Abstract
In 2014, enterovirus D68 (EV-D68), previously associated primarily with mild respiratory illness, caused a large outbreak of severe respiratory illness and, in rare instances, paralysis. We compared the viral binding and replication of eight recent EV-D68 clinical isolates collected both before and during the 2014 outbreak and the prototype Fermon strain from 1962 in cultured HeLa cells and differentiated human primary bronchial epithelial cells (BEC) to understand the possible reasons for the change in virus pathogenicity. We selected pairs of closely related isolates from the same phylogenetic clade that were associated with severe vs. asymptomatic infections. We found no significant differences in binding or replication in HeLa cell cultures between the recent clinical isolates. However, in HeLa cells, Fermon had significantly greater binding (2–3 logs) and virus progeny yields (2–4 logs) but a similar level of replication (1.5–2 log increase in viral RNA from 2 h to 24 h post infection) compared to recent isolates. In differentiated BECs, Fermon and the recent EV-D68 isolates had similar levels of binding; however, the recent isolates produced 1.5–2-log higher virus progeny yields than Fermon due to increased replication. Interestingly, no significant differences in replication were identified between the pairs of genetically close recent EV-D68 clinical isolates despite the observed differences in associated disease severity. We then utilized RNA-seq to define the transcriptional responses in BECs infected with four recent EV-D68 isolates, representing major phylogenetic clades, and the Fermon strain. All the tested clinical isolates induced similar responses in BECs; however, numerous upregulated genes in antiviral and pro-inflammatory response pathways were identified when comparing the response to clinical isolates versus Fermon. These results indicate that the recent emergence in severe EV-D68 cases could be explained by an increased replication efficiency and enhanced inflammatory response induced by newly emerged clinical isolates; however, host factors are likely the main determinants of illness severity.
1. Introduction
EV-D68, a member of the genus Enterovirus, family Picornaviridae, was first identified in 1962 in a child with pneumonia-like symptoms [1]. EV-D68 infections can cause respiratory illnesses ranging from asymptomatic to upper respiratory symptoms to more severe illnesses including bronchitis, wheezing, and/or shortness of breath. In rare instances, EV-D68 infection has also been associated with polio-like neurological symptoms [2]. Its severity has been further demonstrated in multiple epidemiological studies [3,4] with higher rates detected in a hospital compared to outpatient settings. In 2014, reports of an upper respiratory tract infection that was associated with severe outcomes including hospitalization for respiratory distress and paralysis emerged in the United States. The 2014 outbreak is the largest and most widespread outbreak recognized to date, resulting in significant morbidity and mortality [5], which created a need to understand the mechanisms underlying enhanced EV-D68 pathogenicity.
What makes EV-D68 unique from other enteroviruses are some biologic characteristics in common with rhinoviruses (RV), such as its optimal growth at 33 °C and acid lability. In fact, this similarity of EV-D68 with RV led to it being incorrectly classified as an RV in the past [6,7]. EV-D68 can infect the upper respiratory tract epithelium potentially through multiple different receptors [8,9,10]. However, the physiological relevance of some of these receptors has been questioned [10,11].
While EV-D68 was identified 50 years ago, it has rarely been associated with severe illnesses, with only a total of 26 reported cases from 1970 to 2005, with 75% of them occurring in children [12]. In the late 2000s, the prevalence of EV-D68 infections began to increase [3,4]. In recent years, EV-D68 isolates have been grouped into three major phylogenetic clades designated A, B, and C [13]. Multiple clades have circulated with each outbreak of EV-D68 illnesses, including a novel sub-clade (B1) that emerged in the 2014 outbreak. This trend of EV-D68 evolution has continued in 2019–2020, where multiple clades co-circulated and a novel B3 clade emerged [14]. This has led to speculation that the increased prevalence of EV-D68 relates to mutations that altered its antigenicity [4]. This change in antigenicity could sidestep antiviral immunity, leading to increased EV-D68 infection rates.
The innate immune response is critical to controlling EV-D68 and other respiratory viral infections. An impaired innate immune response can increase the risk of severe illness. Both epithelial and mononuclear cells contribute to innate antiviral responses, which include the secretion of interferons (IFN) and pro-inflammatory cytokines, such as IL-6 and TNF. EV-D68 can inhibit the antiviral response by preventing type I interferon activation in HeLa cells [15], but there is less evidence of IFN inhibition in primary epithelial cells.
We hypothesized that recently emerged isolates of EV-D68 might be more virulent because of enhanced binding and/or replication within epithelial cells or induced unique patterns of gene expression. To test this hypothesis, we compared the properties of eight recent EV-D68 isolates to those of the Fermon strain, which was isolated in 1962, in differentiated cultures of human primary bronchial epithelial cells (BEC).
2. Materials and Methods
2.1. Virus Isolate Selection
The EV-D68 clinical isolates selected for this study were identified by RT-PCR and partial sequencing in nasal samples from children enrolled in the Childhood Origins of Asthma (COAST) study, the Urban Environment and Childhood Asthma (URECA) study, and the Preventative Omalizumab or Step-up Therapy for Fall Exacerbations (PROSE) study [16,17,18]. The EV-D68 clinical isolates were then fully sequenced and grouped into 3 major clades by phylogenetic analysis of the full-length genomic sequences (Figure 1). Associated symptom data on illness severity were used to select viruses based on the severity of symptoms. We classified EV-D68 strains into those that caused a severe wheezing episode or asthma exacerbation or asymptomatic infections. We selected pairs of closely related viruses from the same clade that were associated with severe vs. asymptomatic infections. Based upon these criteria, we selected 8 different EV-D68 isolates that belonged to 3 different viral clades: A, B, and C (Figure 1). Within clade B, we included two isolates (C7787 and C7788) from the B1 subclade, which was responsible for many severe EV-D68 cases in the 2014 outbreak [19].
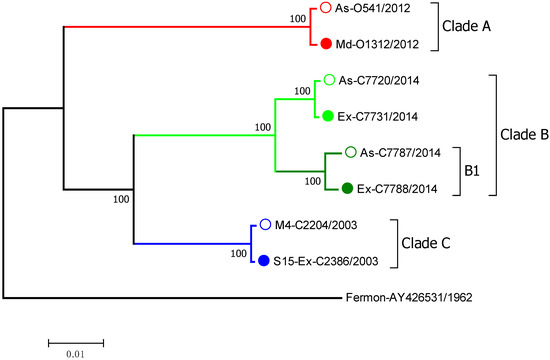
Figure 1.
The phylogenetic tree of EV-D68 clinical isolates selected for cloning. Full-length genome sequences were analyzed. The numbers next to each tree branch represent bootstrap values showing the percentage of replicate trees that the viral strain clustered together. The branch lengths represent the evolutionary distance in units of base pair differences per viral sequence, with the scale bar denoting difference. Severity of symptoms designations: As—asymptomatic; Ex—exacerbation; M—mild; S—severe; numbers following the designations indicate symptom score where available. Filled and empty circles of the same color show symptomatic and asymptomatic infections, respectively.
2.2. Construction of the Full-Length EV-D68 cDNA Infectious Clones
We generated cDNA infectious clones of 8 EV-D68 clinical isolates using the published sequences (Table 1). To construct the full-length cDNA copy of the viral genome, we used 6 overlapping cDNA fragments (gBlocks) for each isolate made by Integrated DNA Technologies (Coralville, IA, USA). The viral genome of each isolate was preceded by the hammerhead ribozyme sequence [20] and followed by the poly(A) tail. We cloned complete viral genomes in two steps using a vector backbone from pR16.11 plasmid [21]. First, we assembled (Gibson Assembly Master Mix, NEB) two large cDNAs corresponding to 5′ and 3′ halves of the viral genome from 3 overlapping gBlocks each. We then linked these two cDNAs by restriction digestion (Table 1) followed by ligation (T4 DNA ligase, NEB) to create a full-length cDNA copy of the viral genome. We also used two Fermon strains, one that was provided to us by Dr. Frank van Kuppeveld (Utrecht University), which we referred to as “Fermon”, and another one that was made using the reverse genetics methods described above and designated as “Lab Fermon”. We sequenced the complete genome of the “Fermon” strain and found three non-synonymous mutations in non-structural proteins, R1179Q and T1410S in 2C, and I2009V in 3D, compared to “Lab Fermon” that was based on published sequence AY426531. For all the experiments comparing Fermon to clinical isolates, we used the recombinant virus produced from a “Fermon” infectious clone.

Table 1.
Sequence accession numbers and restriction enzymes used for cDNA cloning.
2.3. Recombinant Virus Production in WisL Cells
RNA transcripts were synthesized from the linearized plasmids (Table 1) with a RiboMax large-scale RNA production system T7 (Promega) for 3 h. The reaction products were then treated with RQ1 DNase I (Promega) and analyzed by agarose gel electrophoresis. We used 50 μL of RNA directly from this reaction with 50 μL of Lipofectamine 2000 (Invitrogen) for transfection of two T-75 flasks of WisL cells (human embryonic lung fibroblasts). We diluted 50 μL of Lipofectamine in 4 mL of Opti-MEM (Invitrogen) and incubated it for 5 min. Next, we added 50 μL of viral RNA to the diluted liposomes and then gently mixed and incubated it for 20 min at room temperature. Then, two T-75 flasks of WisL cells were washed with PBS, and 2 mL Opti-MEM was added to the cells. Finally, 2 mL of the RNA–Lipofectamine mixture was added and incubated for 2 h at 34 °C. After 2 h, the transfection medium was replaced with complete cell culture medium (10 mL per flask), and the cells were incubated at 34 °C for 24 h. After 24 h, the transfected cells were collected and stored at −80 °C until purification.
2.4. Virus Purification
Transfected WisL cells were frozen and thawed 3 times and scraped. The cell lysate was centrifuged for 10 min at 10,000× g and 4 °C to remove cell debris. The clarified lysate was then treated with RNase A (10 μg/mL; Qiagen) for 10 min at 37 °C to remove free RNA. Next, 1 mL of 10% N-lauroylsarcosine and 20 µL β-mercaptoethanol were added per 10 mL of clarified lysate, and ~11 mL of this mixture was layered over 1 mL of 30% (w/v) sucrose (in PBS) in Ultra-Clear (14 × 89 mm, Beckman) centrifuge tubes. The samples were then centrifuged at 24,200 rpm (SW41 rotor, Beckman L-60) and 10 °C for 4 h.
After 4 h, the supernatant and sucrose were aspirated, 100 µL of cold PBS with 0.01% BSA was added, and the tube was incubated overnight at 4 °C. The virus pellet was then resuspended by pipetting multiple times. Virus suspension was collected and centrifuged at 10,000× g for 3 min to remove any remaining debris. Total RNA was extracted from 2 μL of purified viral prep with an RNeasy Mini Kit (Qiagen) as per the manufacturer’s instructions. Next, EV-D68 RNA concentrations were determined by RT-qPCR with a standard curve from 102–107 plaque-forming unit equivalents (PFUe).
2.5. Reverse Transcription
A TaqMan Reverse Transcription Reagents kit (Life Technologies) was used for reverse transcription (RT) of viral RNA with virus-specific reverse primer R848 (5′-AAACACGGACACCCAAAGTAGT-3′). The RT master mix was made as per the manufacturer’s recommendation. A total of 3.85 µL of RNA with 6.15 µL of reaction mixture was used, and reverse transcription was performed with a thermal profile of 25 °C for 10 min, 48 °C for 30 min, 95 °C for 8 min, and 4 °C. After this, 40 µL of RNase-free water was added, and the viral RNA concentration was assessed by qPCR.
2.6. Quantitative PCR
Quantitative (q) PCR was performed using a Power SYBR green PCR master mix (Applied Biosystems) and 7300 real-time PCR system (Applied Biosystems). The forward (5′-CCTCCGGCCCCTGAAT-3′) and reverse (5′-AAACACGGACACCCAAAGTAGT-3′) primers were used in a 25 µL reaction in duplicate. The thermal profile used was 50 °C for 3 min, 95 °C for 5 min followed by 40 cycles at 95 °C for 15 s and at 60 °C for 30 s. The levels of EV-D68 RNA were determined by a standard curve from serial 10-fold dilutions of virus with a known titer (102–107 PFUe).
2.7. Differentiated BEC Cultures Grown at an Air–Liquid Interface
BECs were obtained from de-identified donors. Frozen stocks of BECs were thawed and washed with PneumaCult-Ex medium (Stemcell Technologies). Then, cells were seeded in T-75 CellBind flasks (Costar; Corning Inc., Corning, NY, USA) and cultured in PneumaCult-Ex medium. At 90% confluence, the BECs were washed with PBS and treated with 0.25% Trypsin-EDTA to dislodge cells. The cells were counted and seeded (105 cells per well) in Transwell inserts (0.4 µM pore size, Costar) coated in 5% human placental collagen type VI solution and placed in 24-well plates. The cells were grown in a PneumaCult-Ex medium with antimicrobials (50 mg/mL gentamicin and 2 mg fluconazole) overnight or until confluence at 37 °C. Then, the PneumaCult-Ex medium was replaced with PneumaCult-ALI maintenance medium in the basal chamber. The BEC cultures were grown at an air–liquid interface (ALI) at 37 °C until fully differentiated (approximately 4 weeks).
2.8. Infection of BEC-ALI Cultures
Prior to viral infection, the complete growth medium was removed from the BEC-ALI cultures and replaced with a PneumaCult-ALI maintenance medium without hydrocortisone for 24 h. After 24 h, the BECs were infected with 105 plaque-forming unit equivalents (PFUe) per well of purified EV-D68 (two wells per each isolate), which was diluted in 50 µL of PneumaCult-ALI maintenance medium without hydrocortisone. The cultures were gently shaken for 15 min at room temperature and then incubated at 34 °C for 2 h. After 2 h, the cells were washed apically 3 times with PBS, and then 350 μL of the RLT lysis buffer was added to one well (to measure virus binding at 2 h p.i.) and 500 µL of ALI growth medium was added basally to the second well. At 24 h p.i., 350 μL of the RLT lysis buffer was added to cells, and then total RNA was extracted with an RNeasy mini kit (Qiagen) as previously described [22] to measure the virus progeny yield and replication.
2.9. HeLa Cell Infections
HeLa cells were grown in T-75 flasks until confluence. The cells were then trypsinized (3 min at 37 °C), spun (500× g, 5 min), resuspended in Medium A (EMEM (Lonza 12-611F) supplemented with non-essential amino acids (Gibco 11140), penicillin/streptomycin (Gibco 15140), and 10% fetal bovine serum (Gemini 100-500)) and plated in 12-well plates at a density of 5 × 105 cells per well. After overnight incubation, the cells were infected with different isolates of EV-D68 (2 wells each) at 2.5 × 106 PFUe per well. The viruses were allowed to bind for 2 h and then washed 3 times with PBS. After the wash step, 350 µL of the RLT buffer was added to one well of cells (to measure virus binding at 2 h), and fresh Medium A was added to the second well (to measure virus progeny yield and replication at 24 h). After 24 h, the growth medium was removed, and 350 µL of the RLT buffer was added to the cells. Total RNA was extracted with an RNeasy mini kit (Qiagen) as per the manufacturer’s instructions.
2.10. RNA-Sequencing Library Construction and Sequencing of Directional Libraries
BEC cultures from 5 donors were differentiated at ALI. We selected one isolate from each EV-D68 phylogenetic clade: O541a (clade A), C7731 (clade B), C7788 (clade B1), C2386 (clade C), and the Fermon strain. The total RNA samples from the BEC cultures collected 24 h p.i. were submitted to the University of Wisconsin-Madison Biotechnology Center and verified for purity and integrity via a NanoDrop One Spectrophotometer and Agilent 2100 BioAnalyzer, respectively. Then, using Illumina® TruSeq® Stranded mRNA Sample Preparation kits (Illumina Inc., San Diego, CA, USA), each library was prepared and standardized according to Illumina Inc. protocol. For each library preparation, mRNA was purified from 200 ng of total RNA using poly-T oligo-attached magnetic beads. Subsequently, each poly-A-enriched sample was fragmented using divalent cations under elevated temperature. The RNA was synthesized into double-stranded cDNA using SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and random primers for first strand cDNA synthesis followed by second strand synthesis. Double-stranded cDNA was purified by paramagnetic beads (Agencourt AMPure XP beads, Beckman Coulter). The cDNA products were incubated with Klenow DNA Polymerase to add an ‘A’ base (Adenine) to the 3′ end of the blunt DNA fragments. DNA fragments were ligated to Illumina adapters. The adapter-ligated DNA products were purified by paramagnetic beads. Adapter-ligated DNA was amplified in a linker-mediated PCR reaction (LM-PCR) for 11 cycles using a PhusionTM DNA Polymerase and Illumina’s PE genomic DNA primer set and then purified by paramagnetic beads. Quality and quantity of the finished libraries were assessed using an Agilent HS DNA or DNA1000 chip (Agilent Technologies, Inc., Santa Clara, CA, USA) and a Qubit® dsDNA HS Assay Kit (Invitrogen, Carlsbad, CA, USA), respectively. Libraries were standardized to 2 nM. Cluster generation was performed using standard Cluster Kits (v4) and the Illumina cBot. Single-end 100 bp sequencing was performed using standard SBS chemistry (v4) on an Illumina HiSeq2500 sequencer. Images were analyzed using the standard Illumina Pipeline, version 1.8.2.
2.11. Statistical Analysis
Viral quantities were compared among the EV-D68 strains using linear mixed-effect models, with fixed-effect terms for strain and random-effect terms for BEC donor. The Tukey adjustment was used to control the family-wise error rate for multiple comparisons between strains. Viral quantities were log-transformed for analysis. Statistical analysis was conducted using R 3.3.1 [23].
RNA sequencing data were analyzed in R using Bioconductor tools. Sequencing reads were aligned to the UCSC hg19 Homo sapiens genome, and gene counts were assigned using the Rsubread 1.22.3 package [24]. Gene annotation was performed using the org.Hs.eg.db 3.3.0 [25] and annotate 1.50.1 packages. Genes were filtered to include genes with >1 reads per million mapped reads in at least 2 libraries. The edgeR 3.14.0 package [26] was used to scale raw library sizes, estimate gene-wise dispersion using the empirical likelihood Bayes method, and fit quasi-likelihood negative binomial models with covariates for the BEC donor and virus strain to assess differential gene expression. Linear contrasts were used to compare gene expression patterns between the Fermon strain and the average expression of the other 4 EV-D68 strains. The false discovery rate was used to account for multiple testing.
3. Results
3.1. EV-D68 Binding and Replication in HeLa Cells
We first assessed binding and replication of recent EV-D68 isolates and Fermon using HeLa cells. This cell line was selected for its susceptibility to EV-D68 infection and genetic uniformity, allowing a comparison of viral binding and replication without the additional genetic variation observed in the primary cells from different donors. The results showed only minor differences among the recent clinical isolates of EV-D68 in either the binding or progeny yields (Figure 2A). In contrast, the Fermon strain demonstrated consistently greater binding (2–3 logs) and progeny yields (2–4 logs) at 2 and 24 h post infection (p.i.), respectively, when compared to the recent EV-D68 isolates. The ratio of 24 h over 2 h p.i. showed that the greater amount of virus progeny produced by Fermon was due to increased binding rather than enhanced viral RNA replication (Figure 2B).
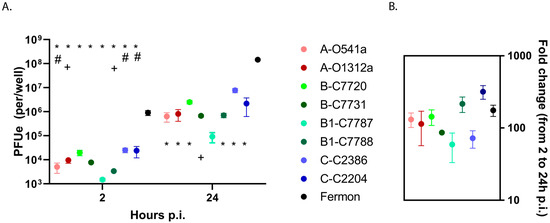
Figure 2.
Viral binding and replication in HeLa cells. (A) Cell-associated input virus (2 h p.i.) and progeny yields (24 h p.i.) were determined by RT-qPCR and (B) viral replication was calculated and compared for each isolate. The first letters in isolate designations represent the phylogenetic clade, and the following letters and numbers indicate the clinical isolate ID. The * represents a difference (p < 0.05) in the clinical isolates vs. Fermon, + represents a difference (p < 0.05) in isolate 7787 vs. other isolates, and # represents a difference (p < 0.05) in isolates C2386, C2204, and C7720 vs. other isolates. Data are means and SEM from five independent experiments. The viruses are color-coded according to clade, with darker color indicating severe illness and lighter color indicating asymptomatic infection.
3.2. Time Course of EV-D68 Binding and Replication in Primary BECs
To further examine the differences in binding and replication between the EV-D68 isolates, we used differentiated cultures of BEC, which are natural host cells for this virus. We first performed time-course experiments to identify the optimal time point p.i. for viral replication assessment. We used a recent EV-D68 clinical isolate from clade B1 (C7788) and Fermon. Both viruses had similar binding at 2 h; however, at 24 h, the C7788 strain demonstrated greater replication than Fermon (p < 0.05, Figure 3). There was no further increase in viral replication at 48 or 72 h p.i. in either strain, so we chose the 24 h p.i. time-point for further studies.
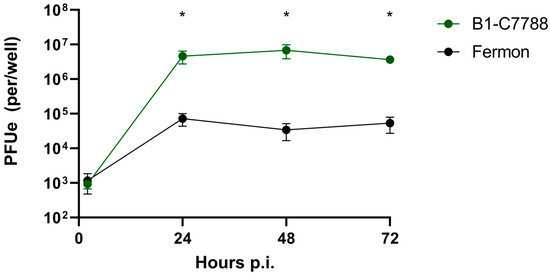
Figure 3.
Time course of infection with isolate C7788 and Fermon in differentiated human primary BEC-ALI cultures. The * represents difference (p < 0.05) between Fermon and B1-7788 isolate. Data are means and SEM from five independent experiments.
3.3. EV-D68 Binding and Replication in Primary BEC-ALI Cultures
Our preliminary experiments with two clinical isolates from the B1 clade (C7787 and C7788) and two variants of Fermon showed similar levels of binding between all four tested viruses but significantly higher (>one-log, p < 0.05) virus progeny yields and replication of clinical isolates compared to the Fermon variants in BEC-ALI cells (Figure S1). We next infected differentiated BEC-ALI cultures from five different donors with all available EV-D68 isolates and Fermon to examine the potential differences in virus binding and replication. In these experiments, there were no significant differences in the bound virus (2 h post-inoculation) between the recent isolates and Fermon (Figure 4A). In contrast, the recent isolates all had a 1.5–2 log greater viral yield and fold increase in EV RNA compared to the Fermon strain (p < 0.05) at 24 h p.i. Interestingly, the binding and replication levels were similar among the recent EV-D68 isolates that were obtained from asymptomatic infections vs. lower respiratory illnesses (Figure 4A,B).
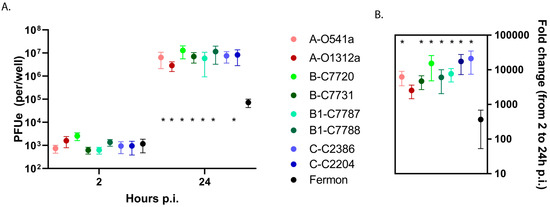
Figure 4.
Viral binding and replication in differentiated human primary BEC-ALI. (A) Cell-associated input virus (2 h p.i.) and progeny yields (24 h p.i.) were determined by RT-qPCR and (B) viral replication was calculated and compared for each isolate. The first letters in isolate designations represent the phylogenetic clade, and the following letters and numbers indicate the clinical isolate ID. The * represents a difference (p < 0.05) between Fermon and the recent clinical isolates. Data are means and SEM from five independent experiments. The viruses are color-coded according to clade with darker color indicating severe illness and lighter color indicating asymptomatic infection.
3.4. Assessment of Host Immune Response to EV-D68 by RNA-Seq Analysis
We next tested for differences in the transcriptional responses elicited by EV-D68 infection in differentiated human primary BECs from five donors. We compared transcript abundances induced by the infection of BEC-ALI cultures with four EV-D68 isolates representing each phylogenetic clade (A: 0541a, B: C7731, B1: C7788, C: C2386, and Fermon), while controlling for differences among BEC donors. Differentially expressed (DE) genes were identified through pairwise comparisons of each isolate using a false discovery rate (FDR) of 0.2.
We first assessed genes that were differentially expressed after infection with clinical isolates from clades A, B, B1, and C compared to mock-infected control cells (Figure S2). We found a robust response in the infected BECs characterized by strong upregulation of antiviral and pro-inflammatory response genes such as OASL, DDX60, INFL1-3, CXCL10, and CXCL11 (Table S1). There were no significant differences in gene expression among the clinical isolates (FDR < 0.2) (Table S1). The Fermon strain induced a number of genes including heavy-metal-associated isoprenylated plant protein (HIPP), which is involved in the Hedgehog signaling pathway, and potassium two-pore domain channel subfamily K member 3 (KCNK3), which encodes a member of the two-pore potassium channel. In contrast to the recent clinical isolates, Fermon did not induce significant differences (FDR < 0.2) in the expression of antiviral and pro-inflammatory genes (Table S1).
Given the similarity in the transcriptional response to the clinical isolates, we compared the transcriptional responses induced by Fermon to the average expression of genes induced by the four clinical isolates. This grouped analysis identified 90 DE genes (FDR = 0.2, Figure 5), and nearly all (88 of 90) of these genes were upregulated. Of the 35 genes (all upregulated) with an FDR < 0.05 and >2-fold change in expression (Table 2), most were related to host defense and antiviral responses (e.g., IFNL1, IFNL3, IFIT1, IFIT2, IFIT3, CXCL10, and CXCL11).
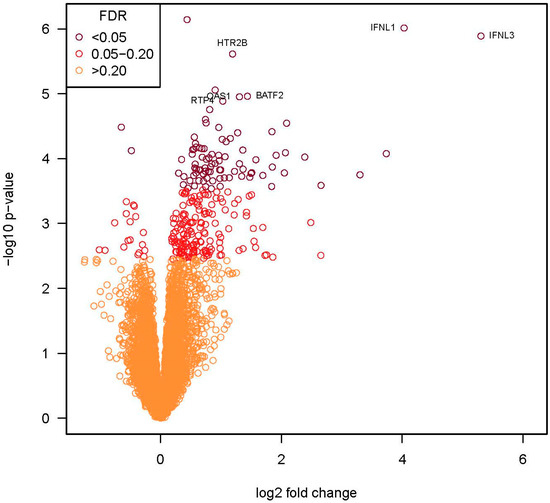
Figure 5.
RNA-seq volcano plot identifying differentially expressed genes in BEC-ALI cultures after infection with recent EV-D68 isolates vs. Fermon. The x-axis represents the log2 values of the fold change observed for each mRNA transcript, and the y-axis represents the log10 of the p-values of the significance tests between replicates for each transcript. The orange circles represent genes that had an FDR of >0.2, circles in red are genes with an FDR between 0.05–0.2, and the circles in burgundy are genes with an FDR of <0.05. The labeled genes (n = 6) had an FDR of <0.03 and a fold change >2.

Table 2.
Differentially expressed genes * from comparison of recent EV-D68 isolates vs. Fermon.
Given that Fermon replicated less efficiently than the recent isolates of EV-D68 in BEC, we next fit a model that included virus isolate, viral replication, and BEC donor. The results showed that isolate-specific differences in gene expression were attributable to differences in viral replication.
4. Discussion
The underlying mechanisms of the recent increase in frequency and severity of illnesses caused by EV-D68 are incompletely understood. This study utilized clinical isolates obtained over the past two decades as well as the prototype Fermon strain isolated in 1962 to assess whether changes in virus binding, replication, and/or the induction of immune response of recent EV-D68 isolates could provide potential explanations for these clinical observations. These studies identified distinct differences in viral replication and the epithelial immune response, as assessed by transcriptomics, between the prototype Fermon strain and the recent clinical isolates from clades A, B (including the B1 subclade), and C. Interestingly, no significant differences were identified in vitro among the recent EV-D68 clinical isolates despite the observed differences in associated disease severity, suggesting that individual factors are the main determinants of illness severity. The recent EV-D68 clinical isolates had similar binding and replication to one another but differed from the Fermon strain, which replicated better in HeLa cells but less well in BEC-ALI cultures. We further investigated the possible differences in host defense pathways by RNA-seq analysis and found an increase in some key host defense transcripts, such as IFNL1, IFNL3, CXCL10, and CXCL11, in response to infection with recent EV-D68 isolates compared to Fermon. Type-III interferons were also strongly induced by the contemporary EV-D68 isolates in PBE-ALI cells in a recent study [27]. These differences related closely to the degree of viral replication observed in these primary epithelial cell cultures. These findings suggest that an increased replication efficiency and enhanced inflammatory response induced by recent isolates in vitro could be responsible for the increased disease severity compared to the Fermon strain representing the past EV-D68 isolates from 1960s.
The recent isolates selected from children with either severe or asymptomatic infections had similar binding, replication, and immune responses in vitro. These findings suggest that host factors could be important in determining the severity of illness. In addition, changes in antigenicity leading to a loss of antibody protection [4] could have contributed to the recent severe outbreaks. The predilection for EV-D68 to more significantly impact young children suggests that the antibody response is likely important to the illness severity. Further, its cell specificity may have changed such that current strains can infect a broader range of cell types [28]. Indeed, some EV-D68 strains are capable of more effectively infecting cells through some unknown glycan-independent mechanism compared to Fermon [11]. The possible receptors that could allow EV-D68 to infect cells more effectively could be sulfated glycosaminoglycans, heparan sulfate proteoglycans, and/or ICAM-5 [9,10,29].
Our study had some strengths and limitations. One strength of our study was the use of differentiated cultures of human primary BEC, which provide a more physiologically relevant model system compared to monolayers of epithelial cells or continuous cell lines. Another important strength was the use of clinical virus isolates, representing all the major phylogenetic clades of EV-D68, that were cloned and produced by reverse genetics rather than ‘laboratory’ strains propagated in cell lines by infection. A limitation of the study was that the full passage history of the Fermon strain is unknown, and so it is possible that the original virus isolated in 1962 could be at least partially adapted to the RD or MRC-5 cell lines used for its propagation [30]. However, we found no significant differences in virus binding or replication between the Fermon variant received from Dr. Frank van Kuppeveld and the variant (Lab Fermon) cloned in our laboratory using the published genome sequence of the original clinical isolate from 1962 (Figure S1). Additionally, the primary human BECs were obtained from de-identified donors, so we do not know if they had underlying asthma or other respiratory conditions.
In conclusion, we identified differences in viral replication and epithelial cell responses to EV-D68 infection in vitro between the Fermon strain isolated in 1960s and the recent clinical isolates from clades A, B (including B1), and C that were obtained over the past two decades. These findings suggest that the enhanced viral replication and cellular inflammatory responses could be responsible for an increase in illness severity associated with recent isolates of EV-D68, and host factors are important in determining the severity of illness. Further studies are needed to fully understand the reasons underlying the EV-D68 outbreak in 2014.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/v15061291/s1: Supplementary Figure S1. Comparison of viral binding and replication of two Fermon strains and two recent EV-D68 clinical isolates in PBE-ALI cells. Supplementary Figure S2. Area-proportional Venn diagram comparing recent EV-D68 isolates differentially expressed genes. Table S1. RNA-seq gene expression analysis. Reference [31] are cited in Supplementary Materials File.
Author Contributions
Conceptualization, D.J.J. and Y.A.B.; methodology, M.K.D.; software, M.D.E.; validation, M.K.D.; formal analysis, M.D.E.; investigation, M.K.D.; resources, D.J.J. and Y.A.B.; data curation, M.K.D.; writing—original draft preparation, M.K.D.; writing—review and editing, J.E.G. and Y.A.B.; visualization, M.K.D.; supervision, Y.A.B. and D.J.J.; project administration, D.J.J.; funding acquisition, D.J.J. All authors have read and agreed to the published version of the manuscript.
Funding
The research reported in this publication was supported by NIH grants U19 opportunity fund U19AI070412 (D.J.J.), R01AI148707 (Y.A.B.), and U19AI104317 (J.E.G.).
Institutional Review Board Statement
Each of the three human studies (COAST, URECA, and PROSE) that contributed clinical samples was approved by the University of Wisconsin-Madison Human Research Subjects Committee, and participants provided written informed consent. Bronchial epithelial tissue samples were obtained from residual surgical specimens of de-identified donors. The protocol was approved by the University of Wisconsin-Madison Human Subjects Committee.
Informed Consent Statement
All nasal samples used from studies were obtained through informed consent. All the BECs used were obtained from de-identified donors.
Data Availability Statement
The RNA-seq data were deposited into the NCBI Gene Expression Omnibus (accession number GSE233630).
Acknowledgments
The authors want to thank Shakher Sijapati for helping with the construction and production of EV-D68 isolates, Suman Das for sequencing the clinical isolates of EV-D68, and Frank van Kuppeveld for kindly providing the Fermon infectious cDNA clone.
Conflicts of Interest
J.E.G. has served as a paid consultant for AstraZeneca, Meissa Vaccines Inc., and Via Nova Therapeutics Inc. and has stock options in Meissa Vaccines Inc. D.J.J. receives grants from GlaxoSmithKline and Regeneron, consulting fees from Avillion, AstraZeneca, Genentech, GlaxoSmithKline, Regeneron, Sanofi, and personal fees for DSMB from Pfizer. The other authors declare no conflict of interest.
References
- Schieble, J.H.; Fox, V.L.; Lennette, E.H. A probable new human picornavirus associated with respiratory diseases. Am. J. Epidemiol. 1967, 85, 297–310. [Google Scholar] [CrossRef]
- Greninger, A.L.; Naccache, S.N.; Messacar, K.; Clayton, A.; Yu, G.; Somasekar, S.; Federman, S.; Stryke, D.; Anderson, C.; Yagi, S.; et al. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the usa (2012–14): A retrospective cohort study. Lancet Infect. Dis. 2015, 15, 671–682. [Google Scholar] [CrossRef]
- Meijer, A.; van der Sanden, S.; Snijders, B.E.; Jaramillo-Gutierrez, G.; Bont, L.; van der Ent, C.K.; Overduin, P.; Jenny, S.L.; Jusic, E.; van der Avoort, H.G.; et al. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in the netherlands in 2010. Virology 2012, 423, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Suzuki, A.; Lupisan, S.; Okamoto, M.; Aniceto, R.; Egos, R.J.; Daya, E.E.; Tamaki, R.; Saito, M.; Fuji, N.; et al. Molecular evolution of enterovirus 68 detected in the philippines. PLoS ONE 2013, 8, e74221. [Google Scholar] [CrossRef] [PubMed]
- Oermann, C.M.; Schuster, J.E.; Conners, G.P.; Newland, J.G.; Selvarangan, R.; Jackson, M.A. Enterovirus D68. A focused review and clinical highlights from the 2014 U.S. Outbreak. Ann. Am. Thorac. Soc. 2015, 12, 775–781. [Google Scholar] [CrossRef]
- Hamparian, V.V.; Colonno, R.J.; Cooney, M.K.; Dick, E.C.; Gwaltney, J.M., Jr.; Hughes, J.H.; Jordan, W.S., Jr.; Kapikian, A.Z.; Mogabgab, W.J.; Monto, A.; et al. A collaborative report: Rhinoviruses--extension of the numbering system from 89 to 100. Virology 1987, 159, 191–192. [Google Scholar]
- Ishiko, H.; Miura, R.; Shimada, Y.; Hayashi, A.; Nakajima, H.; Yamazaki, S.; Takeda, N. Human rhinovirus 87 identified as human enterovirus 68 by vp4-based molecular diagnosis. Intervirology 2002, 45, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Okamoto, M.; Nakakita, S.; Suzuki, A.; Saito, M.; Tamaki, R.; Lupisan, S.; Roy, C.N.; Hiramatsu, H.; Sugawara, K.E.; et al. Antigenic and receptor binding properties of enterovirus 68. J. Virol. 2014, 88, 2374–2384. [Google Scholar] [CrossRef]
- Wei, W.; Guo, H.; Chang, J.; Yu, Y.; Liu, G.; Zhang, N.; Willard, S.H.; Zheng, S.; Yu, X.F. ICAM-5/telencephalin is a functional entry receptor for enterovirus D68. Cell Host Microbe 2016, 20, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Baggen, J.; Liu, Y.; Lyoo, H.; van Vliet, A.L.W.; Wahedi, M.; de Bruin, J.W.; Roberts, R.W.; Overduin, P.; Meijer, A.; Rossmann, M.G.; et al. Bypassing pan-enterovirus host factor pla2g16. Nat. Commun. 2019, 10, 3171. [Google Scholar] [CrossRef]
- Rosenfeld, A.B.; Warren, A.L.; Racaniello, V.R. Neurotropism of enterovirus D68 isolates is independent of sialic acid and is not a recently acquired phenotype. mBio 2019, 10, e02370-19. [Google Scholar] [CrossRef]
- Khetsuriani, N.; Lamonte-Fowlkes, A.; Oberst, S.; Pallansch, M.A. Enterovirus surveillance--united states, 1970–2005. MMWR Surveill. Summ. 2006, 55, 1–20. [Google Scholar] [PubMed]
- Tokarz, R.; Firth, C.; Madhi, S.A.; Howie, S.R.; Wu, W.; Sall, A.A.; Haq, S.; Briese, T.; Lipkin, W.I. Worldwide emergence of multiple clades of enterovirus 68. J. Gen. Virol. 2012, 93, 1952–1958. [Google Scholar] [CrossRef]
- Midgley, S.E.; Benschop, K.; Dyrdak, R.; Mirand, A.; Bailly, J.L.; Bierbaum, S.; Buderus, S.; Böttcher, S.; Eis-Hübinger, A.M.; Hönemann, M.; et al. Co-circulation of multiple enterovirus D68 subclades, including a novel b3 cluster, across europe in a season of expected low prevalence, 2019/20. Eurosurveillance 2020, 25, 1900749. [Google Scholar] [CrossRef]
- Xiang, Z.; Li, L.; Lei, X.; Zhou, H.; Zhou, Z.; He, B.; Wang, J. Enterovirus 68 3c protease cleaves trif to attenuate antiviral responses mediated by toll-like receptor 3. J. Virol. 2014, 88, 6650–6659. [Google Scholar] [CrossRef]
- Lemanske, R.F. The childhood origins of asthma (coast) study. Pediatr. Allergy Immunol. 2002, 13 (Suppl. S15), 38–43. [Google Scholar] [CrossRef]
- Teach, S.J.; Gill, M.A.; Togias, A.; Sorkness, C.A.; Arbes, S.J.; Calatroni, A.; Wildfire, J.J.; Gergen, P.J.; Cohen, R.T.; Pongracic, J.A.; et al. Preseasonal treatment with either omalizumab or an inhaled corticosteroid boost to prevent fall asthma exacerbations. J. Allergy Clin. Immunol. 2015, 136, 1476–1485. [Google Scholar] [CrossRef]
- Gern, J.E.; Visness, C.M.; Gergen, P.J.; Wood, R.A.; Bloomberg, G.R.; O’Connor, G.T.; Kattan, M.; Sampson, H.A.; Witter, F.R.; Sandel, M.T.; et al. The urban environment and childhood asthma (ureca) birth cohort study: Design, methods, and study population. BMC Pulm. Med. 2009, 9, 17. [Google Scholar] [CrossRef]
- Huang, W.; Wang, G.; Zhuge, J.; Nolan, S.M.; Dimitrova, N.; Fallon, J.T. Whole-genome sequence analysis reveals the enterovirus D68 isolates during the united states 2014 outbreak mainly belong to a novel clade. Sci. Rep. 2015, 5, 15223. [Google Scholar] [CrossRef]
- Herold, J.; Andino, R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 2000, 74, 6394–6400. [Google Scholar] [CrossRef]
- Lee, W.M.; Wang, W. Human rhinovirus type 16: Mutant v1210a requires capsid-binding drug for assembly of pentamers to form virions during morphogenesis. J. Virol. 2003, 77, 6235–6244. [Google Scholar] [CrossRef]
- Nakagome, K.; Bochkov, Y.A.; Ashraf, S.; Brockman-Schneider, R.A.; Evans, M.D.; Pasic, T.R.; Gern, J.E. Effects of rhinovirus species on viral replication and cytokine production. J. Allergy Clin. Immunol. 2014, 134, 332–341. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Liao, Y.; Smyth, G.K.; Shi, W. The r package rsubread is easier, faster, cheaper and better for alignment and quantification of rna sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Carlson, M. Org.Hs.Eg.Db: Genome Wide Annotation for Human. R package 3.3.0. 2019. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html (accessed on 22 May 2023).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Freeman, M.C.; Wells, A.I.; Ciomperlik-Patton, J.; Myerburg, M.M.; Yang, L.; Konopka-Anstadt, J.; Coyne, C.B. Respiratory and intestinal epithelial cells exhibit differential susceptibility and innate immune responses to contemporary EV-D68 isolates. eLife 2021, 10, e66687. [Google Scholar] [CrossRef]
- Baggen, J.; Thibaut, H.J.; Staring, J.; Jae, L.T.; Liu, Y.; Guo, H.; Slager, J.J.; de Bruin, J.W.; van Vliet, A.L.; Blomen, V.A.; et al. Enterovirus d68 receptor requirements unveiled by haploid genetics. Proc. Natl. Acad. Sci. USA 2016, 113, 1399–1404. [Google Scholar] [CrossRef]
- Sridhar, A.; Depla, J.A.; Mulder, L.A.; Karelehto, E.; Brouwer, L.; Kruiswijk, L.; Vieira de Sá, R.; Meijer, A.; Evers, M.M.; van Kuppeveld, F.J.M.; et al. Enterovirus D68 infection in human primary airway and brain organoids: No additional role for heparan sulfate binding for neurotropism. Microbiol. Spectr. 2022, 10, e0169422. [Google Scholar] [CrossRef]
- Blomqvist, S.; Savolainen, C.; Raman, L.; Roivainen, M.; Hovi, T. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J. Clin. Microbiol. 2002, 40, 4218–4223. [Google Scholar] [CrossRef]
- Hulsen, T.; de Vlieg, J.; Alkema, W. Biovenn—A web application for the comparison and visualization of biological lists using area-proportional venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).