Unbiased Virus Detection in a Danish Zoo Using a Portable Metagenomic Sequencing System
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Processing of the Samples
2.2. Microarray for the Validation of mNGS Findings
2.3. Sample Pretreatment and Nucleic Acid Extraction
2.4. Whole-Transcriptome and Whole-Genome Amplification for RNA and DNA Viruses
2.5. Metagenomic Sequencing
2.6. Data Analysis
3. Results and Discussion
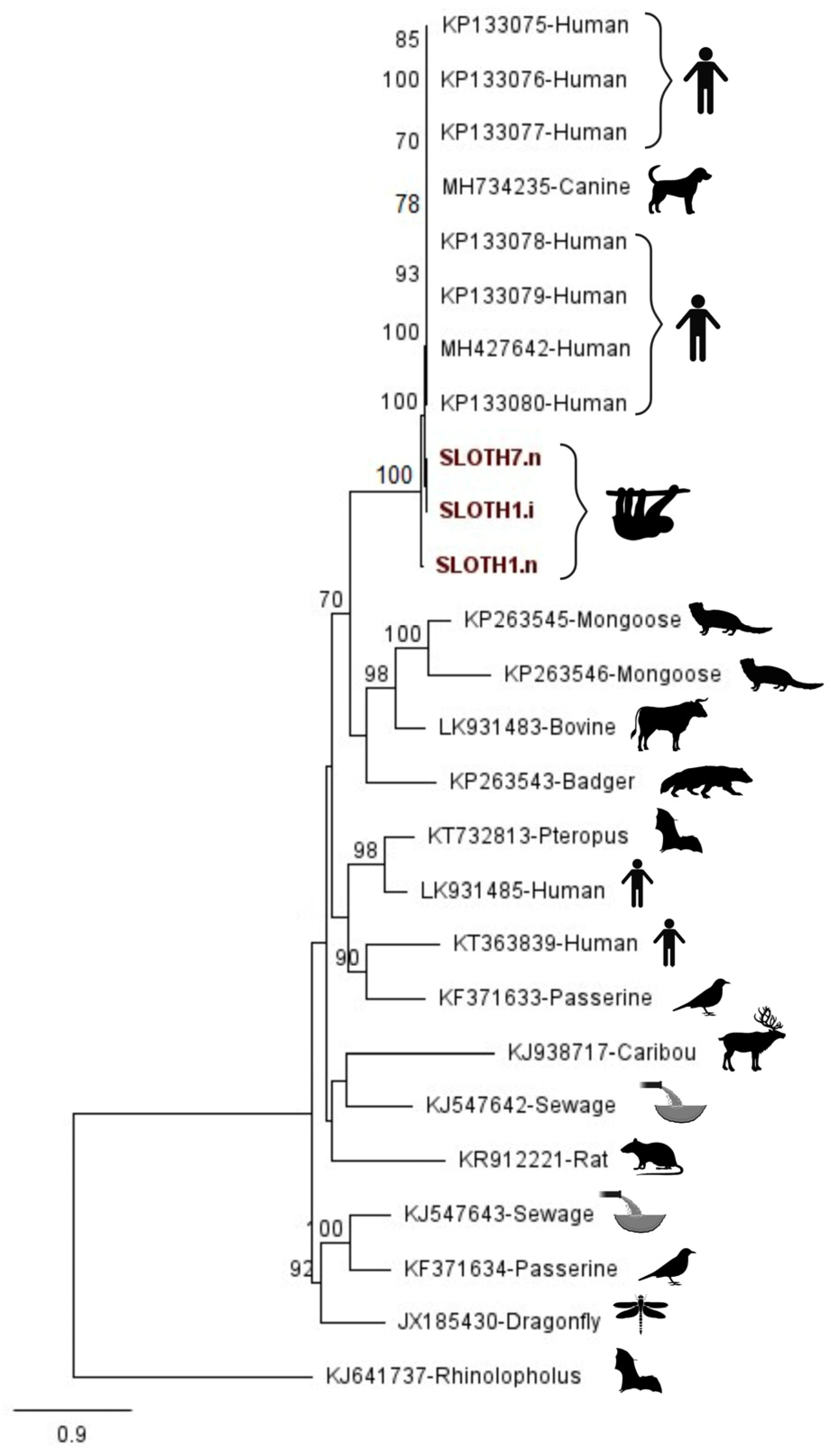
3.1. Case: Human-Associated Gemykibivirus 2 Found in Sloth
3.2. Diverse Viruses Identified from Animals at the Copenhagen Zoo
3.2.1. RNA Viruses
3.2.2. Retroviruses
3.2.3. DNA Viruses
3.2.4. CRESS DNA Viruses
| Virus Detected in This Study | Family | Baltimore Group | Genome Structure | First Reported Host Species | Reference |
|---|---|---|---|---|---|
| Avian leukosis virus | Retroviridae | ssRNA-RT | Non-segmented | Gallus gallus | [52] |
| Elephant endotheliotropic herpesvirus 1 | Herpesviridae | dsDNA | Non-segmented | Elephantidae | [53] |
| Elephant endotheliotropic herpesvirus 4 | Herpesviridae | dsDNA | Non-segmented | Elephantidae | [53] |
| Enzootic nasal tumour virus of goats | Retroviridae | ssRNA-RT | Non-segmented | Capra hircus | [54,55,56] |
| Faeces-associated gemycircularvirus 17 | Genomoviridae | ssDNA | Circular, Non-segmented | Gallus gallus | [57] |
| Gorilla-associated porprismacovirus 1 | Smacoviridae | ssDNA | Circular, Non-segmented | Gorilla gorilla | [49] |
| Horse-associated cyclovirus 1 | Circoviridae | ssDNA | Circular, Non-segmented | Equus caballus | [46] |
| Human-associated gemykibivirus 2 | Genomoviridae | ssDNA | Circular, Non-segmented | Homo sapiens | [28] |
| Jaagsiekte sheep retrovirus | Retroviridae | ssRNA-RT | Non-segmented | Ovis aries | [58] |
| Picornavirus 4 | Picornaviridae | ssRNA(+) | Non-segmented | Gallus gallus | [34,59] |
| Picornavirus 5/Megrivirus | Picornaviridae | ssRNA(+) | Non-segmented | Gallus gallus | [32] |
| MAG: Genomoviridae sp. isolate ctba76 | Genomoviridae | ssDNA | Circular, Non-segmented | Wild mouse | [Accession No. MK032755] |
| Sewage-derived gemycircularvirus 4 | Genomoviridae | ssDNA | Circular, Non-segmented | Acquired from environmental sample(s) | [47] |
| Year of Death | Sample Material | EEHV-1 | EEHV-4 | EEHV-5 | |
|---|---|---|---|---|---|
| Elephant 1 | 2003 | Serum | 30.5 | 28.8 | 33.3 |
| Pericardial fluid | 30.7 | 28.8 | 33.3 | ||
| Elephant 2 | 2014 | Serum | 21.2 | No CT | 32.8 |
| Elephant 3 | 2022 (August) | Heart | 19.2 | No CT | No CT |
3.3. Considerations and Future Perspectives
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pekar, J.E.; Magee, A.; Parker, E.; Moshiri, N.; Izhikevich, K.; Havens, J.L.; Gangavarapu, K.; Malpica Serrano, L.M.; Crits-Christoph, A.; Matteson, N.L. The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2. Science 2022, 377, 960–966. [Google Scholar] [PubMed]
- Vora, N.M.; Hannah, L.; Walzer, C.; Vale, M.M.; Lieberman, S.; Emerson, A.; Jennings, J.; Alders, R.; Bonds, M.H.; Evans, J. Interventions to Reduce Risk for Pathogen Spillover and Early Disease Spread to Prevent Outbreaks, Epidemics, and Pandemics. Emerg. Infect. Dis. 2023, 29, 1–9. [Google Scholar] [PubMed]
- Schrenzel, M.D.; Tucker, T.A.; Donovan, T.A.; Busch, M.D.; Wise, A.G.; Maes, R.K.; Kiupel, M. New hosts for equine herpesvirus 9. Emerg. Infect. Dis. 2008, 14, 1616. [Google Scholar] [PubMed]
- Greenwood, A.D.; Tsangaras, K.; Ho, S.Y.; Szentiks, C.A.; Nikolin, V.M.; Ma, G.; Damiani, A.; East, M.L.; Lawrenz, A.; Hofer, H. A potentially fatal mix of herpes in zoos. Curr. Biol. 2012, 22, 1727–1731. [Google Scholar] [CrossRef]
- Amman, B.R.; Pavlin, B.I.; Albariño, C.G.; Comer, J.A.; Erickson, B.R.; Oliver, J.B.; Sealy, T.K.; Vincent, M.J.; Nichol, S.T.; Paddock, C.D. Pet rodents and fatal lymphocytic choriomeningitis in transplant patients. Emerg. Infect. Dis. 2007, 13, 719. [Google Scholar]
- Schlottau, K.; Jenckel, M.; van den Brand, J.; Fast, C.; Herden, C.; Höper, D.; Homeier-Bachmann, T.; Thielebein, J.; Mensing, N.; Diender, B. Variegated squirrel bornavirus 1 in squirrels, Germany and the Netherlands. Emerg. Infect. Dis. 2017, 23, 477. [Google Scholar]
- Palacios, G.; Lowenstine, L.J.; Cranfield, M.R.; Gilardi, K.V.; Spelman, L.; Lukasik-Braum, M.; Kinani, J.-F.; Mudakikwa, A.; Nyirakaragire, E.; Bussetti, A.V. Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerg. Infect. Dis. 2011, 17, 711. [Google Scholar]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016, 17, 239. [Google Scholar]
- Hoenen, T.; Groseth, A.; Rosenke, K.; Fischer, R.J.; Hoenen, A.; Judson, S.D.; Martellaro, C.; Falzarano, D.; Marzi, A.; Squires, R.B. Nanopore sequencing as a rapidly deployable Ebola outbreak tool. Emerg. Infect. Dis. 2016, 22, 331. [Google Scholar]
- Quick, J.; Loman, N.J.; Duraffour, S.; Simpson, J.T.; Severi, E.; Cowley, L.; Bore, J.A.; Koundouno, R.; Dudas, G.; Mikhail, A. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016, 530, 228–232. [Google Scholar] [PubMed]
- Cui, S.; Liu, Y.; Zhao, J.; Peng, X.; Lu, G.; Shi, W.; Pan, Y.; Zhang, D.; Yang, P.; Wang, Q. An updated review on SARS-CoV-2 infection in animals. Viruses 2022, 14, 1527. [Google Scholar] [CrossRef] [PubMed]
- Kurth, A.; Wibbelt, G.; Gerber, H.-P.; Petschaelis, A.; Pauli, G.; Nitsche, A. Rat-to-elephant-to-human transmission of cowpox virus. Emerg. Infect. Dis. 2008, 14, 670. [Google Scholar] [PubMed]
- Nagarajan, S.; Kumar, M.; Murugkar, H.V.; Tripathi, S.; Shukla, S.; Agarwal, S.; Dubey, G.; Nagi, R.S.; Singh, V.P.; Tosh, C. Novel reassortant highly pathogenic avian influenza (H5N8) virus in zoos, India. Emerg. Infect. Dis. 2017, 23, 717. [Google Scholar] [CrossRef] [PubMed]
- Nolting, J.M.; Dennis, P.; Long, L.; Holtvoigt, L.; Brown, D.; King, M.J.; Shellbarger, W.; Hanley, C.; Killian, M.L.; Slemons, R.D. Low pathogenic influenza A virus activity at avian interfaces in Ohio zoos, 2006–2009. Avian Dis. 2013, 57, 657–662. [Google Scholar] [CrossRef]
- Shriner, S.A.; Root, J.J. A review of avian influenza A virus associations in synanthropic birds. Viruses 2020, 12, 1209. [Google Scholar]
- Delahay, R.J.; de la Fuente, J.; Smith, G.C.; Sharun, K.; Snary, E.L.; Flores Giron, L.; Nziza, J.; Fooks, A.R.; Brookes, S.M.; Lean, F.Z. Assessing the risks of SARS-CoV-2 in wildlife. One Health Outlook 2021, 3, 7. [Google Scholar]
- Baxby, D.; Ashton, D.; Jones, D.; Thomsett, L. An outbreak of cowpox in captive cheetahs: Virological and epidemiological studies. Epidemiol. Infect. 1982, 89, 365–372. [Google Scholar]
- Rosenstierne, M.W.; McLoughlin, K.S.; Olesen, M.L.; Papa, A.; Gardner, S.N.; Engler, O.; Plumet, S.; Mirazimi, A.; Weidmann, M.; Niedrig, M. The microbial detection Array for detection of emerging viruses in clinical samples-a useful Panmicrobial diagnostic tool. PLoS ONE 2014, 9, e100813. [Google Scholar]
- Fomsgaard, A.S.; Rasmussen, M.; Spiess, K.; Fomsgaard, A.; Belsham, G.J.; Fonager, J. Improvements in metagenomic virus detection by simple pretreatment methods. J. Clin. Virol. Plus 2022, 2, 100120. [Google Scholar]
- Rosenstierne, M.W.; Karlberg, H.; Bragstad, K.; Lindegren, G.; Stoltz, M.L.; Salata, C.; Kran, A.-M.B.; Dudman, S.G.; Mirazimi, A.; Fomsgaard, A. Rapid bedside inactivation of Ebola virus for safe nucleic acid tests. J. Clin. Microbiol. 2016, 54, 2521–2529. [Google Scholar] [CrossRef] [PubMed]
- Erlandsson, L.; Rosenstierne, M.W.; McLoughlin, K.; Jaing, C.; Fomsgaard, A. The microbial detection array combined with random Phi29-amplification used as a diagnostic tool for virus detection in clinical samples. PLoS ONE 2011, 6, e22631. [Google Scholar] [CrossRef] [PubMed]
- Rosenstierne, M.W.; Jensen, C.E.; Fomsgaard, A. Rapid, safe, and simple manual bedside nucleic acid extraction for the detection of virus in whole blood samples. JoVE J. Vis. Exp. 2018, 136, e58001. [Google Scholar]
- Gleizes, A.; Laubscher, F.; Guex, N.; Iseli, C.; Junier, T.; Cordey, S.; Fellay, J.; Xenarios, I.; Kaiser, L.; Mercier, P.L. Virosaurus a reference to explore and capture virus genetic diversity. Viruses 2020, 12, 1248. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudabadi, G.; Phillips, R. A comprehensive and quantitative exploration of thousands of viral genomes. eLife 2018, 7, e31955. [Google Scholar] [CrossRef]
- Bezerra, R.S.; Bitencourt, H.T.; Covas, D.T.; Kashima, S.; Slavov, S.N. Metagenomic identification of human Gemykibivirus-2 (HuGkV-2) in parenterally infected blood donors from the Brazilian Amazon. Int. J. Infect. Dis. 2020, 98, 249–251. [Google Scholar] [CrossRef]
- Phan, T.G.; Mori, D.; Deng, X.; Rajindrajith, S.; Ranawaka, U.; Ng, T.F.F.; Bucardo-Rivera, F.; Orlandi, P.; Ahmed, K.; Delwart, E. Small circular single stranded DNA viral genomes in unexplained cases of human encephalitis, diarrhea, and in untreated sewage. Virology 2015, 482, 98–104. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; He, X.; Ma, J.; Hong, W.; Hu, F.; Zhao, L.; Li, Q.; Zhang, J.; Zhang, C. Gemykibivirus genome in lower respiratory tract of elderly woman with unexplained acute respiratory distress syndrome. Clin. Infect. Dis. 2019, 69, 861–864. [Google Scholar] [CrossRef]
- Zucherato, V.S.; Giovanetti, M.; Costa, L.O.A.; Krause, L.M.F.; Alves, D.C.C.; Moreira, R.M.A.; Pimentel, B.M.S.; Haddad, R.; Bitencourt, H.T.; Ciccozzi, M. Molecular identification of the emerging Human Gemykibivirus-2 (HuGkV-2) among Brazilian blood donors. Transfus. Apher. Sci. 2022, 62, 103516. [Google Scholar] [CrossRef]
- Weber, M.; Cibulski, S.; Olegario, J.; Da Silva, M.; Puhl, D.; Mosena, A.; Alves, C.; Paim, W.; Baumbach, L.; Mayer, F. Characterization of dog serum virome from Northeastern Brazil. Virology 2018, 525, 192–199. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Yip, C.C.; Li, K.S.; Fan, R.Y.; Bai, R.; Huang, Y.; Chan, K.-H.; Yuen, K.-Y. Chickens host diverse picornaviruses originated from potential interspecies transmission with recombination. J. Gen. Virol. 2014, 95, 1929–1944. [Google Scholar] [CrossRef]
- Devaney, R.; Trudgett, J.; Trudgett, A.; Meharg, C.; Smyth, V. A metagenomic comparison of endemic viruses from broiler chickens with runting-stunting syndrome and from normal birds. Avian Pathol. 2016, 45, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Boros, Á.; Pankovics, P.; Adonyi, Á.; Fenyvesi, H.; Day, J.M.; Phan, T.G.; Delwart, E.; Reuter, G. A diarrheic chicken simultaneously co-infected with multiple picornaviruses: Complete genome analysis of avian picornaviruses representing up to six genera. Virology 2016, 489, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Fadly, A.M. Isolation and identification of avian leukosis viruses: A review. Avian Pathol. 2000, 29, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Ortín, A.; Cousens, C.; Minguijon, E.; Pascual, Z.; Villarreal, M.P.d.; Sharp, J.M.; Heras, M.D.l. Characterization of enzootic nasal tumour virus of goats: Complete sequence and tissue distribution. J. Gen. Virol. 2003, 84, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Leroux, C.; Girard, N.; Cottin, V.; Greenland, T.; Mornex, J.-F.; Archer, F. Jaagsiekte Sheep Retrovirus (JSRV): From virus to lung cancer in sheep. Vet. Res. 2007, 38, 211–228. [Google Scholar] [CrossRef]
- Rajya, B.; Singh, C. The pathology of pneumonia and associated respiratory disease of sheep and goats. I. Occurrence of jagziekte and maedi in sheep and goats in India. Am. J. Vet. Res. 1964, 25, 61–67. [Google Scholar]
- Griffiths, D.; Martineau, H.; Cousens, C. Pathology and pathogenesis of ovine pulmonary adenocarcinoma. J. Comp. Pathol. 2010, 142, 260–283. [Google Scholar] [CrossRef]
- Long, S.Y.; Latimer, E.M.; Hayward, G.S. Review of elephant endotheliotropic herpesviruses and acute hemorrhagic disease. ILAR J. 2016, 56, 283–296. [Google Scholar] [CrossRef]
- Perrin, K.L.; Nielsen, S.S.; Martinussen, T.; Bertelsen, M.F. Quantification and risk factor analysis of elephant endotheliotropic herpesvirus-haemorrhagic disease fatalities in Asian elephants (Elephas maximus) in Europe (1985–2017). J. Zoo Aquar. Res. 2021, 9, 8–13. [Google Scholar]
- Zhao, L.; Rosario, K.; Breitbart, M.; Duffy, S. Eukaryotic circular rep-encoding single-stranded DNA (CRESS DNA) viruses: Ubiquitous viruses with small genomes and a diverse host range. Adv. Virus Res. 2019, 103, 71–133. [Google Scholar] [PubMed]
- Firth, C.; Charleston, M.A.; Duffy, S.; Shapiro, B.; Holmes, E.C. Insights into the evolutionary history of an emerging livestock pathogen: Porcine circovirus 2. J. Virol. 2009, 83, 12813–12821. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Duffy, S.; Breitbart, M. A field guide to eukaryotic circular single-stranded DNA viruses: Insights gained from metagenomics. Arch. Virol. 2012, 157, 1851–1871. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Li, L.; Giannitti, F.; Low, J.; Keyes, C.; Ullmann, L.S.; Deng, X.; Aleman, M.; Pesavento, P.A.; Pusterla, N.; Delwart, E. Exploring the virome of diseased horses. J. Gen. Virol. 2015, 96 Pt 9, 2721. [Google Scholar] [CrossRef] [PubMed]
- Kraberger, S.; Argüello-Astorga, G.R.; Greenfield, L.G.; Galilee, C.; Law, D.; Martin, D.P.; Varsani, A. Characterisation of a diverse range of circular replication-associated protein encoding DNA viruses recovered from a sewage treatment oxidation pond. Infect. Genet. Evol. 2015, 31, 73–86. [Google Scholar] [CrossRef]
- Rosshart, S.P.; Vassallo, B.G.; Angeletti, D.; Hutchinson, D.S.; Morgan, A.P.; Takeda, K.; Hickman, H.D.; McCulloch, J.A.; Badger, J.H.; Ajami, N.J. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 2017, 171, 1015–1028.e13. [Google Scholar] [CrossRef]
- Ng, T.F.F.; Zhang, W.; Sachsenröder, J.; Kondov, N.O.; Da Costa, A.C.; Vega, E.; Holtz, L.R.; Wu, G.; Wang, D.; Stine, C.O. A diverse group of small circular ssDNA viral genomes in human and non-human primate stools. Virus Evol. 2015, 1, vev017. [Google Scholar] [CrossRef]
- Kim, H.-R.; Yoon, S.-J.; Lee, H.-S.; Kwon, Y.-K. Identification of a picornavirus from chickens with transmissible viral proventriculitis using metagenomic analysis. Arch. Virol. 2015, 160, 701–709. [Google Scholar] [CrossRef]
- Sachsenröder, J.; Twardziok, S.; Hammerl, J.A.; Janczyk, P.; Wrede, P.; Hertwig, S.; Johne, R. Simultaneous identification of DNA and RNA viruses present in pig faeces using process-controlled deep sequencing. PLoS ONE 2012, 7, e34631. [Google Scholar] [CrossRef]
- Eilerman, V.; Bang, O. Experimental leukemia in chickens. Zent. Bakteriol. Parasitenkd. Infekt. Hyg. 1908, 46, 595–609. [Google Scholar]
- McCully, R.; Basson, P.; Pienaar, J.; Erasmus, B.; Young, E. Herpes nodules in the lung of the African elephant, Loxodonta africana (Blumenbach, 1797). Onderstepoort J. Vet. Res. 1971, 38, 225–235. [Google Scholar] [PubMed]
- Camy, M. Papillome granuleux dês cavités nasales du mouton. Bull. L’acad. Vét. Fr. 1955, 108, 31–34. [Google Scholar] [CrossRef]
- Drieux, H.; Glaunes, J.; Courtehoux, P. Epithelioma des premieres voies respiratoires d’llure contagiose ou hereditaire chez le mouton. Acta Unio Int. Contrra Cancrum 1952, 8, 444–446. [Google Scholar]
- Lombard, C.; Cabanie, P.; Crespin, J. Adénopapillome de la muqueuse pituitaire chez la chèvre. Bull. L’acad. Vet. Fr. 1966, 39, 199–202. [Google Scholar] [CrossRef]
- Steel, O.; Kraberger, S.; Sikorski, A.; Young, L.M.; Catchpole, R.J.; Stevens, A.J.; Ladley, J.J.; Coray, D.S.; Stainton, D.; Dayaram, A. Circular replication-associated protein encoding DNA viruses identified in the faecal matter of various animals in New Zealand. Infect. Genet. Evol. 2016, 43, 151–164. [Google Scholar] [CrossRef] [PubMed]
- York, D.F.; Querat, G. A history of ovine pulmonary adenocarcinoma (jaagsiekte) and experiments leading to the deduction of the JSRV nucleotide sequence. Curr. Top. Microbiol. Immunol. 2003, 275, 1–23. [Google Scholar]
- Farkas, T.; Fey, B.; Hargitt, E.; Parcells, M.; Ladman, B.; Murgia, M.; Saif, Y. Molecular detection of novel picornaviruses in chickens and turkeys. Virus Genes 2012, 44, 262–272. [Google Scholar] [CrossRef]
- Hansen, S.; Faye, O.; Sanabani, S.S.; Faye, M.; Böhlken-Fascher, S.; Faye, O.; Sall, A.A.; Bekaert, M.; Weidmann, M.; Czerny, C.-P. Combination random isothermal amplification and nanopore sequencing for rapid identification of the causative agent of an outbreak. J. Clin. Virol. 2018, 106, 23–27. [Google Scholar] [CrossRef]
- Russell, J.A.; Campos, B.; Stone, J.; Blosser, E.M.; Burkett-Cadena, N.; Jacobs, J.L. Unbiased strain-typing of arbovirus directly from mosquitoes using nanopore sequencing: A field-forward biosurveillance protocol. Sci. Rep. 2018, 8, 5417. [Google Scholar] [CrossRef] [PubMed]
- ZOO. Copenhagen Zoo Animal Inventory. 2021. Available online: https://www.zoo.dk/s/dyr/forvaltning-af-dyrebestanden?language=da (accessed on 2 March 2023).
- Schottstedt, V.; Blümel, J.; Burger, R.; Drosten, C.; Gröner, A.; Gürtler, L.; Heiden, M.; Hildebrandt, M.; Jansen, B.; Montag-Lessing, T. Human cytomegalovirus (HCMV)–revised. Transfus. Med. Hemotherapy 2010, 37, 365. [Google Scholar]
- Sah, P.; Fitzpatrick, M.C.; Zimmer, C.F.; Abdollahi, E.; Juden-Kelly, L.; Moghadas, S.M.; Singer, B.H.; Galvani, A.P. Asymptomatic SARS-CoV-2 infection: A systematic review and meta-analysis. Proc. Natl. Acad. Sci. USA 2021, 118, e2109229118. [Google Scholar] [CrossRef] [PubMed]
| Sample Type | Material | Species | Samples | Sub-Pool | Population |
|---|---|---|---|---|---|
| Swabs | Enclosure | American flamingo (Phoenicopterus ruber) | 1 | 8 | 80 |
| Black-capped squirrel monkey (Saimiri boliviensis) | 1 | 4 | 12 | ||
| Blesbok (Damaliscus pygargus phillipsi) | 1 | 1 | 2 | ||
| Chicken (Gallus gallus domesticus) | 1 | 5 | 13 | ||
| Grant’s zebra (Equus quagga boehmi) | 1 | 2 | 3 | ||
| Humboldt penguin (Spheniscus humboldti) | 1 | 8 | 40 | ||
| Kea (Nestor notabilis) | 1 | 4 | 3 | ||
| Llama (Lama glama) | 1 | 2 | 2 | ||
| Ostrich (Struthio camelus camelus) | 2 | 1 | 2 | ||
| Reticulated giraffe (Giraffa camelopardalis reticulata) | 1 | 4 | 5 | ||
| Sable antilope (Hippotragus niger) | 1 | 3 | 5 | ||
| Linnaeus’s two-toed sloth (Choloepus didactylus) | 1 | 1 | 3 | ||
| Southern white rhinoceros (Ceratotherium simum simum) | 1 | 1 | 4 | ||
| Faecal | Amur Leopard (Panthera pardus orientalis) | 1 | 1 | 3 | |
| Amur tiger (Panthera tigris altaica) | 1 | 1 | 2 | ||
| Caracal (Caracal caracal) | 2 | 1 | 2 | ||
| Eurasian brown bear (Ursus arctos arctos) | 1 | 1 | 3 | ||
| Golden marmoset (Leontopithecus rosalia) | 1 | 1 | 2 | ||
| Linnaeus’s two-toed sloth (Choloepus didactylus) | 1 | 1 | 4 | ||
| Hamaydrian baboon (Papio hamadryas) | 1 | 3 | 17 | ||
| Lion (Panthera leo) | 1 | 1 | 4 | ||
| Muskox (Ovibos moschatus) | 1 | 1 | 3 | ||
| Polar bear (Ursus maritimus) | 1 | 1 | 4 | ||
| Rock wallaby (Petrogale xanthopus) | 1 | 1 | 4 | ||
| Giant panda (Ailuropoda melanoleuca) | 2 | 1 | 2 | ||
| Gibbon (Hylobates lar) | 1 | 1 | 5 | ||
| Hippopotamus (Hippopotamus amphibius) | 1 | 1 | 5 | ||
| Nasal | Linnaeus’s two-toed sloth (Choloepus didactylus) | 2 | 1 | 3 | |
| Hippopotamus (Hippopotamus amphibius) | 1 | 1 | 5 | ||
| Goat (Capra hircus) | 1 | 3 | 26 | ||
| Horse (Equus ferus caballus) | 4 | 1 | 5 | ||
| Pig (Sus scrofa domesticus) | 1 | 1 | 1 | ||
| Southern white rhinoceros (Ceratotherium simum simum) | 2 | 1 | 4 | ||
| Oral | Chimpanzee (Pan troglodytes) | 1 | 1 | 11 | |
| Giant panda (Ailuropoda melanoleuca) | 2 | 1 | 2 | ||
| Linnaeus’s two-toed sloth (Choloepus didactylus) | 2 | 1 | 3 | ||
| Hippopotamus (Hippopotamus amphibius) | 1 | 1 | 3 | ||
| Egyptian fruit bat (Rousettus aegyptiacus) | 2 | 4 | 150 | ||
| Nasal/oral | Linnaeus’s two-toed sloth (Choloepus didactylus) | 2 | 1 | 3 | |
| Gibbon (Hylobates lar) | 1 | 1 | 5 | ||
| Cloacal | Crested Pigeon (Ocyphaps lophotes) | 2 | 3 | 6 | |
| Galah parrot (Eolophus roseicapilla) | 1 | 2 | 2 | ||
| Tasmanian devil (Sarcophilus harrisii) | 1 | 1 | 5 | ||
| Rectal | Egyptian fruit bat (Rousettus aegyptiacus) | 2 | 4 | 150 | |
| Linnaeus’s two-toed sloth (Choloepus didactylus) | 1 | 1 | 3 | ||
| Vari (Varecia variegata) | 1 | 1 | 7 | ||
| Fluids | Urine | Egyptian fruit bat (Rousettus aegyptiacus) | 1 | 2 | 150 |
| Full blood * | Ball python (Python regius) | 2 | 1 | NA | |
| Serum | Egyptian fruit bat (Rousettus aegyptiacus) | 2 | 4 | 150 | |
| Serum * | Asian Elephant (Elephas maximus) | 2 | 1 | NA | |
| Pericardial fluid * | Asian Elephant (Elephas maximus) | 1 | 1 | NA | |
| Tissue | Heart * | Asian Elephant (Elephas maximus) | 1 | 1 | NA |
| Kidney | Linnaeus’s two-toed sloth (Choloepus didactylus) | 1 | 1 | NA | |
| Liver | Linnaeus’s two-toed sloth (Choloepus didactylus) | 1 | 1 | 3 |
| Sample | Sample Material | Virus Species Detected | Virus Accession Number | Reference Genome Size | Reference Genome Coverage (%) | Mean Depth | Viral Reads Mapped | Total Reads | Pairwise Identity% Range between Contigs and NCBI | Microarray Confirmed |
|---|---|---|---|---|---|---|---|---|---|---|
| Elephant 1 | Serum | Elephant endotheliotropic herpesvirus 4 | KT832477 | 205,896 bp | 8.9 | 0.4 | 557 | 87,808 | 86.7–100.0 | No |
| Pericardial fluid | 0.6 | 0.1 | 46 | 112,835 | 90.3–100.0 | No | ||||
| Elephant 2 | Serum | Elephant endotheliotropic herpesvirus 1 | KC462165 | 180,421 bp | 0.3 | 0.0 | 25 | 89,629 | 95.4–99.1 | Yes |
| Elephant 3 | Heart | 3.6 | 0.3 | 212 | 58,492 | 86.6–100.0 | Yes | |||
| Chicken | Enclosure swab | Avian leukosis virus | KU937324 | 7669 nt | 2.8 | 0.0 | 2 | 107,628 | 96.0–98.3 | Yes |
| Picornavirus 4 | KF979335 | 9564 nt | 26.9 | 0.9 | 38 | 107,666 | 89.6–91.7 | Yes | ||
| Picornavirus 5/megrivirus | MH806866 | 9567 nt | 10.5 | 0.5 | 25 | 107,656 | 90.5–94.4 | Yes | ||
| Gibbon | Faecal swab | Gorilla associated porprismacovirus 1 | KP233191 | 2532 bp | 53.4 | 2.6 | 26 | 61,534 | 90.3 | NA |
| Goat | Nasal swab | Enzootic nasal tumour virus of goats | MK164400 | 7279 nt | 27.7 | 4.2 | 92 | 95,465 | 86.3–98.1 | Yes |
| Jaagsiekte sheep like-retrovirus | DQ838494 | 7430 nt | 48.4 | 38.5 | 536 | 95,710 | 90.4 | Yes | ||
| Horse associated cyclovirus 1 | KR902499 | 1843 bp | 100 | 13.4 | 37 | 95,419 | 97.8 | No | ||
| Sewage derived gemycircularvirus 4 | KJ547634 | 2115 bp | 100 | 12.1 | 103 | 95,463 | 96.8 | NA | ||
| Horse 1 | Nasal swab | Faeces associated gemycircularvirus 17 | KT862242 | 2230 bp | 100 | 18.0 | 76 | 68,170 | 92.2 | NA |
| MAG: Genomoviridae sp. isolate ctba76 | MK032755 | 2176 bp | 100 | 27.7 | 133 | 68,170 | 92.1 | NA | ||
| Horse 2 | Nasal swab | 100 | 42.7 | 1959 | 92,294 | 91.5 | ||||
| Horse 3 | Nasal swab | 100 | 60.8 | 3047 | 112,632 | 92.3 | ||||
| Pig | Nasal swab | 100 | 71.9 | 539 | 142,068 | 97.2 | ||||
| Sloth 1 | Nasal swab | Human associated gemykibivirus 2 | MH734235 | 2210 bp | 96.6 | 11.3 | 88 | 23,782 | 91.3–96.0 | Yes |
| Sloth 7 | Nasal swab | 84.3 | 16.4 | 133 | 177,374 | 98.9 | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fomsgaard, A.S.; Tahas, S.A.; Spiess, K.; Polacek, C.; Fonager, J.; Belsham, G.J. Unbiased Virus Detection in a Danish Zoo Using a Portable Metagenomic Sequencing System. Viruses 2023, 15, 1399. https://doi.org/10.3390/v15061399
Fomsgaard AS, Tahas SA, Spiess K, Polacek C, Fonager J, Belsham GJ. Unbiased Virus Detection in a Danish Zoo Using a Portable Metagenomic Sequencing System. Viruses. 2023; 15(6):1399. https://doi.org/10.3390/v15061399
Chicago/Turabian StyleFomsgaard, Anna S., Stamatios A. Tahas, Katja Spiess, Charlotta Polacek, Jannik Fonager, and Graham J. Belsham. 2023. "Unbiased Virus Detection in a Danish Zoo Using a Portable Metagenomic Sequencing System" Viruses 15, no. 6: 1399. https://doi.org/10.3390/v15061399
APA StyleFomsgaard, A. S., Tahas, S. A., Spiess, K., Polacek, C., Fonager, J., & Belsham, G. J. (2023). Unbiased Virus Detection in a Danish Zoo Using a Portable Metagenomic Sequencing System. Viruses, 15(6), 1399. https://doi.org/10.3390/v15061399







