| Thousand grain weight | I-variant | 208 | 25.5 | 5 | 48.3 | 8.7 | 34.1 |
| C-variant | 229 | 43.5 | 25.37 | 64.8 | 7.8 | 17.9 |
| Trait | Effect | Degrees of Freedom | F-Value | Pr > F |
|---|---|---|---|---|
| Relative virus titer | Genotype (G) | 190 | 2.26 | <0.0001 |
| Total grain weight | Genotype (G) | 190 | 1.05 | 0.49 |
| Treatment (T) | 1 | 364.08 | <0.0001 | |
| G × T | 184 | 0.84 | 0.74 | |
| Plant height | Genotype (G) | 190 | 5.43 | <0.0001 |
| Treatment (T) | 1 | 3651.82 | <0.0001 | |
| G × T | 186 | 9.26 | <0.0001 | |
| Number of ears per plant | Genotype (G) | 190 | 1.04 | 0.50 |
| Treatment (T) | 1 | 443.86 | <0.0001 | |
| G × T | 186 | 1.83 | <0.0001 | |
| Thousand grain weight | Genotype (G) | 190 | 3.05 | <0.0001 |
| Treatment (T) | 1 | 1233.26 | <0.0001 | |
| G × T | 184 | 2.19 | <0.0001 |
| Trait | Marker Name | Chr a | Pos b | p Value | Identified Genes in QTL Region | ||
|---|---|---|---|---|---|---|---|
| Gapit c | Tassel c | FarmCPU | |||||
| Relative virus titer | JHI−Hv50k−2016−202912 | 3H | 562,758,917 | 3.3 × 10−4 | 2.6 × 10−4 | 2.2 × 10−4 | |
| Relative total grain weight | JHI−Hv50k−2016−196649 | 3H | 534,052,013 | 6.8 × 10−5 | 7.7 × 10−4 | 2.8× 10−6 | |
| Relative total grain weight | BOPA1_2955−452 | 4H | 552,300,974 | 9.5 × 10−4 | 2.4 × 10−5 | 9.4 × 10−5 | Cysteine proteinase inhibitor |
| Relative total grain weight | BOPA2_12_10333 | 5H | 554,416,618 | 3.4 × 10−4 | 1.1 × 10−4 | 1.7 × 10−4 | |
| Relative plant height | BOPA2_12_21049 | 2H | 31,329,721 | 3.3 × 10−5 | 3.5 × 10−5 | 6.6 × 10−4 | Dihydrofolate reductase |
| Relative plant height | JHI−Hv50k−2016−435708 | 7H | 1,402,273 | 1.5 × 10−5 | 7.4 × 10−5 | 1.1 × 10−6 | |
| Relative number of ears per plant | JHI−Hv50k−2016−123144 | 2H | 631,278,948 | 1.8 × 10−4 | 8 × 10−4 | 2.7 × 10−6 | NBS−LRR disease resistance protein |
| Relative number of ears per plant | JHI−Hv50k−2016−142550 | 2H | 666,139,797 | 6.4 × 10−5 | 1.0 × 10−4 | 1.7 × 10−7 | Dihydroflavonol 4−reductase |
| Relative thousand grain weight | JHI−Hv50k−2016−435708 | 7H | 1402273 | 4.8 × 10−6 | 1.7 × 10−5 | 8.7 × 10−9 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soleimani, B.; Lehnert, H.; Trebing, S.; Habekuß, A.; Ordon, F.; Stahl, A.; Will, T. Identification of Markers Associated with Wheat Dwarf Virus (WDV) Tolerance/Resistance in Barley (Hordeum vulgare ssp. vulgare) Using Genome-Wide Association Studies. Viruses 2023, 15, 1568. https://doi.org/10.3390/v15071568
Soleimani B, Lehnert H, Trebing S, Habekuß A, Ordon F, Stahl A, Will T. Identification of Markers Associated with Wheat Dwarf Virus (WDV) Tolerance/Resistance in Barley (Hordeum vulgare ssp. vulgare) Using Genome-Wide Association Studies. Viruses. 2023; 15(7):1568. https://doi.org/10.3390/v15071568
Chicago/Turabian StyleSoleimani, Behnaz, Heike Lehnert, Sarah Trebing, Antje Habekuß, Frank Ordon, Andreas Stahl, and Torsten Will. 2023. "Identification of Markers Associated with Wheat Dwarf Virus (WDV) Tolerance/Resistance in Barley (Hordeum vulgare ssp. vulgare) Using Genome-Wide Association Studies" Viruses 15, no. 7: 1568. https://doi.org/10.3390/v15071568
APA StyleSoleimani, B., Lehnert, H., Trebing, S., Habekuß, A., Ordon, F., Stahl, A., & Will, T. (2023). Identification of Markers Associated with Wheat Dwarf Virus (WDV) Tolerance/Resistance in Barley (Hordeum vulgare ssp. vulgare) Using Genome-Wide Association Studies. Viruses, 15(7), 1568. https://doi.org/10.3390/v15071568







