1. Introduction
Hantaviruses (Order
Bunyavirales, Family
Hantaviridae) comprise a group of negative-sense RNA viruses that are distributed worldwide. While rodents and eulipotyphlans (shrews and moles) have been advanced as the primary hosts of hantaviruses globally, new strains have recently been detected in chiropterans [
1], reptiles, and fish [
2]. As a result, four subfamilies are recognized within Hantaviridae: Mammantavirinae (
Loanvirus,
Mobatvirus,
Orthohantavirus, and
Thottimvirus), the bat-, mole-, shrew-, and rodent-borne viruses; Repantavirinae (
Reptillovirus), the reptile-borne viruses; Actantavirinae (
Actinovirus), the ray-finned fish-borne viruses; and Agantavirinae (
Agnathovirus), the jawless fish-borne viruses [
3]. In general, hantaviruses trend towards host specificity (one virus, one host [
4]); however, there have been instances of multiple strains detected in a single host or the same strain found in multiple different host species [
5,
6,
7,
8]. To date, only rodent-borne orthohantaviruses have been associated with human disease.
Zoonotic infection of humans by hantaviruses occurs through the inhalation of aerosols contaminated by the urine or feces of an infected host [
9]. Some New World hantaviruses cause hantavirus pulmonary syndrome (HPS) or hantavirus cardiopulmonary syndrome (HCPS), which can lead to fatal cardiac shock in humans [
10,
11], whereas Old World hantaviruses cause hemorrhagic fever with renal syndrome (HFRS). In the Americas, HCPS is characterized as a “flu-like illness” with gastrointestinal symptoms that can range from mild to severe. Orthohantavirus diseases have a mortality rate ranging from 12% (HFRS) to 40% (HCPS) [
12], although many milder cases may go unreported [
13]. In contrast, rodent hosts appear to be relatively unaffected [
14], making them both reservoirs and zoonotic vectors. Establishment of laboratory animal models for hantavirus is challenging [
15,
16] due to the risks to personnel, which necessitates expanded investigation of wild hosts and positions hantaviruses as a model for a One Health (human–animal–environment) approach towards emerging infectious diseases.
In the early 2000s, an acute outbreak of HCPS on the Azuero Peninsula in the province of Los Santos in central Panama led to the first documentation of hantavirus in the country [
17,
18]. The causal agent,
Orthohantavirus chocloense [
19], has since been isolated and sequenced from both humans and its primary wildlife reservoir,
Oligoryzomys fulvescens (=
costaricensis) [
20,
21], the Costa Rican pygmy rice rat [
22,
23]. Previous ecological studies of CHOV and
Oligoryzomys, along with human epidemiological surveillance, have characterized a “CHOV-endemic region” in central-western Panama [
18,
24,
25,
26,
27]. Yet until recent years, the eastern and northern regions of the country had been undersampled and understudied, so it was unclear whether CHOV was not present there or simply undetected. Understanding where the host and virus are distributed across the landscape is a necessary first step in the identification of risk areas for emergence that forms future public health guidance and future surveillance efforts of both wildlife and people.
Understanding how viral prevalence changes over time is critical and hinges on the availability of temporally deep wildlife archives. In the case of CHOV, the wild host is known to experience seasonal population cycles [
28] that may also affect the prevalence and distribution of the disease [
29]. In areas unaltered by agriculture, both the spatial distribution and abundance of
O. costaricensis vary seasonally and inter-annually, with abundance generally lowest in December during the transition from wet to dry season and populations reestablishing and then increasing in abundance during the dry season [
28]. Such demographic fluctuations are less prominent in agriculturally modified areas where host reproductive output can occur year-round unrestricted by the availability of food [
30] This pattern suggests that agricultural development may have driven CHOV emergence in Panama [
5,
22,
31]. Increased abundance of
O. costaricensis in response to excess agricultural food resources may increase host density, elevate pathogen prevalence, and ultimately increase risk of zoonotic spillover and human infection [
22,
26,
28,
32,
33]. Agricultural development also reduces biodiversity, which can unintentionally lead to higher prevalence of a disease in remaining host species [
34]. To this end, it is important to understand associations between host distribution and abundance, viral prevalence, and environments across both space and time.
We conducted holistic mammal surveys from 2000 to 2019. We obtained mitochondrial barcodes and seroprevalence data from the rodent hosts to document the spatial extent of
O. costaricensis and CHOV. We explored host distribution and geographic variation in Panama and assessed the viral presence across five microhabitat categories (cropland, pasture, secondary vegetation, shrub, and peridomestic [
26]) within five of the seven ecoregions in Panama to better understand CHOV prevalence across space and time.
2. Materials and Methods
2.1. Study Area and Small Mammal Surveys
A survey of non-volant small mammals in Panama was undertaken from February 2000 to December 2019. Panama supports diverse habitats with an elevational range from sea level to 3475 m, annual average precipitation varies from 1200 to 7000 mm, and temperatures span 7–27 °C. The total terrestrial area of the country is 74,177 km
2 and approximately 40% of that is agricultural or pastureland [
35,
36]. Generally, Panama is hot and humid along the coasts, while the interior experiences greater environmental variation dependent on elevation [
37].
Survey methods followed animal care and use procedures as outlined by the American Society of Mammalogists [
38,
39]. Specimens were holistically collected [
40], morphologically identified to species level, and collaboratively preserved at the Museum of Southwestern Biology at the University of New Mexico, the Vertebrate Museum of the University of Panama, or the Zoological Collection of the Gorgas Memorial Institute for Health Studies to maximize the utility of the collected samples to the extended biodiversity and public health communities. Standard measurements, sex, reproductive status, and GPS coordinates associated with the collection locality (WGS 1984) were recorded at the time of collection for all animals [
41]. These data are publicly available through the Arctos database (
https://arctosdb.org, accessed on 30 April 2023). To maximize consistency, one investigator recorded 80% of the measurements over the 19 years of sampling (MA). We emphasize the value of long-term collection efforts in enabling temporally deep and geographically broad public health perspectives on emerging zoonotic diseases [
42].
2.2. Sequencing and Serology
To verify morphological host species determinations and assess the potential geographic variation of
O. costaricensis within Panama, we sequenced part of the mitochondrial cytochrome b (
cytb, 1140 bp) gene as a molecular barcode for host identification. Representatives from populations spanning its Panamanian distribution were selected for molecular analysis. DNA was extracted from the spleen, liver, or kidney from 33 host specimens (
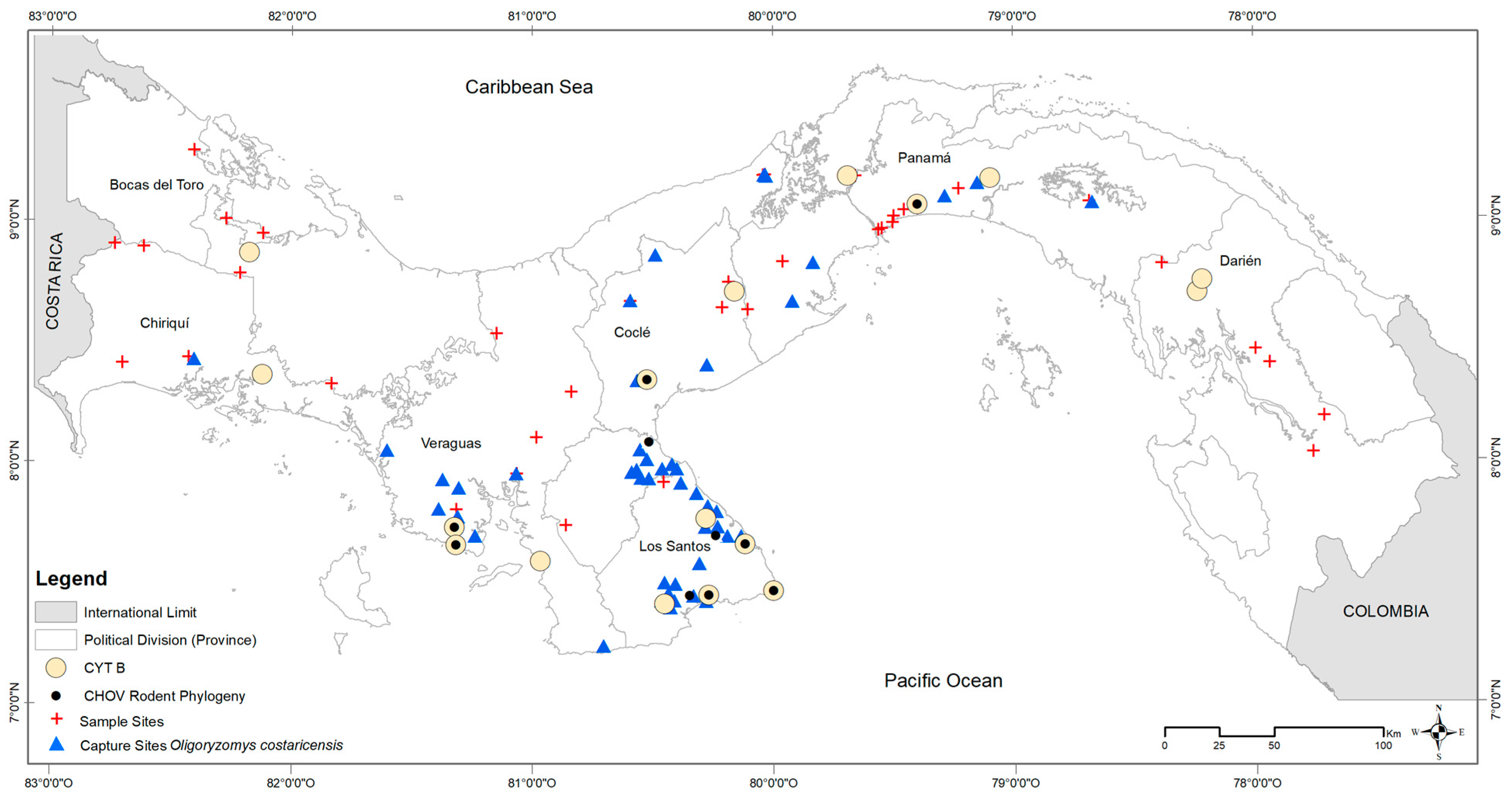
Supplementary Table S1) using a QIAamp DNA Mini Kit (Qiagen Inc., Valencia, CA, USA) following the manufacturer’s protocols. We included at least two individuals from each of seven provinces of Panama: Cañazas, Bocas del Toro; Boca del Monte, Chiriquí; Aguas Claras, Colón; Tamarindo and Zimba, Darién; El Bebedero, Cañas, Barriada 8 de Noviembre, San José, Pocrí, and Punta Mala, Los Santos; Santa Rosa Abajo and Tocumen, Panamá; Malena, La Zumbona, and Punta San Lorenzo, Veraguas (see
Figure 1).
Partial
cytb was PCR-amplified for each host using primers MVZ05 (5′- CGAAGCTTGATATGAAAAACCATCGTTG—3′ [
43]) and MVZ14 (5′—GGTCTTCATCTYHGGYTTACAAGA—3′ [
44]). PCR reactions were performed using Taq PCR Master Mix (Qiagen Inc.) with 1.5 mM MgCl
2, 0.2 mM dNTPs, 0.4 ρmol of forward and reverse primers, 2.5 units of Taq polymerase, and 1 μL template DNA (50 ng/μL) for a final volume of 25 μL with the following thermal cycling conditions: 94 °C for 3 min, 30 cycles at 94 °C for 30 s, 45 °C for 90 s, 72 °C for 90 s, and a final extension for 10 min at 72 °C. Products were visualized on 1.5% agarose gel. Amplicons were cleaned using the QIAquick PCR Purification Kit (Qiagen). Sequencing reactions were performed using the ABI BigDye Terminator v. 3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA) with an ABI PRISM 3130xl Genetic Analyzer (Life Technologies, Carlsbad, CA, USA). All sequences were assembled and aligned using Sequencher v. 4.6 (GeneCodes, Ann Arbor, MI, USA).
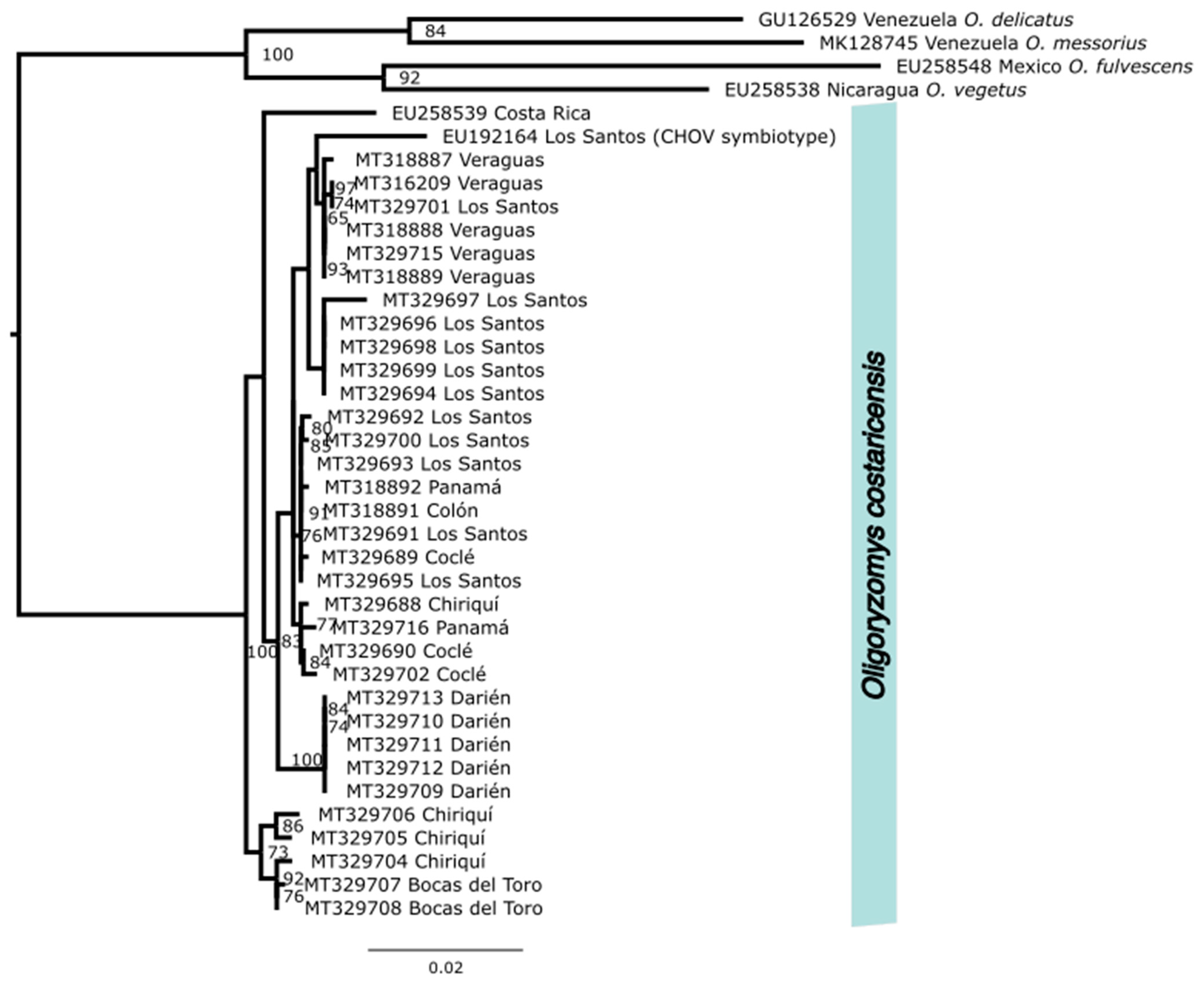
We used NCBI’s (National Center for Biotechnology Information) Basic Local Alignment Search Tool (BLAST [
45]) to identify two additional
cytb sequences from
O. costaricensis (EU192164, EU258539), which were available on GenBank. Those 35 sequences were combined with
cytb sequences from four outgroup species also from the Neotropics,
O. fulvescens (EU258548),
O. vegetus (EU258538),
O. delicatus (GU126529), and
O. messorius (MK128745), which were used to root the phylogeny. Sequences were aligned using MUSCLE [
46] and maximum likelihood phylogeny was inferred using IQ-TREE, V.2.2.2.6 (Nguyen et al. 2018) with automated model selection, which was conducted through ModelFinder [
47]. Trees were generated with 1000 ultrafast bootstrap alignments repeated a maximum of 1000 times, and the consensus species tree was visualized using Fig Tree v 4.2 (
http://tree.bio.ed.ac.uk/software/figtree/) with text adjustments in InkScape (InkScape Project 2020;
inkscape.org/, accessed on 10 May 2023) (see
Figure 2).
To test for current or prior hantavirus infection in the host, blood samples from 778 of the 883 captured
O. costaricensis were screened for antibodies using an IgG strip immunoblot assay [
48]. Immature individuals (<10 g) were removed from analysis to avoid inflating prevalence estimates with potential transovarial transmission [
5,
49,
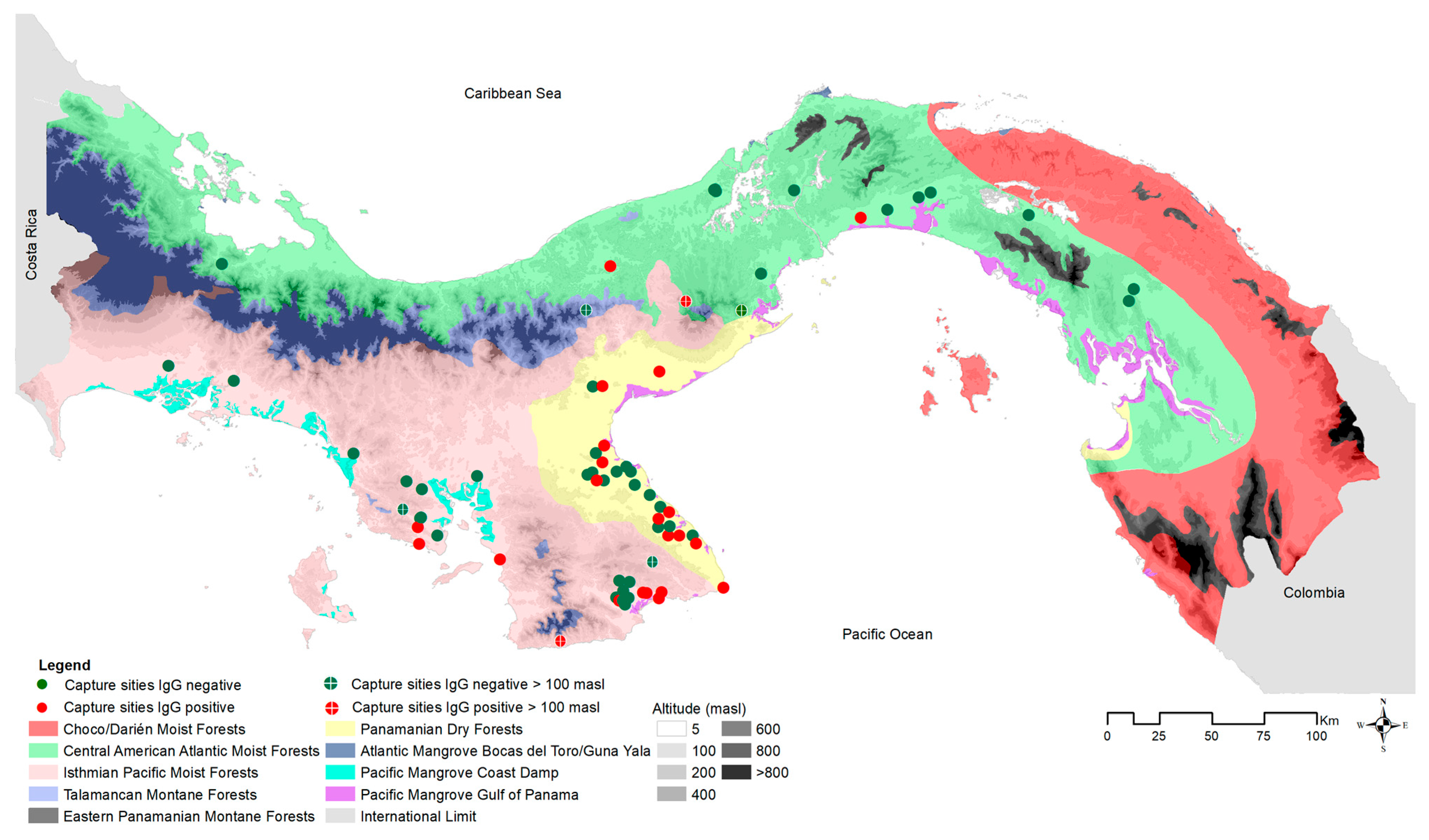
50], leaving 626 adults. Positive and negative results were visualized in geographic space using ArcGIS v. 10.7 (ESRI 2019), compared against microhabitat (cropland, peridomestic [e.g., households, adjacent outbuildings, gardens, livestock enclosures], pasture, shrub, and secondary vegetation), and ecoregion (see
Figure 3 and
Figure 4). Positive cases were summed by microhabitat category within each ecoregion [
26].
2.3. Environmental Associations
About 44% of Panama is wooded, 39% is rural agricultural (crops and pasture, [
26,
35], and 2% is multi-use, which includes towns and industrial centers [
35,
51]. To explore the association between host, viral presence, and general environmental composition, each site was classified at two coarse spatial scales: (1) microhabitat, (cropland, pasture, peridomestic [e.g., households, adjacent outbuildings, gardens, livestock enclosures], secondary vegetation, and shrub), and (2) ecoregion [
51]. There are 9 distinct ecoregions (7 terrestrial, 2 marine) in Panama. We sampled one coastal ecoregion (Pacific Mangrove South America) and six other terrestrial ecoregions: (1) Central American Atlantic Moist Forests, which includes Ngäbe-Buglé, one of Panama’s five ‘comarcas indígenas’ formerly belonging to the provinces of Bocas del Toro, Chiriquí, and Veraguas; (2) Talamancan Montane Forests; (3) Isthmian-Pacific Moist Forests; (4) Panamanian Dry Forests; (5) Choco/Darién Moist Forests; and (6) Eastern Panamanian Montane Forest (see
Supplementary Table S2). Within the last 80 years [
52], much of the historical Central American Atlantic Moist Forests, Isthmian-Pacific Moist Forests, and Panamanian Dry Forest ecoregions have been converted to agricultural lands (67% cattle pasture and 5% crops [rice, corn, and cane sugar]). To account for sampling biases across ecoregions, the total number of captures in each environmental type (microhabitat categories, ecoregion) was scaled by the total number captured in that environment.
2.4. Statistical Analyses
Continuous and categorical variables were analyzed using EPIINFO Version 7.2.4.0 (Centers for Disease Control and Prevention, Atlanta, GA, USA) and assessed using parametric and nonparametric techniques. A p-value with alpha <0.05 was considered significant.
3. Results
3.1. Biorepository Development & Spatial Distributions of Host & Pathogen
This 20-year surveillance project generated >10,500 specimens representing 110 species of non-volant small mammals distributed throughout Panama. This biodiversity archive provided critical biological material for the two recognized hosts of two orthohantaviruses in Panama,
O. costaricensis (CHOV) and
Zygodontomys brevicauda, the primary host of Calabazo virus [
17]. The archive also built comprehensive sampling of the associated mammalian communities for diverse other studies. All specimens were holistically prepared, including heart, lung, kidney, spleen, and blood samples, cryogenically preserved, and archived at the Gorgas Memorial Institute, the Vertebrate Museum of the University of Panama, and Museum of Southwestern Biology (MSB) in Albuquerque, New Mexico. Traditional host voucher specimens were archived in the MSB Division of Mammals. Tissues (heart, liver, kidney, spleen) were cryogenically preserved in nitrogen in the field and permanently archived in ultracold freezers (−80 °C) in the Gorgas Memorial Institute or in nitrogen vapor tanks (−190 °C) in the MSB Division of Genomic Resources. Data associated with each specimen are openly available through the Arctos museum database and physical specimens can be loaned from the institutions.
In total, 833 wild
O. costaricensis were collected from 2000 to 2019. Of those, 380 specimens collected between 2000 and 2006 formed the basis for earlier investigations [
22,
24,
26,
33]. Here, we add an additional 453 samples to extend the geographic extent and temporal span of earlier samples to include 2007 to 2019 (see
Figure 1). Collection localities were widespread, reaching from the southwestern border of Panama and Costa Rica to 40 km east of the Panama Canal. Of the 155 sites sampled for rodents,
O. costaricensis were detected at 71 sites. Occurrences were consistent with the previously described distribution of
O. costaricensis in Panama [
53], but we document important new records in Ngäbe-Buglé to the north and also eastward into Darién (see
Supplementary Figure S1), potentially related to agricultural development and host range expansion. About 93% of
O. costaricensis captures occurred below 100 m elevation, with only six exceptions, each a single capture event: Veraguas (El Jagua, Cerro Hoya), Coclé (El Cope National Park, San Miguel Centro), Panama (Altos de Campana National Park), and Los Santos (Oria) (see
Figure 4). The highest elevational record of
O. costaricensis was at 797 m.
Costa Rican pygmy rice rats were frequently detected in anthropogenically transformed rural areas (91%; 65 of 71 sites) or highly-disturbed urban areas (3%; 2 sites), as opposed to intact natural areas (6%; 4 sites). Among ecoregions (see
Figure 1), 42% of all sites where
O. costaricensis were recorded were in the Isthmian-Pacific Moist Forest (30 of 71 sites) and 34% were in the Panamanian Dry Forest (24 of 71 sites there). There was only one record each from the Talamancan Montane Forest and Pacific Mangrove South America ecoregions. Although there were four sampling sites in Choco/Darién, no
O. costaricensis were detected there. Only 21% of all sites in Panama where
O. costaricensis were detected were located in the Central American Atlantic Moist Forest ecoregion (15 of 71 sites), the largest ecoregion in Panama. Two sites (Zimba, Tamarindo) in this ecoregion located in Darién province had
O. costaricensis, representing the most eastern records for the species. These extend the known distribution of
O. costaricensis [
53] by ca. 120 km to the east (see
Supplementary Figure S1). Finally, for the total number of individual captures of
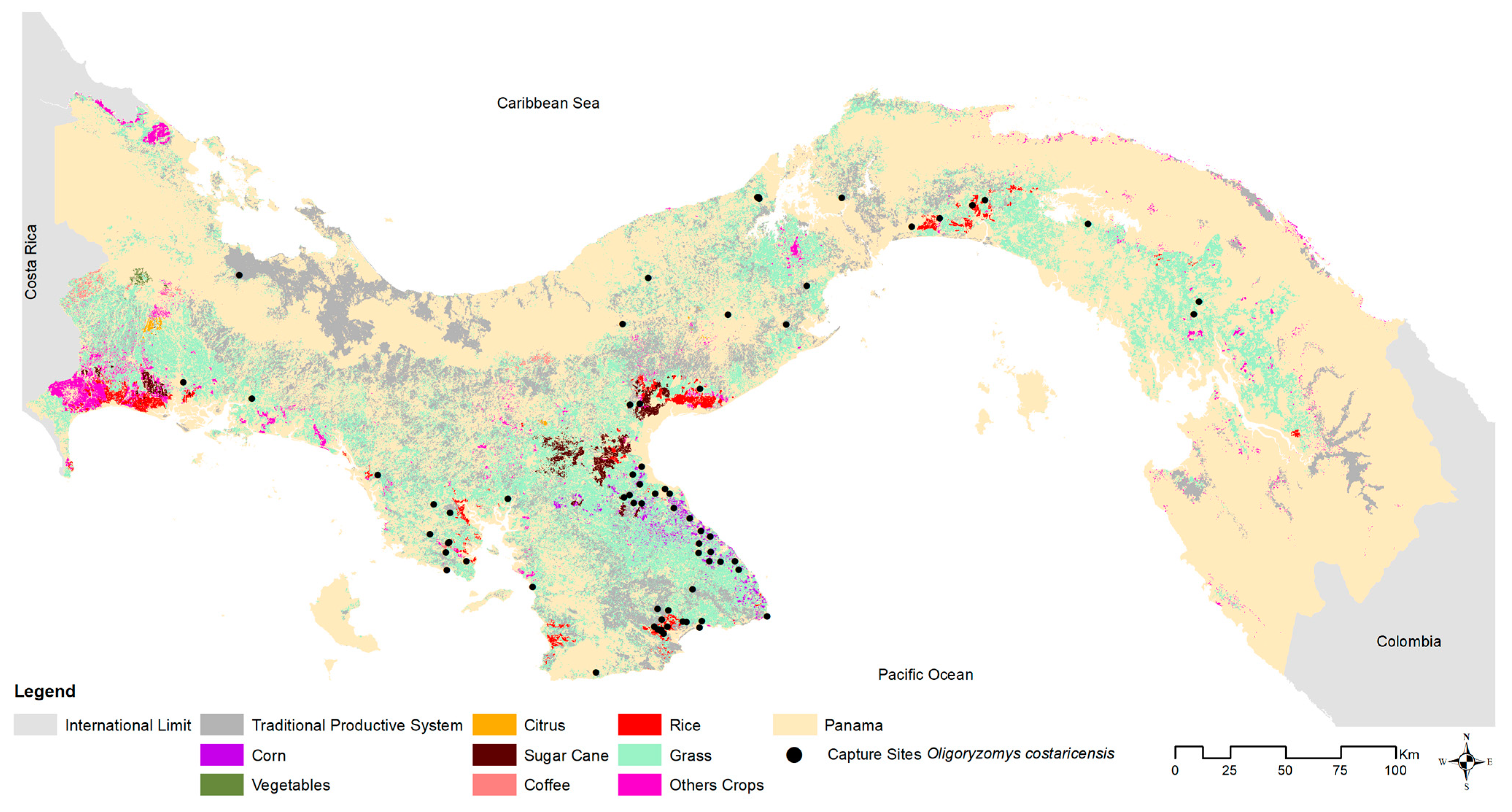
O. costaricensis across microhabitat categories, 28% (234) of
O. costaricensis captures occurred in croplands, 26% (217) in peridomestic sites, and 39% (322) in pastures.
3.2. Sequencing and Serology
We generated high-quality sequence data from host tissues cryogenically preserved in museum collections. Partial
cytb sequences from
O. costaricensis formed a single monophyletic group with 100% bootstrap support, consistent with morphological species diagnoses that a single species of pygmy rice rat occurs in Panama (see
Figure 2) with generally minimal substructure. Within the Panamanian clade, however, potential geographic substructure was identified with a well-supported Darien clade. Our species records extend the documented range of pygmy rice rats east into Darién (see
Supplementary Figure S1) and north into Ngäbe-Buglé. Preliminary mitochondrial sequence data identified pygmy rice rats from Costa Rica as ancestral to those in Panama. Two major mitochondrial clades were detected within Panama, one distributed in the eastern provinces of Chiriquí and Bocas del Toro and another distributed throughout the rest of Panama.
Hantavirus seroprevalence in
O. costaricensis was 16% (122/778) overall. After removing immature individuals, adult seroprevalence levels increased to 18% (111/626), with significant differences between male (22.5% [87/386]; 95% CI = 19.0, 27.0) and female (10.00% [24/240]; 95% CI = 7.0–15.0%) prevalence across sites (X
2 = 15.10;
p = 0.0001). Seroprevalence data were summarized by ecoregion and microhabitat (see
Table 1).
The highest prevalence coincided with the historically recognized endemic area of the hantavirus disease [
24,
26,
54], but 3 of 15
O. costaricensis individuals sampled outside the area of endemism tested positive.
4. Discussion
We summarize two decades of hantavirus field studies in Panama that were initiated following the emergence of CHOV in late 1999. We update the known distributional limits of O. costaricensis, the primary host of CHOV, and demonstrate limited molecular variation across Panama, with the exception of Darien specimens, based on a single mitochondrial DNA barcode. Additional independent nuclear markers and more comprehensive geographic sampling of the host in Panama, Costa Rica, and Colombia, with a focus on undisturbed areas, should now be developed. Such molecular investigations could explore in more detail potential geographic variations in the host, demographic and phylodynamic history (e.g., expansion and contraction), and biogeographic origins.
We then associated the geographic distribution and relative abundance of
O. costaricensis in Panama with the prevalence of CHOV antibody detection across multiple ecoregions. Human CHOV cases, reported outside of this investigation, primarily occur in agricultural communities in the central region of Panama (Los Santos, Herrera, Coclé, and Veraguas provinces [
55]) likely due to local abundance of granivorous rodents in response to excess food availability. A greater abundance of rodents increases the opportunity for contact with and zoonotic transmission to humans, especially for those working and living close to agricultural fields. Through our long-term screening efforts, we have extended the understanding of the Costa Rican pygmy rice rat as the primary wild reservoir of CHOV in Panama. Consistent with previous investigations, we document a wide geographic distribution of the host [
18,
22,
24,
26,
31,
53], with pygmy rice rats found in four of seven terrestrial and coastal ecoregions. Seropositive hosts were identified across four ecoregions and five distinct microhabitats. All habitats where
O. costaricensis were detected have been modified somewhat by human perturbation (e.g., cropland, pasture, shrub, secondary vegetation, and peridomestic). New host records in the Central American Atlantic Moist Forests (Ngäbe-Buglé) and Eastern Panamanian Montane Forest (Darién) ecoregions suggest that the host species may have expanded northward and eastward coincident with regional agricultural expansion [
56,
57]; however, the species may have simply been undetected until now.
A pathogen always has a broader spatial distribution than that of the disease itself [
58] and a narrower distribution than that of its hosts. Consistent with this hypothesis, we did not record CHOV-positive mice throughout the entire geographic range of
O. costaricensis. Instead, we found higher prevalence of CHOV in disturbed rural areas compared to natural areas, although our sampling was primarily focused on the former. Agricultural proximity is associated with sustained, year-round reproduction in
Oligoryzomys, presumably because such areas provide an abundance of supplementary and stable food resources [
30]. We hypothesize that higher host densities translate to higher pathogen densities. Support for this scenario has been documented for other rodent–hantavirus systems in South America, where host population density and prevalence of hantavirus infection were significantly higher in peridomestic habitats [
59].
Understanding the natural history of the host, in addition to the distribution of the pathogen, is essential to refining our understanding of the spatial distribution of the disease and forecasting how that distribution might change in the future. In the case of orthohantaviruses in Panama, expanded agricultural development may have facilitated an expanded geographic distribution of the host (e.g., captures west of the Panama Canal, where previous surveys from 1970–1977 had not detected the species) and also led to increased host population sizes through elevated reproductive output. These possibilities should be further monitored and rigorously tested. Previous longitudinal studies of small mammals and
Sin Nombre orthohantavirus in the southwestern United States [
41] demonstrated a positive relationship between increased resource availability due to natural environmental change (El Niño Southern Oscillation [ENSO] events), rodent density, and subsequent changes in hantavirus prevalence. Human-induced habitat changes have the potential to artificially mirror naturally occurring events, particularly when the reservoir species involved are well-adapted for rapid response to favorable conditions (e.g., cricetid rodents). Hosts for zoonotic viruses are more likely to be opportunistic, generalist species that frequently inhabit anthropogenically disturbed habitats [
60]. Wildlife surveillance that targets agricultural interfaces near human city centers may be most effective at detecting and subsequently mitigating regional hantavirus outbreaks. Further, forest restoration was shown to decrease the abundance of hantavirus reservoir rodents, including
Oligoryzomys (from 89% to 43%), thereby decreasing the chance of zoonotic transmission by ~45% [
57]. Expanded sampling and regular resurveys across representative environments remains critical, especially as anthropogenic impact on the environment induces change in natural communities. Substantial environmental heterogeneity in western Panama is associated with elevated endemism in rodents, which may also contribute to patterns of zoonotic transmission, but sampling remains too limited to effectively explore this possibility.
This work was limited by several assumptions that require further exploration. First, many pathogens, including hantaviruses [
61], are capable of infecting multiple host species [
8] but being able to infect a host is not the same as being productively infectious. Second, seroprevalence is not a measure of active infection and, although serological data for hosts <10 g were excluded from analysis, the possibility and frequency of transovarial transmission of the virus or maternal antibodies remain unknown. That knowledge gap challenges our ability to extend these test results into environmentally and spatially explicit models [
5,
50]. We only generated sequence data for a subset of seropositive hosts and only from a single mitochondrial marker; therefore, additional genetic structure within the host may be yet undetected. More comprehensive assessment of host genomic variation might provide key insight into the host colonization dynamics of newly disturbed habitats in Panama. Host phylogeography also can provide insights into mechanisms underlying viral evolution [
62]. Given that these specimen collections were accumulated over the last two decades, insight into temporal aspects of both host neutral and immune-related genes [
63] as well as
Orthohantavirus evolution can now be pursued. Full genome sequencing of the hosts and viruses should be a priority as we aim to understand issues such as mutation rates, reassortment, and potential interactions with other co-circulating orthohantaviruses in these mammalian communities.
5. Future Directions
The Gorgas Memorial Institute’s approach to CHOV field studies in Panama emphasizes the development of temporally deep and spatially broad biorepositories of both host and pathogen samples. Now, after 20 years of site-intensive sampling, questions related to CHOV evolution and ecology are tractable, reinforcing the value of holistic collecting [
40] of wildlife hosts and their associated parasites, endoparasites, and pathogens. The One Health approach is predicated on building long-term biorepositories to allow for the replication and extension of studies and as technology improves, these historic samples allow us to more fully understand host–pathogen dynamics in addition to the distribution and ecology of emerging zoonotic diseases [
64,
65,
66].
Although not explored in detail here, preliminary sampling in undisturbed natural ecosystems (e.g., Parque Nacional Amistad, Vulcan Baru) has identified other potential seropositive hosts for orthohantavirus not included here, such as
Peromyscus nudipes,
Reithrodontomys mexicanus,
R. sumichrasti,
R. creper,
Sigmodon hirsutus,
Liomys adspersus, and
Transandinomys talamancae, which we are now characterizing. An incomplete understanding of the mammalian communities that may serve as reservoirs for CHOV or other yet unrecognized orthohantaviruses in Panama may complicate interpretations of the spatial distribution of the pathogen and risk landscape. Other orthohantaviruses, such as Calabazo virus that has been found primarily in
Z. brevicauda, are circulating in these mammalian communities but have been only minimally surveyed [
17,
22]. Survey efforts will require viral screening of specimens of other wild mammalian hosts that are now available through these archival collections built over two decades of fieldwork. Understanding potential interactions among the diverse pathogens circulating in these communities may be facilitated by the application of new metatranscriptomic or metagenomic sequencing methods [
67]. More extensive and detailed genetic characterization and analyses of both host and virus are needed to test these preliminary findings using phylodynamic methods and disease modeling, among other approaches [
68,
69]. Such analyses will help to characterize the virus’ evolution and identify the suite of environmental and ecological conditions suitable for the host and the subset of conditions in which the virus is found.
While we use gross correlative methods to characterize broad environmental associations among hosts, pathogens, and their environment, the next step will be to incorporate these data into an ecological niche modeling framework [
70]. Such a framework will allow us to visualize how the suitability of the landscape for each interacting component changes across space and time and to identify potentially causal features useful for building predictive models of emerging disease. Habitat conversion, for example, is now opening the Darién Gap to expansion by commensal mammalian species likely to cause disease outbreaks (e.g., grassland species such as
Oligoryzomys, livestock-fed
Desmodus vampire bats, and edge-exploiting species such as
Didelphis marsupials). Pairing this habitat conversion with increasing human migration through the Panamanian Isthmus suggests the importance of wildlife pathogen surveillance to public health throughout the Americas.
Supplementary Materials
The following supporting information can be downloaded at:
https://www.mdpi.com/article/10.3390/v15061390/s1, Figure S1: Historical and current spatial distribution of
O. costaricensis. The map is created from the collection sites of museum specimens (striped lines) using as reference the map generated by Méndez (1993) [
53]. The Hantavirus Project 2000–2019 distribution map creates a spatial model generated by circular polygons around capture points, assuming a dispersion range of 20 km; Table S1 ([
43,
44,
71,
72]): List of primers used for amplifications and sequencing of
Choclo orthohantavirus S segment and
cytb gene of
Oligoryzomys costaricensis. Table S2: Distribution of ecoregions of Panama according to land use [
35], area, and altitude.
Author Contributions
Conceptualization, B.A., A.G.A., J.A.C. and G.E.G.; methodology, P.G., T.P.S., J.R.S., M.A., J.P.C., J.L.D., G.E.G., G.G., E.J., K.L., E.P., Y.M., J.M.P., A.G.A., J.A.C. and B.A.; formal analysis, P.G., T.P.S., J.R.S., J.P.C., J.L.D., G.E.G., E.J., K.L., E.P., Y.M., J.A.C. and B.A.; investigation, P.G., T.P.S., J.R.S., M.A., J.P.C., J.L.D., G.E.G., G.G., E.J., K.L., E.P., Y.M., J.M.P., A.G.A., J.A.C. and B.A.; resources, M.A., G.G., Y.M., K.L., E.P. and J.M.P.; data curation, J.L.D., P.G., T.P.S., J.R.S., M.A., E.J., Y.M., E.P. and B.A.; writing—original draft preparation, P.G., T.P.S., J.R.S., J.P.C., J.L.D., A.G.A., J.A.C. and B.A.; writing—review and editing, P.G., T.P.S., J.R.S., M.A., J.P.C., J.L.D., G.E.G., G.G., E.J., K.L., E.P., Y.M., J.M.P., A.G.A., J.A.C. and B.A.; supervision, B.A., A.G.A., J.L.D. and J.A.C.; project administration, B.A., J.A.C., J.L.D., K.L. and G.E.G.; funding acquisition, B.A., A.G.A., J.A.C., G.E.G. and J.M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by: the Opportunity Pool Award and supplemented by the International Centers for Infectious Diseases Research Program of the National Institutes of Health (U19-AI 45452) to G.G. and B.A. in 2000–2010, F.G. in 2000–2001, and J.A.C. in 2004–2010; funds from the Gorgas Memorial Institute of Studies of Health, Hantavirus Research Project (04-90-0075-8) to F.G. and B.A. in 2000–2005; funds from the Secretaria Nacional de Ciencia y Tecnologia (Innovation and Technology Program ftd06-089) to B.A. in 2006–2008; Intramural funding from the Climate Change Capacity Building Program: Research and Public Health Action Award from the Centers for Infectious Diseases, “Effect of Anthropogenic Climate Change on the Ecology of Zoonotic and Vector-Borne Diseases (Peridomestic Mammals)” (Project ID 347998861); funds from the Ministry of Economy and Finance of Panama (FPI-MEF-056), “Epidemiología y ecología de Hantavirus, otras enfermedades zoonóticas y transmitidas por vectores (emergentes y re-emergentes) en Panamá” Fase-I y Fase II (PHoEZyTV I-II) to B.A. in 2010–2014; and funds from the Ministry of Economy and Finance of Panama [grant no. 111130150.501.274] to B.A. in 2014–2019.
Institutional Review Board Statement
The animal study protocol was approved by the Panamanian Ministry of Environmental Affairs and the Institutional Review Board of Gorgas Committee for Animal Care and Use (GMI; # 001/05 CIUCAL/ICGES, 2005) for studies involving animals and conducted in accordance with Law No. 23 (Animal Welfare Assurance, 1997) of the Republic of Panama.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data are available through NCBI’s GenBank (Accession numbers for
cytb sequences: MT316209, MT318887, MT318892, MT329688, MT329716). Specimens and tissues are available through the Museum of Southwestern Biology at the University of New Mexico, the Vertebrate Museum of the University of Panama, or the Zoological Collection of the Gorgas Memorial Institute for Health Studies in Panama City, with digital data available through the Arctos database (
arctosdb.org, accessed on 30 April 2023).
Acknowledgments
We thank the Panamanian Ministry of Environmental Affairs, the Gorgas Committee for Animal Care and Use, the International Center for Infectious Disease Research Program of the National Institutes of Health, the University of New Mexico Museum of Southwestern Biology, the Gorgas Memorial Institute for Health Studies (GMISH), PICANTE (Pathogen Informatics Center for Analysis, Networking, Translation, and Education, US National Science Foundation 2155222), the Panamanian Institute of Livestock and Agricultural Research, the Ministry of Health, the Ministry of Agricultural Development, the Smithsonian Tropical Research Institute (Stanley Heckadon, Oris Sanjur, Isis Ochoa, and Allen Herre), and the Ministry of Environment for their support. Terry Yates, Brian Hjelle, and Greg Mertz from the University of New Mexico, and especially Fred Koster, then with the Lovelace Respiratory Research Institute in Albuquerque, were instrumental in ensuring successful collaborations on multiple aspects of this research in Panama on hantaviruses over two decades. We also thank individuals from the communities and areas surveyed, sever state organization, and the rodent ecology team of the Ministry of Health and GMISH, especially Fernando Gracia, Carlos Muñoz, Eustiquio Broce, Omar Vargas (R.I.P), Francisco Crespo, Carlos Falcon, Ricardo Rodriguez, Juan Bosco Navarro, Santiago Bosh, Ricardo Cedeño, Daniel Gonzalez, Carlos Falconet, Jose Miguel Montenegro, Miguel Vergara, Victor Dominguez, Ariel Perez, Joel Gonzalez, Algis Vergara, Heriberto Rivera, Cesar Lombardo, Domingo Archibold, Leonel Castillo, Alexis Ortiz, Donderis Soto, Carmelo Ortiz (R.I.P), Alexis Araúz, Juán De Leon, Juán Díaz, Ramón García, Jose Santizo (R.I.P), Juan Francisco Tello, Jorge Garzón, Gilberto Niño, Ricardo Rodríguez, Dagoberto Olivares, Jony Castillo, Candelario Olivares, Emilio Salazar, Pablo Gutiérrez, Lorenzo Aldobán, Olmedo Castro, José Valencia (R.I.P), Armando Mepaquito, Silvio Bethancourt, Enrique Ramos (R.I.P), and Rafael Figueroa for support during fieldwork. We thank the lab team, Yamitzel Zaldivar, Juan Castillo, Migdalis Ortega, Marla Ramos, Natalia Vega, José Correa, José Cedeño, Jorge Herrera, Jonathan Montenegro, Ricardo Cumbrera, Verónica Ventura, Kirian Miranda, and Sadith Aldrette. We thank Claudia Dominguez and Neida Gonzalez, database manager. Thanks to Rosa de Vargas and Iris Reyes for the significant administrative support given as part of the Department of Research in Emerging and Zoonotic Infectious Diseases, Gorgas Memorial Institute for Health Studies.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Gu, S.H.; Lim, B.K.; Kadjo, B.; Arai, S.; Kim, J.A.; Nicolas, V.; Lalis, A.; Denys, C.; Cook, J.A.; Dominguez, S.R.; et al. Molecular Phylogeny of Hantaviruses Harbored by Insectivorous Bats in Côte d’Ivoire and Vietnam. Viruses 2014, 6, 1897–1910. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Chen, X.; Tian, J.H.; Chen, L.J.; Li, K.; Wang, W.; Eden, J.S.; Shen, J.J.; Liu, L.; et al. The Evolutionary History of Vertebrate RNA Viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef]
- Laenen, L.; Vergote, V.; Calisher, C.H.; Klempa, B.; Klingström, J.; Kuhn, J.H.; Maes, P. Hantaviridae: Current Classification and Future Perspectives. Viruses 2019, 11, 788. [Google Scholar] [CrossRef]
- Hjelle, B.; Torres-Pérez, F. Hantaviruses in the Americas and Their Role as Emerging Pathogens. Viruses 2010, 2, 2559–2586. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, C.B.; Figueiredo, L.T.M.; Vapalahti, O. A Global Perspective on Hantavirus Ecology, Epidemiology, and Disease. Clin. Microbiol. Rev. 2010, 23, 412–441. [Google Scholar] [CrossRef] [PubMed]
- Yanagihara, R.; Gu, S.H.; Arai, S.; Kang, H.J.; Song, J.W. Hantaviruses: Rediscovery and New Beginnings. Virus. Res. 2014, 187, 6–14. [Google Scholar] [CrossRef]
- Ermonval, M.; Baychelier, F.; Tordo, N. What Do We Know about How Hantaviruses Interact with Their Different Hosts? Viruses 2016, 8, 223. [Google Scholar] [CrossRef]
- Goodfellow, S.M.; Nofchissey, R.A.; Schwalm, K.C.; Cook, J.A.; Dunnum, J.L.; Guo, Y.; Ye, C.; Mertz, G.J.; Chandran, K.; Harkins, M.; et al. Tracing Transmission of Sin Nombre Virus and Discovery of Infection in Multiple Rodent Species. J. Virol. 2021, 93, e02185-18. [Google Scholar] [CrossRef]
- Forbes, K.M.; Sironen, T.; Plyusnin, A. Hantavirus Maintenance and Transmission in Reservoir Host Populations. Curr. Opin. Virol. 2018, 28, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hussein, I.T.M.; Haseeb, A.; Haque, A.; Mir, M.A. Recent Advances in Hantavirus Molecular Biology and Disease. Adv. Appl. Microbiol. 2011, 74, 35–75. [Google Scholar] [CrossRef]
- Hallin, G.W.; Simpson, S.Q.; Crowell, R.E.; James, D.S.; Koster, F.T.; Mertz, G.J.; Levy, H. Cardiopulmonary Manifestations of Hantavirus Pulmonary Syndrome. Crit. Care Med. 1996, 24, 252–258. [Google Scholar] [CrossRef]
- Avšič-Županc, T.; Saksida, A.; Korva, M. Hantavirus Infections. Clin. Microbiol. Infect. 2019, 21, e6–e16. [Google Scholar] [CrossRef]
- Mertz, G.J. Bunyaviridae: Bunyaviruses, Phleboviruses, Nairoviruses, and Hantaviruses. In Clinical Virology; ASM Press: Hoboken, NJ, USA, 2009; pp. 977–1007. ISBN 9781683674078. [Google Scholar]
- Childs, J.E.; Glass, G.E.; Korch, G.W.; Leduc3, J.W. Effects of Hantaviral Infection on Survival, Growth and Fertility in Wild Rat (Rattus Norvegicus) Populations of Baltimore, Maryland; Journal of Wildlife Diseases. J. Wildl. Dis. 1989, 25, 469–476. [Google Scholar] [CrossRef]
- Hooper, J.W.; Ferro, A.M.; Wahl-Jensen, V. Immune Serum Produced by DNA Vaccination Protects Hamsters against Lethal Respiratory Challenge with Andes Virus. J. Virol. 2008, 82, 1332–1338. [Google Scholar] [CrossRef]
- Zivcec, M.; Safronetz, D.; Haddock, E.; Feldmann, H.; Ebihara, H. Validation of Assays to Monitor Immune Responses in the Syrian Golden Hamster (Mesocricetus auratus). J. Immunol. Methods 2011, 368, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.J.; Quiroz, E.; Gracia, F.; Sanchez, A.J.; Ksiazek, T.G.; Kitsutani, P.T.; Ruedas, L.A.; Tinnin, D.S.; Caceres, L.; Garcia, A.; et al. Hantavirus Pulmonary Syndrome in Panama: Identification of Novel Hantaviruses and Their Likely Reservoirs. Virology 2000, 277, 14–19. [Google Scholar] [CrossRef]
- Bayard, V.; Kitsutani, P.T.; Barria, E.O.; Ruedas, L.A.; Tinnin, D.S.; Munoz, C.; de Mosca, I.B.; Guerrero, G.; Kant, R.; Garcia, A.; et al. Outbreak of Hantavirus Pulmonary Syndrome, Los Santos, Panama, 1999–2000. Emerg. Infect. Dis. 2004, 10, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Bradfute, S.B.; Calisher, C.H.; Klempa, B.; Klingström, J.; Laenen, L.; Palacios, G.; Schmaljohn, C.S.; Tischler, N.D.; Maes, P. Pending Reorganization of Hantaviridae to Include Only Completely Sequenced Viruses: A Call to Action. Viruses 2023, 15, 660. [Google Scholar] [CrossRef]
- Hurtado, N.; D’Elía, G. An Assessment of Species Limits of the South American Mouse Genus Oligoryzomys (Rodentia, Cricetidae) Using Unilocus Delimitation Methods. Zool. Scr. 2019, 48, 557–570. [Google Scholar] [CrossRef]
- Hanson, J.D.; Utrera, A.; Fulhorst, C.F. The Delicate Pygmy Rice Rat (Oligoryzomys Delicatus) Is the Principal Host of Maporal Virus (Family Bunyaviridae, Genus Hantavirus). Vector-Borne Zoonotic Dis. 2011, 11, 691–695. [Google Scholar] [CrossRef]
- Ruedas, L.; Salazar-Bravo, J.; Tinnin, D.; Armien, B.; Caceres, L.; Garcia, A.; Diaz, M.A.; Gracia, F.; Suzan, G.; Peters, C.; et al. Community Ecology of Small Mammal Populations in Panama Following an Outbreak of Hantavirus Pulmonary Syndrome. J. Vector Ecol. 2004, 29, 177–191. [Google Scholar] [PubMed]
- Vigueras-Galván, A.L.; López-Pérez, A.M.; García-Peña, G.E.; Rico-Chávez, O.; Sarmiento-Silva, R.E. Current Situation and Perspectives on Hantaviruses in Mexico. Viruses 2019, 11, 642. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Bravo, J.; Armién, B.; Suzán, G.; Armién, A.; Ruedas, L.A.; Avila, M.; Zaldívar, Y.; Pascale, J.M.; Gracia, F.; Yates, T.L. Serosurvey of Wild Rodents for Hantaviruses in Panama, 2000–2002. J. Wildl. Dis. 2004, 40, 103–109. [Google Scholar] [CrossRef]
- Armien, B.; Pascale, J.M.; Bayard, V.; Munoz, C.; Mosca, I.; Guerrero, G.; Armien, A.; Quiroz, E.; Castillo, Z.; Zaldivar, Y.; et al. High Seroprevalence of Hantavirus Infection on the Azuero Peninsula of Panama. Am. J. Trop. Med. Hyg. 2004, 70, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Armién, A.G.; Armién, B.; Koster, F.; Pascale, J.M.; Avila, M.; Gonzalez, P.; De La Cruz, M.; Zaldivar, Y.; Mendoza, Y.; Gracia, F.; et al. Hantavirus Infection and Habitat Associations among Rodent Populations in Agroecosystems of Panama: Implications for Human Disease Risk. Am. J. Trop. Med. Hyg. 2009, 81, 59–66. [Google Scholar] [CrossRef]
- Armien, B.; Pascale, J.M.; Munoz, C.; Lee, S.J.; Choi, K.L.; Avila, M.; Broce, C.; Armien, A.G.; Gracia, F.; Hjelle, B.; et al. Incidence Rate for Hantavirus Infections without Pulmonary Syndrome, Panama. Emerg. Infect. Dis. 2011, 17, 1936–1939. [Google Scholar] [CrossRef]
- Armién, B.; Ortiz, P.L.; Gonzalez, P.; Cumbrera, A.; Rivero, A.; Avila, M.; Armién, A.G.; Koster, F.; Glass, G. Spatial-Temporal Distribution of Hantavirus Rodent-Borne Infection by Oligoryzomys Fulvescens in the Agua Buena Region—Panama. PLoS Negl. Trop. Dis. 2016, 10, e0004460. [Google Scholar] [CrossRef]
- Mills, J.N.; Childs, J.E. Ecologic Studies of Rodent Reservoirs: Their Relevance for Human Health. Emerg. Infect. Dis. 1998, 4, 529–537. [Google Scholar] [CrossRef]
- Braga, C.; Sobral, G.; Zeppelini, C.G.; Fagundes, R.; Pires, M.R.S. Cornfield Effects on Breeding and Abundance of Oligoryzomys Nigripes (Rodentia: Sigmodontinae). Mastozool. Neotrop. 2020, 27, 234–240. [Google Scholar] [CrossRef]
- Suzán, G.; Armién, A.; Mills, J.N.; Marcé, E.; Ceballos, G.; Ávila, M.; Salazar-Bravo, J.; Ruedas, L.; Armién, B.; Yates, T.L. Epidemiological Considerations of Rodent Community Composition in Fragmented Landscapes in Panama. J. Mammal. 2008, 89, 684–690. [Google Scholar] [CrossRef]
- Suzán, G.; Giermakowski, J.T.; Marcé, E.; Suzán-Azpiri, H.; Armién, B.; Yates, T.L. Modeling Hantavirus Reservoir Species Dominance in High Seroprevalence Areas on the Azuero Peninsula of Panama. Am. J. Trop. Med. Hyg. 2006, 74, 1103–1110. [Google Scholar] [CrossRef]
- Suzán, G.; Marcé, E.; Giermakowski, J.T.; Armién, B.; Pascale, J.; Mills, J.; Ceballos, G.; Gómez, A.; Aguirre, A.A.; Salazar-Bravo, J.; et al. The Effect of Habitat Fragmentation and Species Diversity Loss on Hantavirus Prevalence in Panama. Ann. N. Y. Acad. Sci. 2008, 1149, 80–83. [Google Scholar] [CrossRef]
- Keesing, F.; Ostfeld, R.S. Impacts of Biodiversity and Biodiversity Loss on Zoonotic Diseases. Proc. Natl. Acad. Sci. USA 2021, 118, e2023540118. [Google Scholar] [CrossRef]
- MiAmbiente Gaceta Oficial Digital N° 28229-A. 2017. Available online: https://www.gacetaoficial.gob.pa/pdfTemp/28229_A/GacetaNo_28229a_20170303.pdf (accessed on 2 February 2023).
- INEC Panamá En Cifras: Años 2014-18. Available online: https://www.inec.gob.pa/publicaciones/Default3.aspx?ID_PUBLICACION=990&ID_CATEGORIA=17&ID_SUBCATEGORIA=45 (accessed on 7 February 2023).
- ANAM. Atlas Ambiental de La República de Panamá; Autoridad Nacional del Ambiente: Panama City, Panama, 2010; ISBN 978-9962-651-49-9.
- Sikes, R.S. 2016 Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research and Education. J. Mammal. 2016, 97, 663–688. [Google Scholar] [CrossRef]
- Colella, J.P.; Greiman, S.E.; Kutz, S.; Lutz, H.L.; Cook, J.A. Genomic Identification and Surveillance of Infectious Diseases in Natural Systems (in Press). Appl. Environ. Genom. CSIRO 2023, in press.
- Galbreath, K.E.; Hoberg, E.P.; Cook, J.A.; Armién, B.; Bell, K.C.; Campbell, M.L.; Dunnum, J.L.; Dursahinhan, A.T.; Eckerlin, R.P.; Gardner, S.L.; et al. Building an Integrated Infrastructure for Exploring Biodiversity: Field Collections and Archives of Mammals and Parasites. J. Mammal. 2019, 100, 382–393. [Google Scholar] [CrossRef]
- Yates, T.L.; Mills, J.N.; Parmenter, C.A.; Ksiazek, T.G.; Parmenter, R.R.; Vande Castle, J.R.; Calisher, C.H.; Nichol, S.T.; Abbott, K.D.; Young, J.C. The Ecology and Evolutionary History of an Emergent Disease: Hantavirus Pulmonary Syndrome. Bioscience 2002, 52, 989–998. [Google Scholar] [CrossRef]
- Thompson, C.W.; Phelps, C.W.; Allard, K.L.; Cook, M.W.; Dunnum, J.A.; Ferguson, J.L.; Gelang, A.W.; Khan, M.; Paul, F.L.; Reeder, D.L.; et al. Preserve a Voucher Specimen! The Critical Need for Integrating Natural History Collections in Infectious Disease Studies. mBio 2021, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.M.; Kocher, T.D.; Wilson, A.C. Evolution of the Cytochrome b Gene of Mammals. J. Mol. Evol. 1991, 32, 128–144. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.F.; Patton, J.L. Variation in Mitochondrial Cytochrome b Sequence in Natural Populations of South American Akodontine Rodents (Muridae: Sigmodontinae). Mol. Biol. Evol. 1993, 8, 85–103. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hjelle, B.; Jenison, S.; Torrez-Martinez, N.; Herring, B.; Quan, S.; Polito, A.; Pichuantes, S.; Yamada, T.; Morris, C.; Elgh, F.; et al. Rapid and Specific Detection of Sin Nombre Virus Antibodies in Patients with Hantavirus Pulmonary Syndrome by a Strip Immunoblot Assay Suitable for Field Diagnosis. J. Clin. Microbiol. 1997, 35, 600–608. [Google Scholar] [CrossRef]
- Voutilainen, L.; Kallio, E.R.; Niemimaa, J.; Vapalahti, O.; Henttonen, H. Temporal Dynamics of Puumala Hantavirus Infection in Cyclic Populations of Bank Voles. Sci. Rep. 2016, 6, 21323. [Google Scholar] [CrossRef]
- Kallio, E.R.; Klingström, J.; Gustafsson, E.; Manni, T.; Vaheri, A.; Henttonen, H.; Vapalahti, O.; Lundkvist, Å. Prolonged Survival of Puumala Hantavirus Outside the Host: Evidence for Indirect Transmission via the Environment. J. Gen. Virol. 2006, 87, 2127–2134. [Google Scholar] [CrossRef]
- Dinerstein, E.; Olson, D.M.; Graham, D.J.; Webster, A.L.; Primm, S.A.; Bookbinder, M.P.O.; Ledec, G. Una Evaluación Del Estado de Conservación de Las Eco-Regiones Terrestres de América Latina y El Caribe; Banco Mundial: Washington, DC, USA, 1995; ISBN 0821332961. [Google Scholar]
- Heckadon, M.S.; Espinoza, G.J. Agonia de La Naturaleza: Ensayos Sobre El Costo Ambiental Del Desarrollo Panameño; Instituto de Investigación Agropecuaria de Panamá: Panama City, Panama, 1985; Available online: https://bdigital.binal.ac.pa/binal/iframes/cldetalle.php?id=132521&from=l (accessed on 25 March 2023).
- Méndez, E. Los Roedores de Panama; Impresora Pacífico: Panama City, Panama, 1993. [Google Scholar]
- Armien, B.; Pascale, J.M.; Munoz, C.; Marinas, J.; Núnez, H.; Herrera, M.; Trujillo, J.; Sánchez, D.; Mendoza, Y.; Hjelle, B.; et al. Hantavirus Fever without Pulmonary Syndrome in Panama. Am. J. Trop. Med. Hyg. 2013, 89, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Armien, B.; Muñoz, C.; Cedeño, H.; Salazar, J.; Salinas, T.; González, P.; Trujillo, J.; Sánchez, D.; Mariñas, J.; Hernández, A.; et al. Hantavirus in Panama: Twenty Years of Epidemiological Surveillance Experience. Viruses 2023, in press. [Google Scholar]
- Medan, D.; Torretta, J.P.; Hodara, K.; de la Fuente, E.B.; Montaldo, N.H. Effects of Agriculture Expansion and Intensification on the Vertebrate and Invertebrate Diversity in the Pampas of Argentina. Biodivers. Conserv. 2011, 20, 3077–3100. [Google Scholar] [CrossRef]
- Prist, P.R.; Prado, A.; Tambosi, L.R.; Umetsu, F.; de Arruda Bueno, A.; Pardini, R.; Metzger, J.P. Moving to Healthier Landscapes: Forest Restoration Decreases the Abundance of Hantavirus Reservoir Rodents in Tropical Forests. Sci. Total Environ. 2021, 752, 141967. [Google Scholar] [CrossRef] [PubMed]
- Audy, J.R. The Localization of Disease with Special Reference to the Zoonoses. Trans. R. Soc. Trop. Med. Hyg. 1958, 52, 308–328. [Google Scholar] [CrossRef] [PubMed]
- Palma, R.E.; Polop, J.J.; Owen, R.D.; Mills, J.N. Ecology of Rodent-Associated Hantaviruses in the Southern Cone of South America: Argentina, Chile, Paraguay, and Uruguay. J. Wildl. Dis. 2012, 48, 267–281. [Google Scholar] [CrossRef]
- Mills, J.N. Biodiversity Loss and Emerging Infectious Disease: An Example from the Rodent-Borne Hemorrhagic Fevers. Biodiversity 2006, 7, 9–17. [Google Scholar] [CrossRef]
- Milholland, M.T.; Castro-Arellano, I.; Suzán, G.; Garcia-Peña, G.E.; Lee, T.E.; Rohde, R.E.; Alonso Aguirre, A.; Mills, J.N. Global Diversity and Distribution of Hantaviruses and Their Hosts. EcoHealth 2018, 15, 163–208. [Google Scholar] [CrossRef] [PubMed]
- Liphardt, S.W.; Kang, H.J.; Arai, S.; Gu, S.H.; Cook, J.A.; Yanagihara, R. Reassortment Between Divergent Strains of Camp Ripley Virus (Hantaviridae) in the Northern Short-Tailed Shrew (Blarina Brevicauda). Front. Cell. Infect. Microbiol. 2020, 10, 460. [Google Scholar] [CrossRef] [PubMed]
- Boric-Bargetto, D.; Rodríguez-Serrano, E.; Hernández, C.E.; Jaksic, F.M.; Palma, R.E. Temporal Variation in Genetic Diversity during an Outbreak of Oligoryzomys Longicaudatus (Rodentia, Sigmodontinae) in a Temperate Forest of Southern Chile. Biochem. Syst. Ecol. 2012, 44, 383–389. [Google Scholar] [CrossRef]
- Dunnum, J.L.; Yanagihara, R.; Johnson, K.M.; Armien, B.; Batsaikhan, N.; Morgan, L.; Cook, J.A. Biospecimen Repositories and Integrated Databases as Critical Infrastructure for Pathogen Discovery and Pathobiology Research. PLoS Negl. Trop. Dis. 2017, 11, e0005133. [Google Scholar] [CrossRef]
- Schindel, D.E.; Cook, J.A. The next Generation of Natural History Collections. PLoS Biol. 2018, 16, e2006125. [Google Scholar] [CrossRef]
- Colella, J.P.; Bates, J.; Burneo, S.F.; Camacho, M.A.; Bonilla, C.C.; Constable, I.; D’Elia, G.; Dunnum, J.L.; Greiman, S.; Hoberg, E.P.; et al. Leveraging Natural History Biorepositories as a Global, Decentralized, Pathogen Surveillance Network. PLoS Pathog. 2021, 17, e1009583. [Google Scholar] [CrossRef]
- Vittor, A.Y.; Armien, B.; Gonzalez, P.; Carrera, J.-P.; Dominguez, C.; Valderrama, A.; Glass, G.E.; Beltran, D.; Cisneros, J.; Wang, E.; et al. Epidemiology of Emergent Madariaga Encephalitis in a Region with Endemic Venezuelan Equine Encephalitis: Initial Host Studies and Human Cross-Sectional Study in Darien, Panama. PLoS Negl. Trop. Dis. 2016, 10, e0004554. [Google Scholar] [CrossRef]
- Volz, E.M.; Romero-Severson, E.; Leitner, T. Phylodynamic Inference across Epidemic Scales. Mol. Biol. Evol. 2017, 34, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Featherstone, L.A.; Zhang, J.M.; Vaughan, T.G.; Duchene, S. Epidemiological Inference from Pathogen Genomes: A Review of Phylodynamic Models and Applications. Viruses Evol. 2022, 8, 1–12. [Google Scholar] [CrossRef]
- Cobos, M.E.; Peterson, A.T. Detecting Signals of Species’ Ecological Niches in Results of Studies with Defined Sampling Protocols: Example Application to Pathogen Niches. Biodivers. Inform. 2022, 17, 50. [Google Scholar] [CrossRef]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Paabo, S.; Villablanca, F.X.; Wilson, A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 1989, 86, 6196–6200. [Google Scholar] [CrossRef]
- Kent, J.R.; Norris, D.E. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am. J. Trop. Med. Hyg. 2005, 73, 336–342. [Google Scholar]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).