Assembly of the Tripartite and RNA Condensates of the Respiratory Syncytial Virus Factory Proteins In Vitro: Role of the Transcription Antiterminator M2-1
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Purification
2.2. Protein Fluorescent Labeling
2.3. Turbidity Experiments
2.4. Bright Field and Fluorescence Microscopy
2.5. Fluorescence Recovery after Photobleaching
2.6. Electro Mobility Shift Assay (EMSA)
2.7. Partition Experiments
2.8. Cell Imaging
2.9. Bioinformatics Analysis of the Intrinsic Disorder Propensity
2.10. RNAs
3. Results
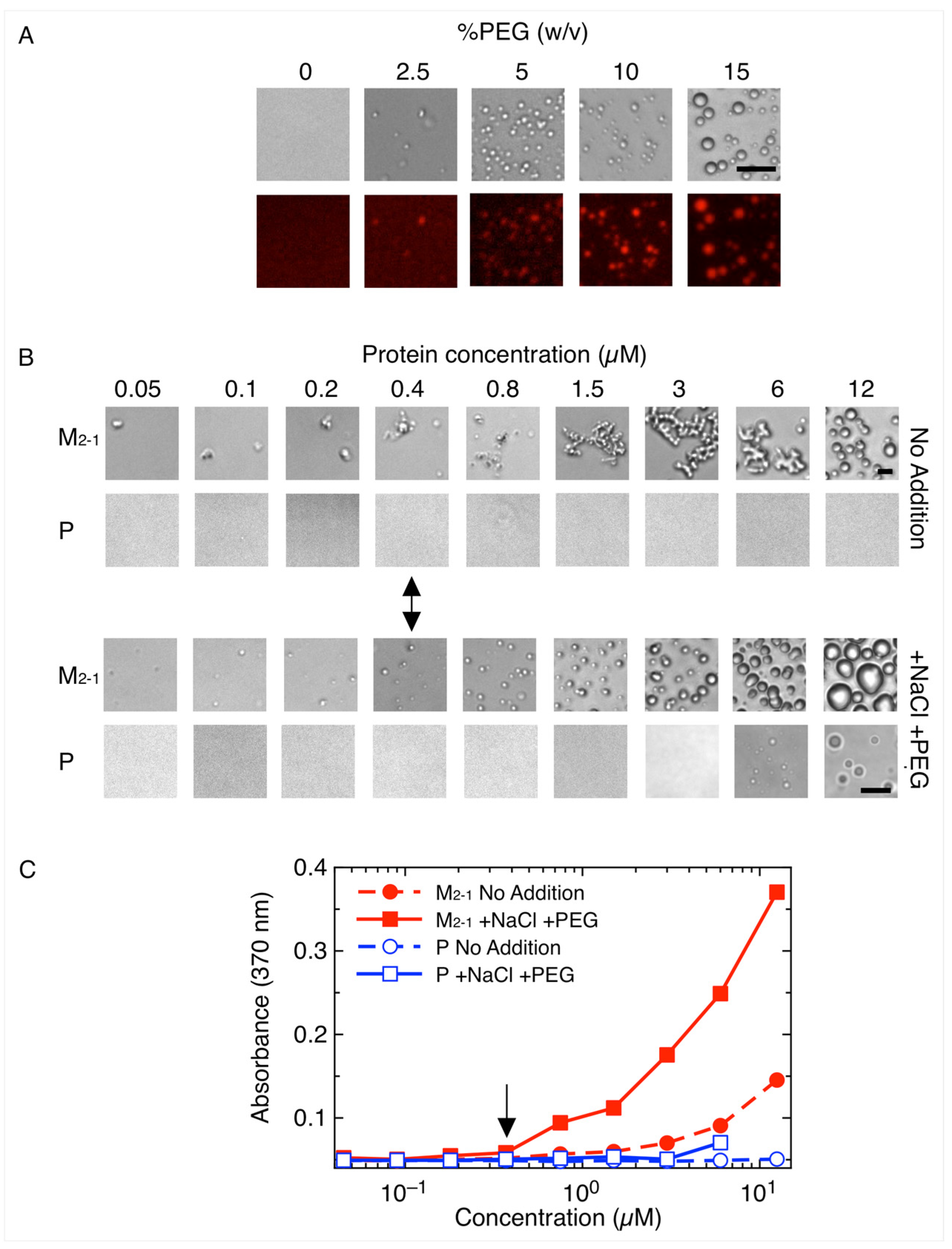
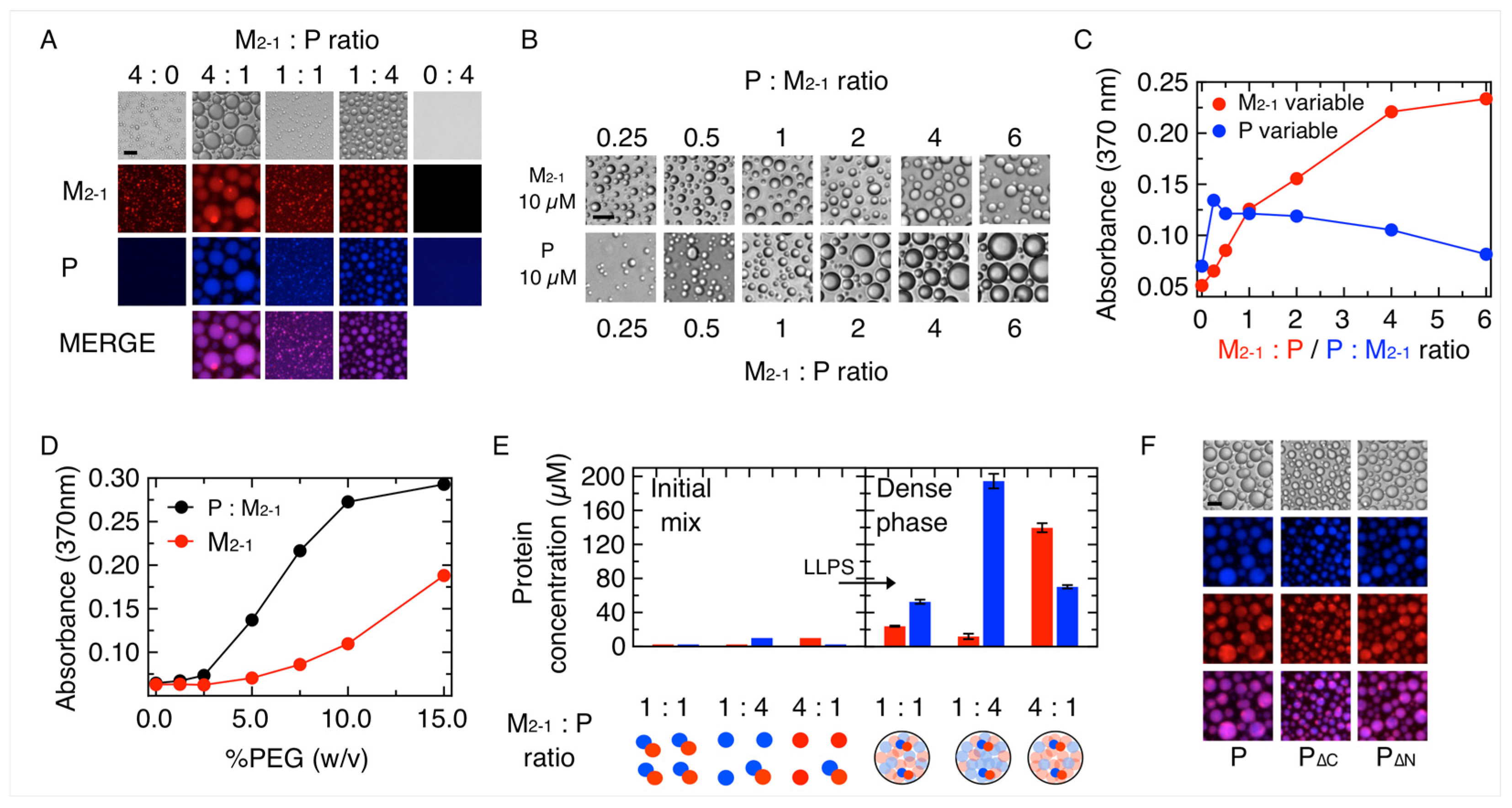
3.1. M2-1 Readily Undergoes Homotypic and Heterotypic Condensation with P
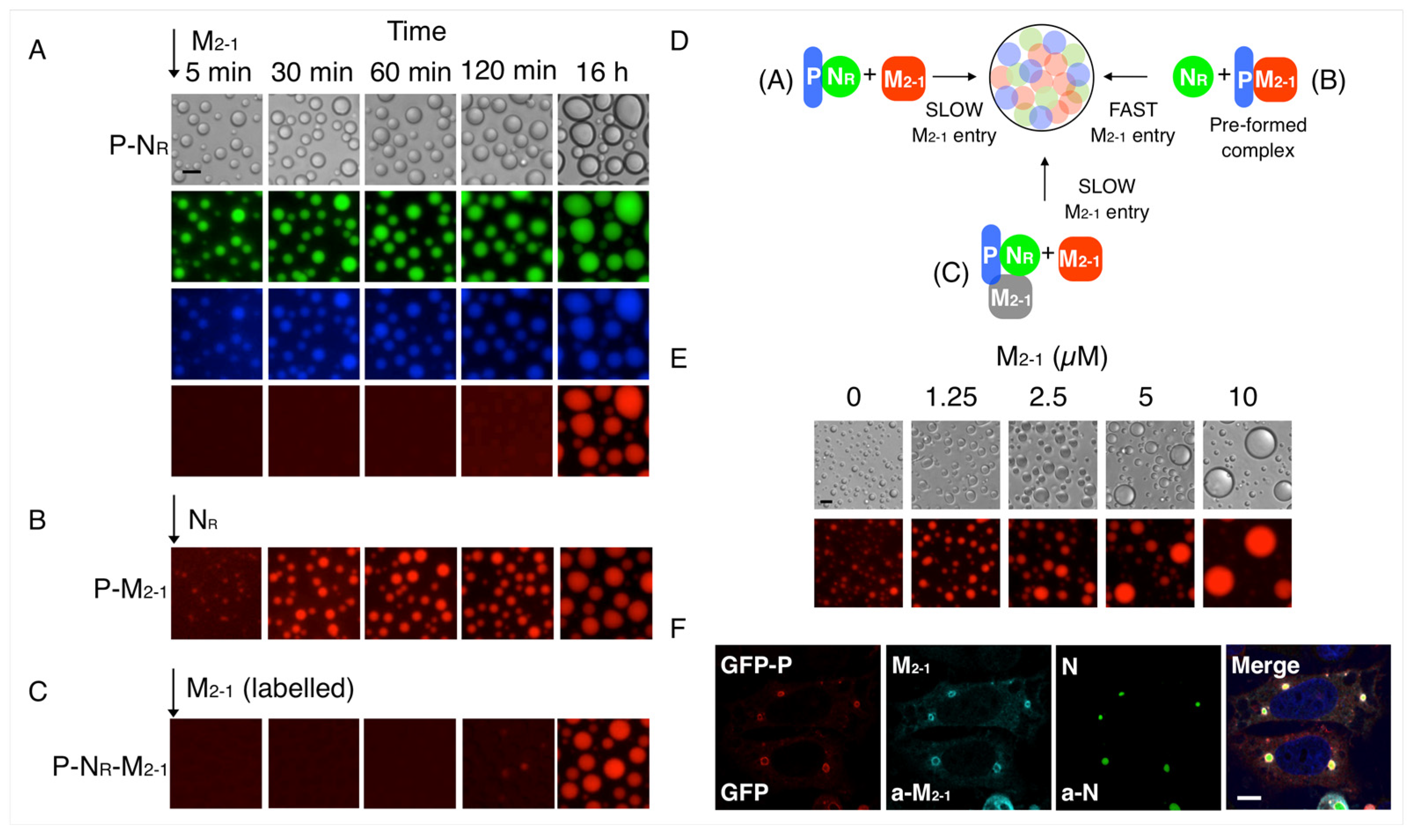
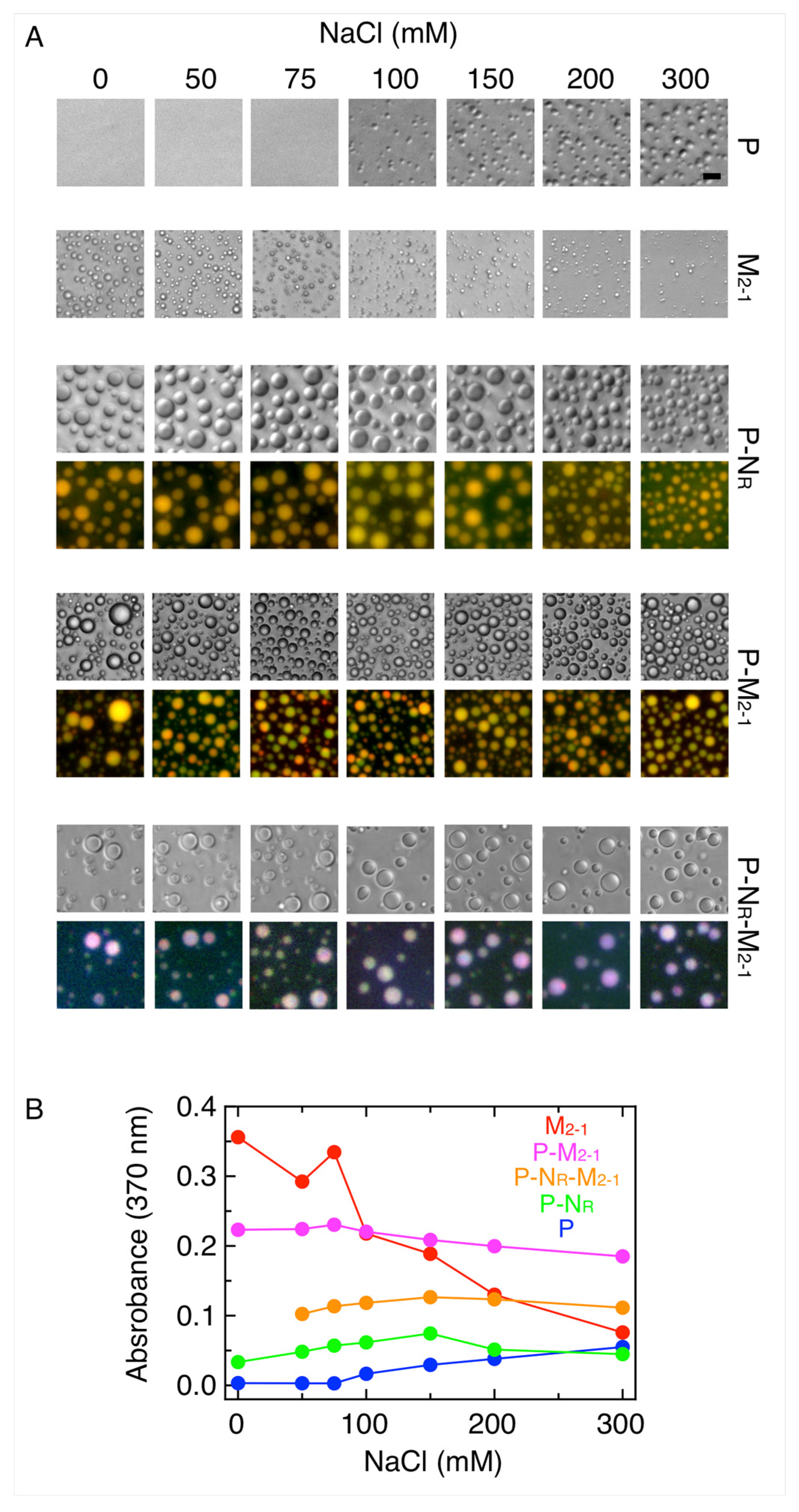
3.2. Formation and Modulation of Tripartite NR-P-M2-1 Condensates
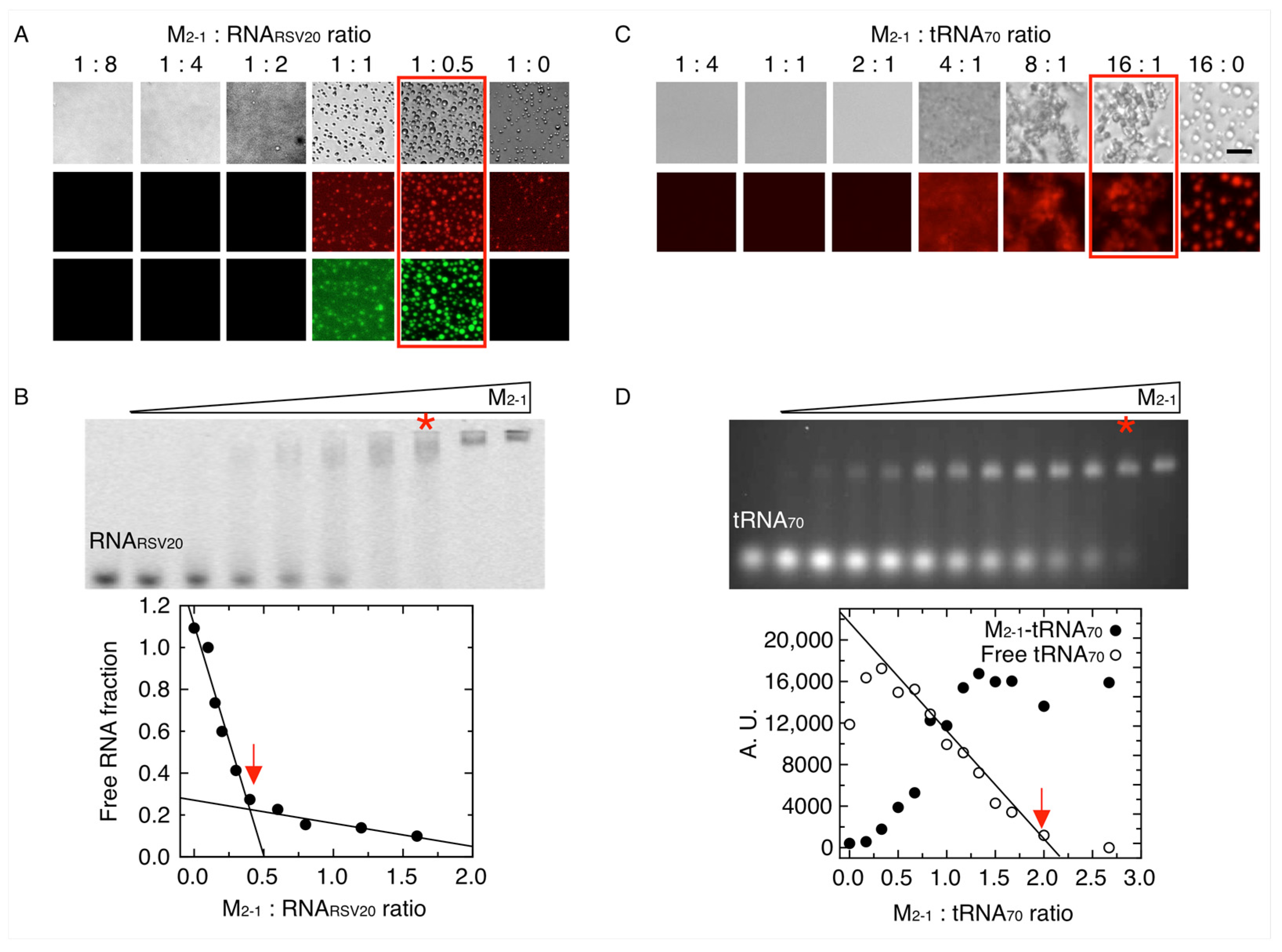
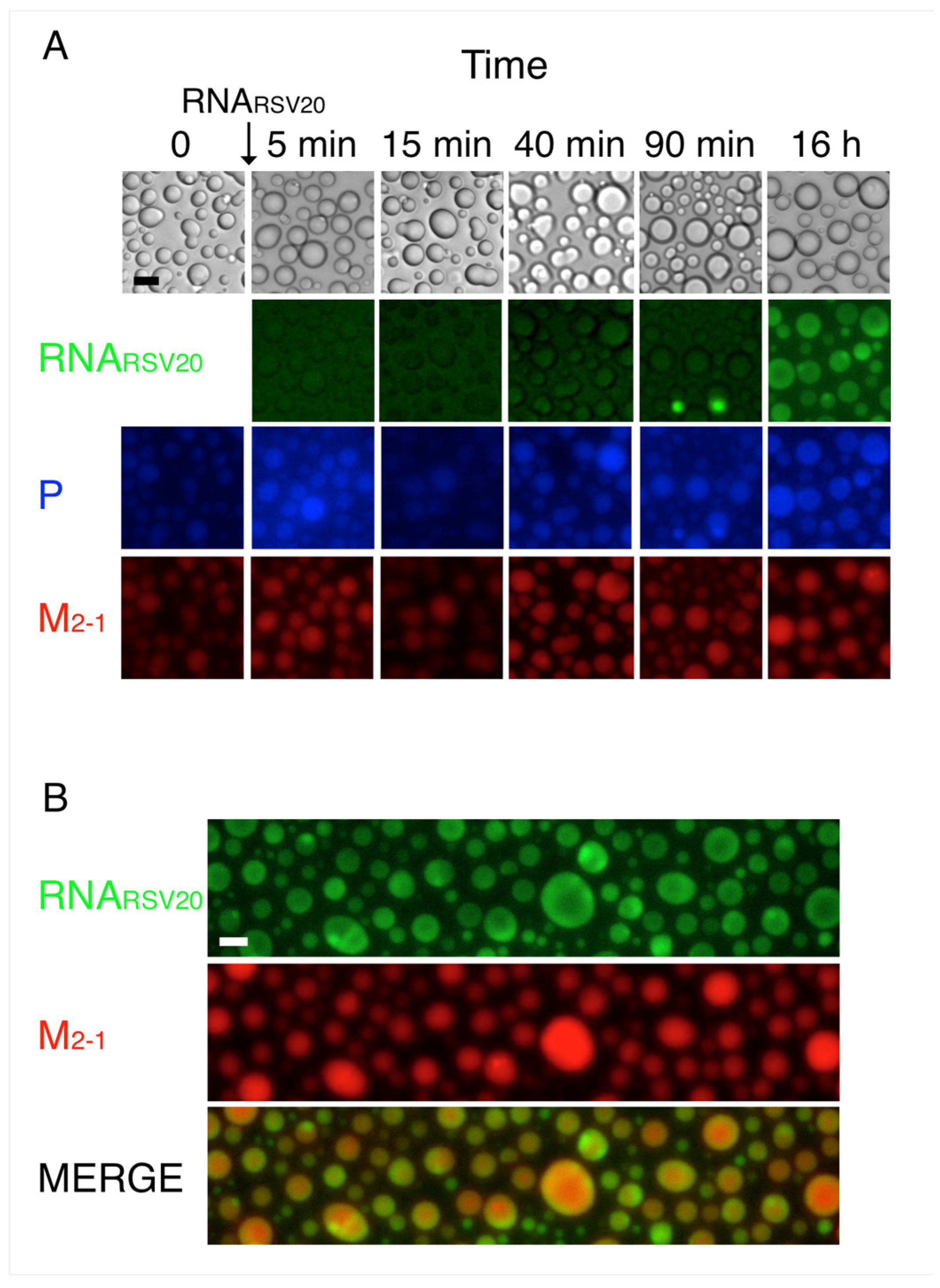
3.3. M2-1-RNA Condensates Form Subcompartments within the In Vitro Tripartite Condensates
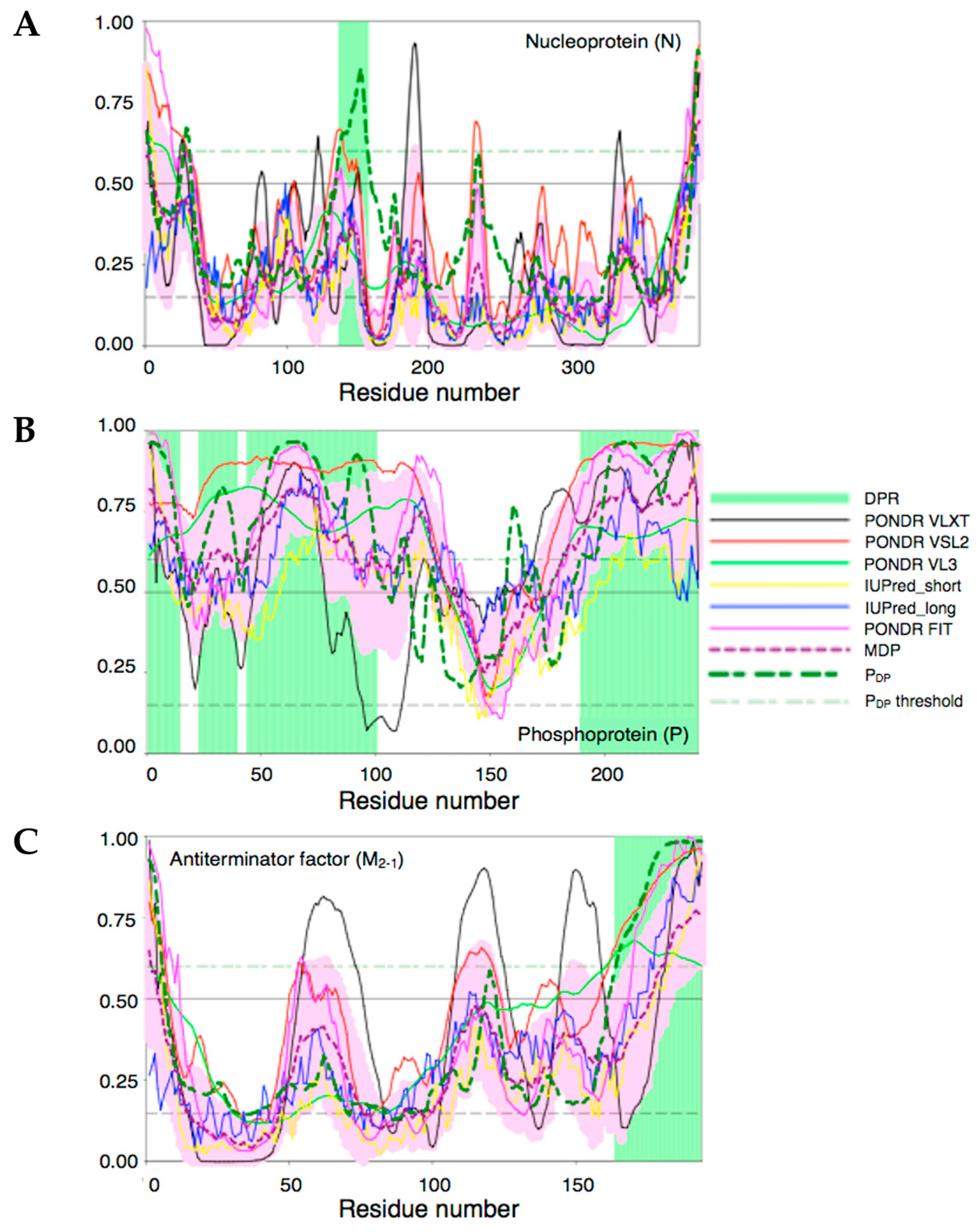
3.4. Bioinformatics Analysis of the Intrinsic Disorder Propensity of the RSV M2-1, P, and N Proteins and Their Predisposition for Phase Separation
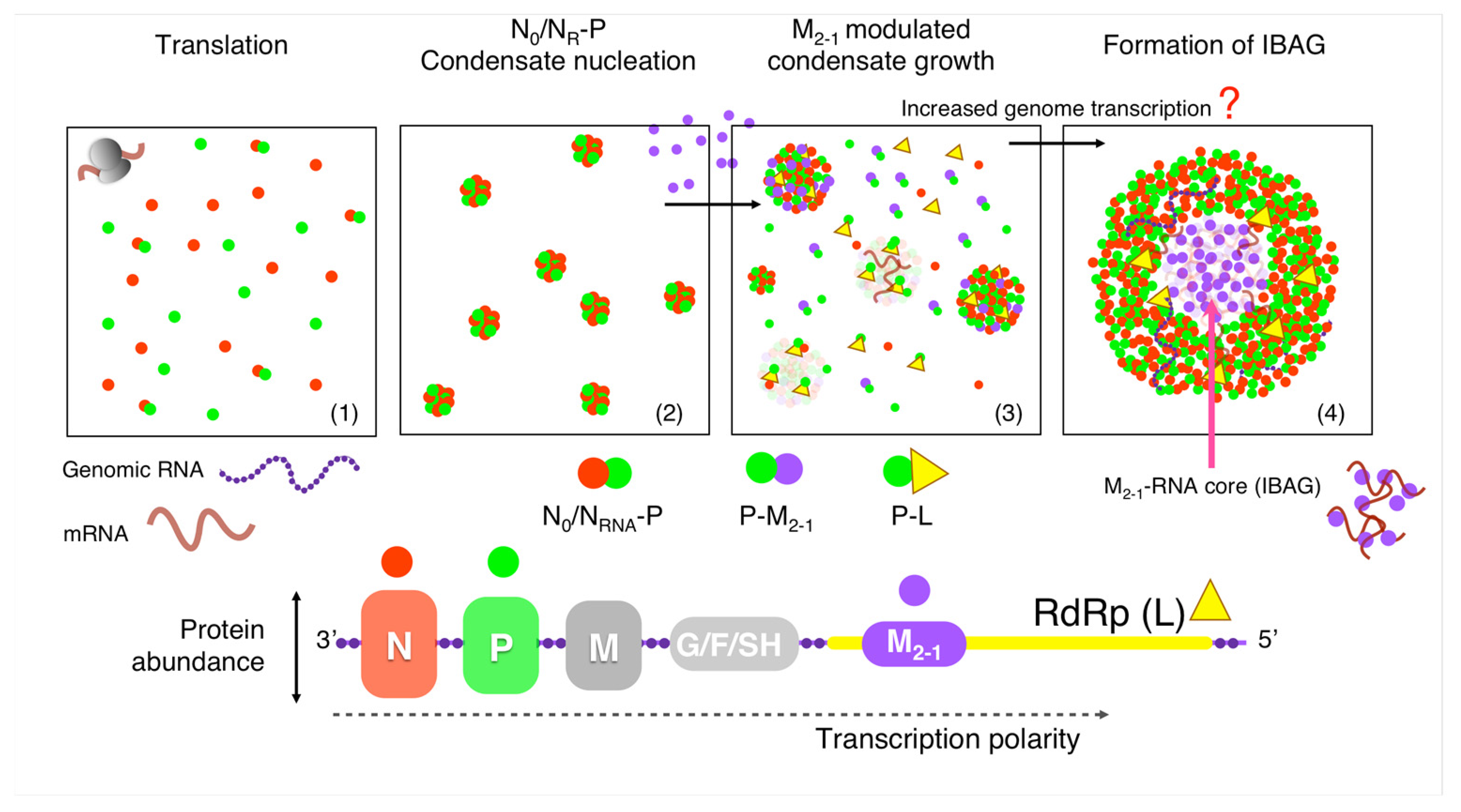
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fields, B.N. Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; ISBN 978-1-4698-7422-7. [Google Scholar]
- Fooks, A.R.; Cliquet, F.; Finke, S.; Freuling, C.; Hemachudha, T.; Mani, R.S.; Müller, T.; Nadin-Davis, S.; Picard-Meyer, E.; Wilde, H.; et al. Rabies. Nat. Rev. Dis. Primer 2017, 3, 17091. [Google Scholar] [CrossRef]
- Wu, C.; Holehouse, A.S.; Leung, D.W.; Amarasinghe, G.K.; Dutch, R.E. Liquid Phase Partitioning in Virus Replication: Observations and Opportunities. Annu. Rev. Virol. 2022, 9, 285–306. [Google Scholar] [CrossRef]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing Protein Intrinsic Disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef]
- Charman, M.; Weitzman, M.D. Replication Compartments of DNA Viruses in the Nucleus: Location, Location, Location. Viruses 2020, 12, 151. [Google Scholar] [CrossRef] [PubMed]
- Lopez, N.; Camporeale, G.; Salgueiro, M.; Borkosky, S.S.; Visentín, A.; Peralta-Martinez, R.; Loureiro, M.E.; De Prat-Gay, G. Deconstructing Virus Condensation. PLoS Pathog. 2021, 17, e1009926. [Google Scholar] [CrossRef] [PubMed]
- Borodavka, A.; Acker, J. Seeing Biomolecular Condensates Through the Lens of Viruses. Annu. Rev. Virol. 2023. [Google Scholar] [CrossRef]
- Caragliano, E.; Brune, W.; Bosse, J.B. Herpesvirus Replication Compartments: Dynamic Biomolecular Condensates? Viruses 2022, 14, 960. [Google Scholar] [CrossRef]
- Buchholz, U.J.; Biacchesi, S.; Pham, Q.N.; Tran, K.C.; Yang, L.; Luongo, C.L.; Skiadopoulos, M.H.; Murphy, B.R.; Collins, P.L. Deletion of M2 Gene Open Reading Frames 1 and 2 of Human Metapneumovirus: Effects on RNA Synthesis, Attenuation, and Immunogenicity. J. Virol. 2005, 79, 6588–6597. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.L.; Wertz, G.W. The Envelope-Associated 22K Protein of Human Respiratory Syncytial Virus: Nucleotide Sequence of the MRNA and a Related Polytranscript. J. Virol. 1985, 54, 65–71. [Google Scholar] [CrossRef]
- Tanner, S.J.; Ariza, A.; Richard, C.-A.; Kyle, H.F.; Dods, R.L.; Blondot, M.-L.; Wu, W.; Trincão, J.; Trinh, C.H.; Hiscox, J.A.; et al. Crystal Structure of the Essential Transcription Antiterminator M2-1 Protein of Human Respiratory Syncytial Virus and Implications of Its Phosphorylation. Proc. Natl. Acad. Sci. USA 2014, 111, 1580–1585. [Google Scholar] [CrossRef]
- Hardy, R.W.; Wertz, G.W. The Cys 3 -His 1 Motif of the Respiratory Syncytial Virus M2-1 Protein Is Essential for Protein Function. J. Virol. 2000, 74, 5880–5885. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.R.; Noton, S.L.; Blanchard, E.L.; Shareef, A.; Santangelo, P.J.; Johnson, W.E.; Fearns, R. Respiratory Syncytial Virus M2-1 Protein Associates Non-Specifically with Viral Messenger RNA and with Specific Cellular Messenger RNA Transcripts. PLoS Pathog. 2021, 17, e1009589. [Google Scholar] [CrossRef] [PubMed]
- Blondot, M.-L.; Dubosclard, V.; Fix, J.; Lassoued, S.; Aumont-Nicaise, M.; Bontems, F.; Eléouët, J.-F.; Sizun, C. Structure and Functional Analysis of the RNA- and Viral Phosphoprotein-Binding Domain of Respiratory Syncytial Virus M2-1 Protein. PLoS Pathog. 2012, 8, e1002734. [Google Scholar] [CrossRef]
- García-Barreno, B.; Delgado, T.; Melero, J.A. Identification of Protein Regions Involved in the Interaction of Human Respiratory Syncytial Virus Phosphoprotein and Nucleoprotein: Significance for Nucleocapsid Assembly and Formation of Cytoplasmic Inclusions. J. Virol. 1996, 70, 801–808. [Google Scholar] [CrossRef]
- Galloux, M.; Gabiane, G.; Sourimant, J.; Richard, C.-A.; England, P.; Moudjou, M.; Aumont-Nicaise, M.; Fix, J.; Rameix-Welti, M.-A.; Eléouët, J.-F. Identification and Characterization of the Binding Site of the Respiratory Syncytial Virus Phosphoprotein to RNA-Free Nucleoprotein. J. Virol. 2015, 89, 3484–3496. [Google Scholar] [CrossRef] [PubMed]
- Noval, M.G.; Esperante, S.A.; Molina, I.G.; Chemes, L.B.; Prat-Gay, G.d. Intrinsic Disorder to Order Transitions in the Scaffold Phosphoprotein P from the Respiratory Syncytial Virus RNA Polymerase Complex. Biochemistry 2016, 55, 1441–1454. [Google Scholar] [CrossRef]
- Pereira, N.; Cardone, C.; Lassoued, S.; Galloux, M.; Fix, J.; Assrir, N.; Lescop, E.; Bontems, F.; Eléouët, J.-F.; Sizun, C. New Insights into Structural Disorder in Human Respiratory Syncytial Virus Phosphoprotein and Implications for Binding of Protein Partners. J. Biol. Chem. 2017, 292, 2120–2131. [Google Scholar] [CrossRef]
- Galloux, M.; Tarus, B.; Blazevic, I.; Fix, J.; Duquerroy, S.; Eléouët, J.-F. Characterization of a Viral Phosphoprotein Binding Site on the Surface of the Respiratory Syncytial Nucleoprotein. J. Virol. 2012, 86, 8375–8387. [Google Scholar] [CrossRef]
- Gilman, M.S.A.; Liu, C.; Fung, A.; Behera, I.; Jordan, P.; Rigaux, P.; Ysebaert, N.; Tcherniuk, S.; Sourimant, J.; Eléouët, J.-F.; et al. Structure of the Respiratory Syncytial Virus Polymerase Complex. Cell 2019, 179, 193–204.e14. [Google Scholar] [CrossRef]
- Cao, D.; Gao, Y.; Roesler, C.; Rice, S.; D’Cunha, P.; Zhuang, L.; Slack, J.; Domke, M.; Antonova, A.; Romanelli, S.; et al. Cryo-EM Structure of the Respiratory Syncytial Virus RNA Polymerase. Nat. Commun. 2020, 11, 368. [Google Scholar] [CrossRef]
- Asenjo, A.; González-Armas, J.C.; Villanueva, N. Phosphorylation of Human Respiratory Syncytial Virus P Protein at Serine 54 Regulates Viral Uncoating. Virology 2008, 380, 26–33. [Google Scholar] [CrossRef]
- Alvarez Paggi, D.; Esperante, S.A.; Salgueiro, M.; Camporeale, G.; De Oliveira, G.A.P.; Prat Gay, G. A Conformational Switch Balances Viral RNA Accessibility and Protection in a Nucleocapsid Ring Model. Arch. Biochem. Biophys. 2019, 671, 77–86. [Google Scholar] [CrossRef]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef]
- Shin, Y.; Brangwynne, C.P. Liquid Phase Condensation in Cell Physiology and Disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Banjade, S.; Cheng, H.-C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase Transitions in the Assembly of Multivalent Signalling Proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, J.; Le Bars, R.; Lama, Z.; Scrima, N.; Lagaudrière-Gesbert, C.; Gaudin, Y.; Blondel, D. Negri Bodies Are Viral Factories with Properties of Liquid Organelles. Nat. Commun. 2017, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Richard, C.-A.; Rincheval, V.; Lassoued, S.; Fix, J.; Cardone, C.; Esneau, C.; Nekhai, S.; Galloux, M.; Rameix-Welti, M.-A.; Sizun, C.; et al. RSV Hijacks Cellular Protein Phosphatase 1 to Regulate M2-1 Phosphorylation and Viral Transcription. PLoS Pathog. 2018, 14, e1006920. [Google Scholar] [CrossRef] [PubMed]
- Rincheval, V.; Lelek, M.; Gault, E.; Bouillier, C.; Sitterlin, D.; Blouquit-Laye, S.; Galloux, M.; Zimmer, C.; Eleouet, J.-F.; Rameix-Welti, M.-A. Functional Organization of Cytoplasmic Inclusion Bodies in Cells Infected by Respiratory Syncytial Virus. Nat. Commun. 2017, 8, 563. [Google Scholar] [CrossRef]
- Galloux, M.; Risso-Ballester, J.; Richard, C.-A.; Fix, J.; Rameix-Welti, M.-A.; Eléouët, J.-F. Minimal Elements Required for the Formation of Respiratory Syncytial Virus Cytoplasmic Inclusion Bodies In Vivo and In Vitro. mBio 2020, 11, e01202-20. [Google Scholar] [CrossRef] [PubMed]
- Salgueiro, M.; Camporeale, G.; Visentin, A.; Aran, M.; Pellizza, L.; Esperante, S.A.; Corbat, A.; Grecco, H.; Sousa, B.; Esperón, R.; et al. Molten Globule Driven and Self-Downmodulated Phase Separation of a Viral Factory Scaffold. J. Mol. Biol. 2023, 168153. [Google Scholar] [CrossRef]
- Nevers, Q.; Scrima, N.; Glon, D.; Le Bars, R.; Decombe, A.; Garnier, N.; Ouldali, M.; Lagaudrière-Gesbert, C.; Blondel, D.; Albertini, A.; et al. Properties of Rabies Virus Phosphoprotein and Nucleoprotein Biocondensates Formed in Vitro and in Cellulo. PLoS Pathog. 2022, 18, e1011022. [Google Scholar] [CrossRef]
- Zhou, Y.; Su, J.M.; Samuel, C.E.; Ma, D. Measles Virus Forms Inclusion Bodies with Properties of Liquid Organelles. J. Virol. 2019, 93, e00948-19. [Google Scholar] [CrossRef]
- Esperante, S.A.; Noval, M.G.; Altieri, T.A.; De Oliveira, G.A.P.; Silva, J.L.; De Prat-Gay, G. Fine Modulation of the Respiratory Syncytial Virus M2–1 Protein Quaternary Structure by Reversible Zinc Removal from Its Cys3-His1 Motif. Biochemistry 2013, 52, 6779–6789. [Google Scholar] [CrossRef] [PubMed]
- Esperante, S.A.; Paris, G.; De Prat-Gay, G. Modular Unfolding and Dissociation of the Human Respiratory Syncytial Virus Phosphoprotein P and Its Interaction with the M2–1 Antiterminator: A Singular Tetramer–Tetramer Interface Arrangement. Biochemistry 2012, 51, 8100–8110. [Google Scholar] [CrossRef] [PubMed]
- Alshareedah, I.; Kaur, T.; Banerjee, P.R. Methods for Characterizing the Material Properties of Biomolecular Condensates. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 646, pp. 143–183. ISBN 978-0-12-821159-5. [Google Scholar]
- Molina, I.G.; Esperante, S.A.; Marino-Buslje, C.; Chemes, L.B.; De Prat-Gay, G. Cooperative RNA Recognition by a Viral Transcription Antiterminator. J. Mol. Biol. 2018, 430, 777–792. [Google Scholar] [CrossRef] [PubMed]
- Dayhoff, G.W.; Uversky, V.N. Rapid Prediction and Analysis of Protein Intrinsic Disorder. Protein Sci. Publ. Protein Soc. 2022, 31, e4496. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence Complexity of Disordered Protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-Dependent Prediction of Protein Intrinsic Disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing Long Intrinsic Disorder Predictors with Protein Evolutionary Information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A Meta-Predictor of Intrinsically Disordered Amino Acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Mészáros, B.; Erdos, G.; Dosztányi, Z. IUPred2A: Context-Dependent Prediction of Protein Disorder as a Function of Redox State and Protein Binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef] [PubMed]
- Hardenberg, M.; Horvath, A.; Ambrus, V.; Fuxreiter, M.; Vendruscolo, M. Widespread Occurrence of the Droplet State of Proteins in the Human Proteome. Proc. Natl. Acad. Sci. USA 2020, 117, 33254–33262. [Google Scholar] [CrossRef] [PubMed]
- Vendruscolo, M.; Fuxreiter, M. Sequence Determinants of the Aggregation of Proteins Within Condensates Generated by Liquid-Liquid Phase Separation. J. Mol. Biol. 2022, 434, 167201. [Google Scholar] [CrossRef]
- Tran, T.-L.; Castagné, N.; Bhella, D.; Varela, P.F.; Bernard, J.; Chilmonczyk, S.; Berkenkamp, S.; Benhamo, V.; Grznarova, K.; Grosclaude, J.; et al. The Nine C-Terminal Amino Acids of the Respiratory Syncytial Virus Protein P Are Necessary and Sufficient for Binding to Ribonucleoprotein Complexes in Which Six Ribonucleotides Are Contacted per N Protein Protomer. J. Gen. Virol. 2007, 88, 196–206. [Google Scholar] [CrossRef]
- Selvaraj, M.; Yegambaram, K.; Todd, E.J.A.A.; Richard, C.-A.; Dods, R.L.; Pangratiou, G.M.; Trinh, C.H.; Moul, S.L.; Murphy, J.C.; Mankouri, J.; et al. The Structure of the Human Respiratory Syncytial Virus M2-1 Protein Bound to the Interaction Domain of the Phosphoprotein P Defines the Orientation of the Complex. mBio 2018, 9, e01554-18. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Protter, D.S.W.; Rosen, M.K.; Parker, R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60, 208–219. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Hottle, G.A.; Morgan, C.; Peers, J.H.; Wyckoff, R.W.G. The Election Microscopy of Rabies Inclusion (Negri) Bodies. Exp. Biol. Med. 1951, 77, 721–723. [Google Scholar] [CrossRef]
- Matsumoto, S. Electron Microscopy of Nerve Cells Infected with Street Rabies Virus. Virology 1962, 17, 198–202. [Google Scholar] [CrossRef]
- Lahaye, X.; Vidy, A.; Pomier, C.; Obiang, L.; Harper, F.; Gaudin, Y.; Blondel, D. Functional Characterization of Negri Bodies (NBs) in Rabies Virus-Infected Cells: Evidence That NBs Are Sites of Viral Transcription and Replication. J. Virol. 2009, 83, 7948–7958. [Google Scholar] [CrossRef] [PubMed]
- Boggs, K.B.; Edmonds, K.; Cifuentes-Munoz, N.; El Najjar, F.; Ossandón, C.; Roe, M.; Wu, C.; Moncman, C.L.; Creamer, T.P.; Amarasinghe, G.K.; et al. Human Metapneumovirus Phosphoprotein Independently Drives Phase Separation and Recruits Nucleoprotein to Liquid-Like Bodies. mBio 2022, 13, e01099-22. [Google Scholar] [CrossRef] [PubMed]
- Neitzel, A.E.; Fang, Y.N.; Yu, B.; Rumyantsev, A.M.; De Pablo, J.J.; Tirrell, M.V. Polyelectrolyte Complex Coacervation across a Broad Range of Charge Densities. Macromolecules 2021, 54, 6878–6890. [Google Scholar] [CrossRef]
- Portz, B.; Shorter, J. Biochemical Timekeeping Via Reentrant Phase Transitions. J. Mol. Biol. 2021, 433, 166794. [Google Scholar] [CrossRef] [PubMed]
- Bailly, B.; Richard, C.-A.; Sharma, G.; Wang, L.; Johansen, L.; Cao, J.; Pendharkar, V.; Sharma, D.-C.; Galloux, M.; Wang, Y.; et al. Targeting Human Respiratory Syncytial Virus Transcription Anti-Termination Factor M2-1 to Inhibit in Vivo Viral Replication. Sci. Rep. 2016, 6, 25806. [Google Scholar] [CrossRef] [PubMed]
- Samal, S.K. The Biology of Paramyxoviruses; Caister Academic Press: Norfolk, UK, 2011; ISBN 978-1-904455-85-1. [Google Scholar]
- Peeples, W.; Rosen, M.K. Mechanistic Dissection of Increased Enzymatic Rate in a Phase-Separated Compartment. Nat. Chem. Biol. 2021, 17, 693–702. [Google Scholar] [CrossRef]
- Risso-Ballester, J.; Galloux, M.; Cao, J.; Le Goffic, R.; Hontonnou, F.; Jobart-Malfait, A.; Desquesnes, A.; Sake, S.M.; Haid, S.; Du, M.; et al. A Condensate-Hardening Drug Blocks RSV Replication in vivo. Nature 2021, 595, 596–599. [Google Scholar] [CrossRef]

| Stoichiometry | Fold Concentration 1 | ||
|---|---|---|---|
| Initial | Dense Phase | ||
| P:M2-1 | 04:01 | 16:01 | 19.5:4.8 |
| P:M2-1 | 01:01 | 01:00.5 | 21.2:9.6 |
| P:M2-1 | 01:04 | 01:02 | 28:14:00 |
| P:NR | 2.5:1 | 01:01 | 19.7:40.7 |
| P:NR:M2-1 | 2.5:1:2.5 | 01:00.5 | 24.2:46.7:12.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Visentin, A.; Demitroff, N.; Salgueiro, M.; Borkosky, S.S.; Uversky, V.N.; Camporeale, G.; de Prat-Gay, G. Assembly of the Tripartite and RNA Condensates of the Respiratory Syncytial Virus Factory Proteins In Vitro: Role of the Transcription Antiterminator M2-1. Viruses 2023, 15, 1329. https://doi.org/10.3390/v15061329
Visentin A, Demitroff N, Salgueiro M, Borkosky SS, Uversky VN, Camporeale G, de Prat-Gay G. Assembly of the Tripartite and RNA Condensates of the Respiratory Syncytial Virus Factory Proteins In Vitro: Role of the Transcription Antiterminator M2-1. Viruses. 2023; 15(6):1329. https://doi.org/10.3390/v15061329
Chicago/Turabian StyleVisentin, Araceli, Nicolás Demitroff, Mariano Salgueiro, Silvia Susana Borkosky, Vladimir N. Uversky, Gabriela Camporeale, and Gonzalo de Prat-Gay. 2023. "Assembly of the Tripartite and RNA Condensates of the Respiratory Syncytial Virus Factory Proteins In Vitro: Role of the Transcription Antiterminator M2-1" Viruses 15, no. 6: 1329. https://doi.org/10.3390/v15061329
APA StyleVisentin, A., Demitroff, N., Salgueiro, M., Borkosky, S. S., Uversky, V. N., Camporeale, G., & de Prat-Gay, G. (2023). Assembly of the Tripartite and RNA Condensates of the Respiratory Syncytial Virus Factory Proteins In Vitro: Role of the Transcription Antiterminator M2-1. Viruses, 15(6), 1329. https://doi.org/10.3390/v15061329






