A Chemical Proteomics Approach to Discover Regulators of Innate Immune Signaling
Abstract
1. Introduction
2. Materials and Methods
3. Results
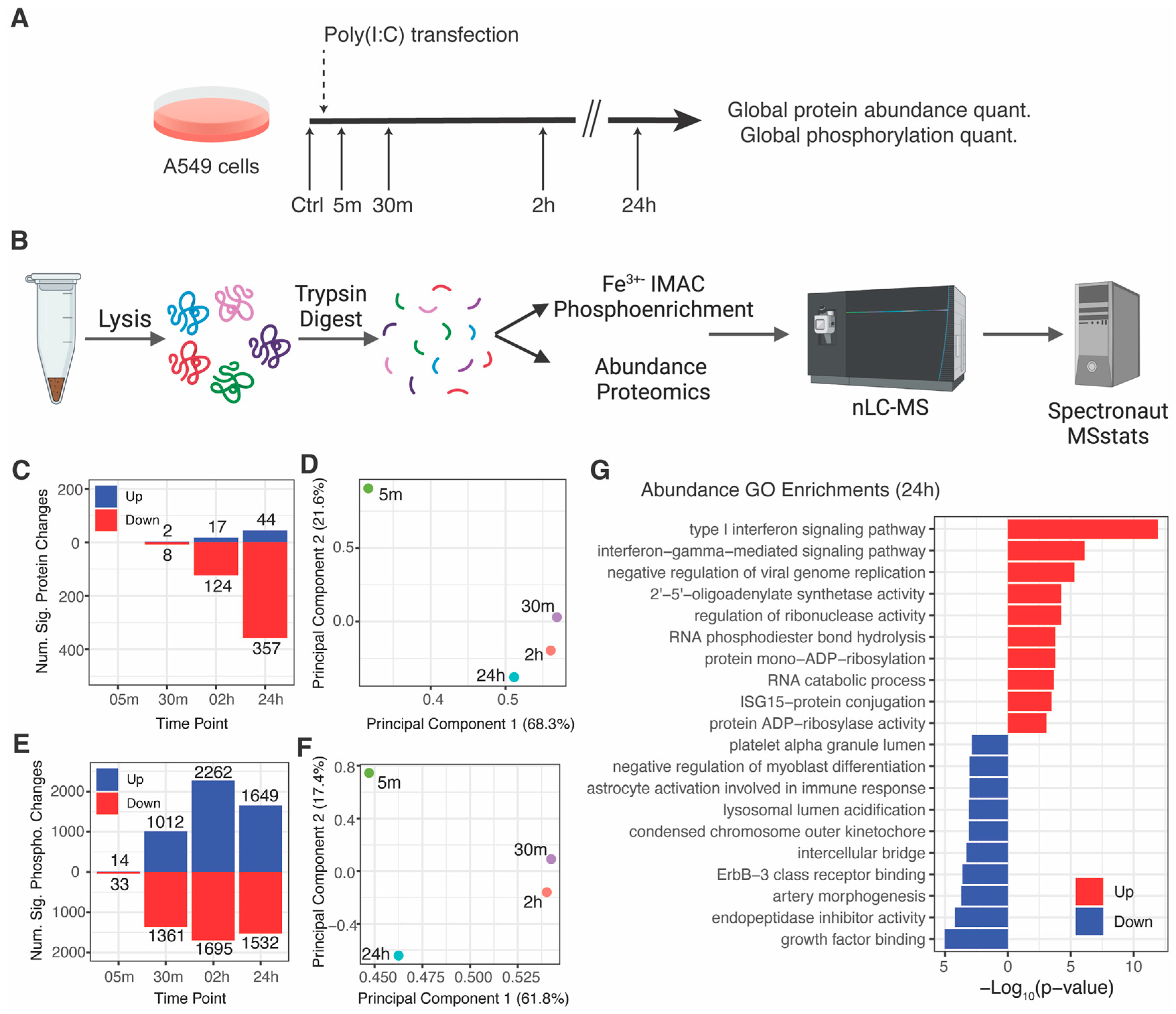
3.1. Quantification of Proteome and Phosphoproteome Responses to RIG-I Activation
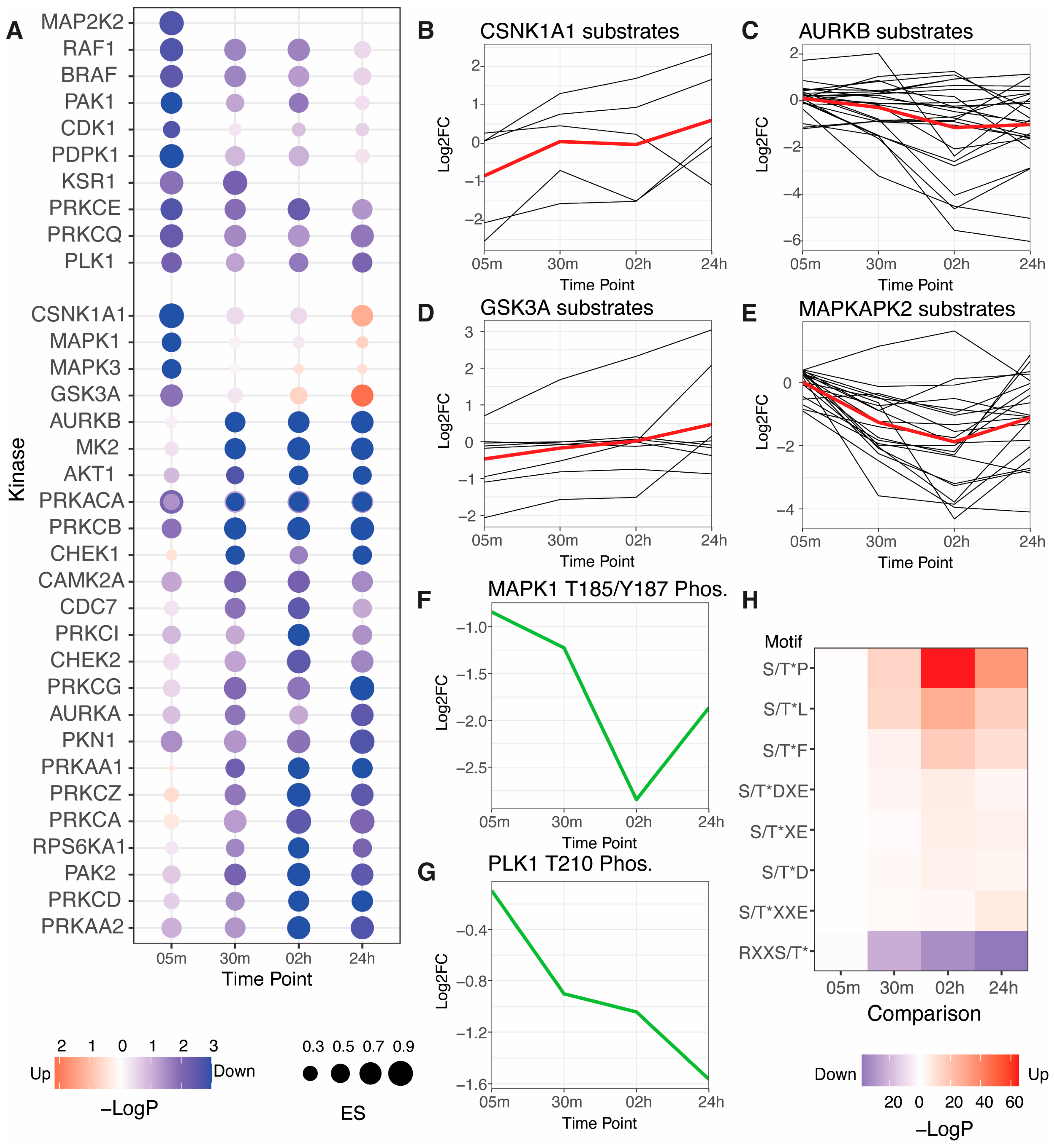
3.2. Kinase Activity Analysis Reveals a Decrease in Cellular Kinase Activities in Response to RLR Pathway Activation
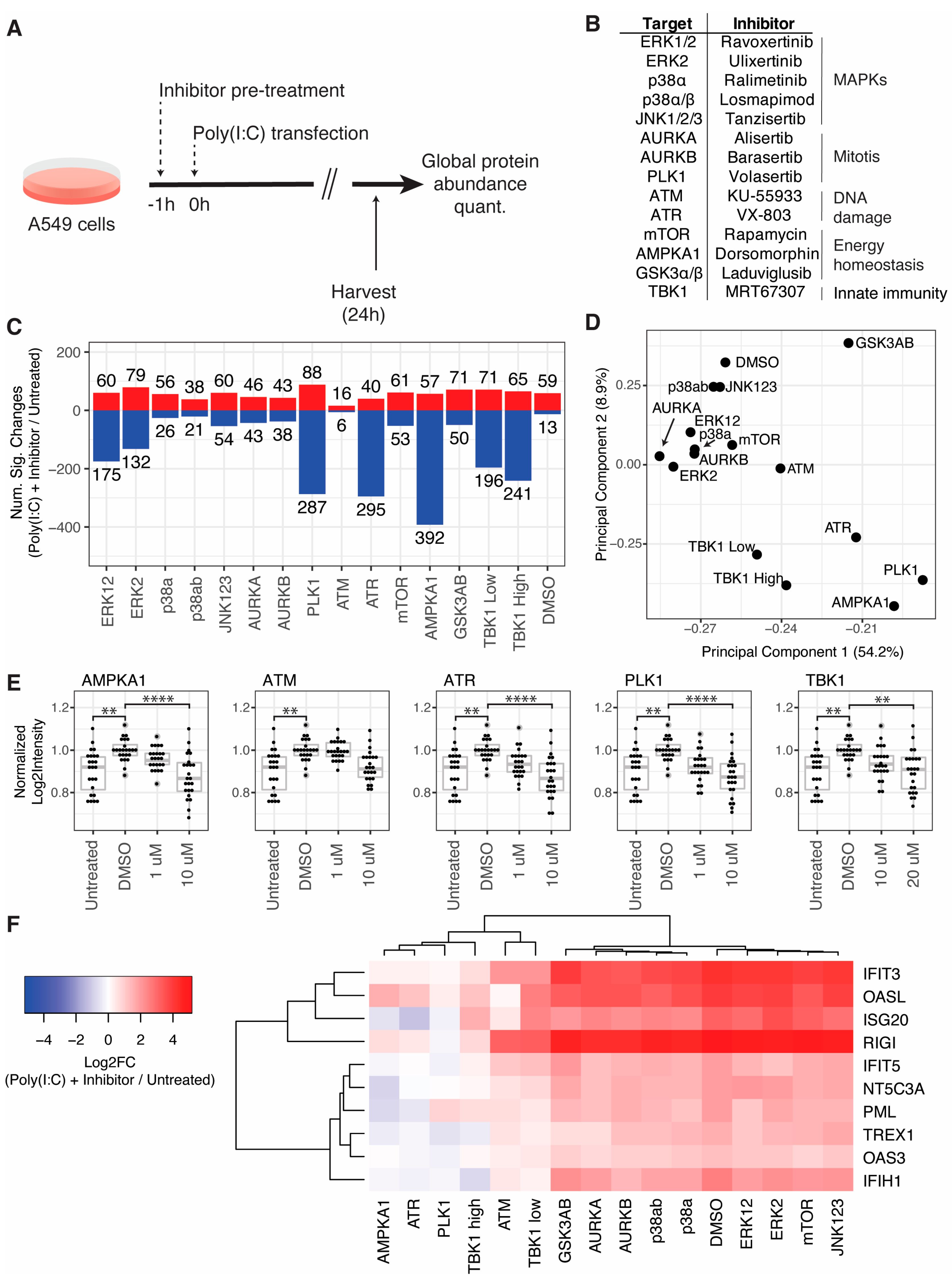
3.3. Kinase Inhibitors Targeting ATM, ATR, PLK1, and AMPKA1 Inhibit ISG Induction in Response to RLR Activation
3.4. Genetic Kinase Depletion Does Not Confirm Effects of Kinase Inhibitors in Response to RLR or IFN Pathway Activation
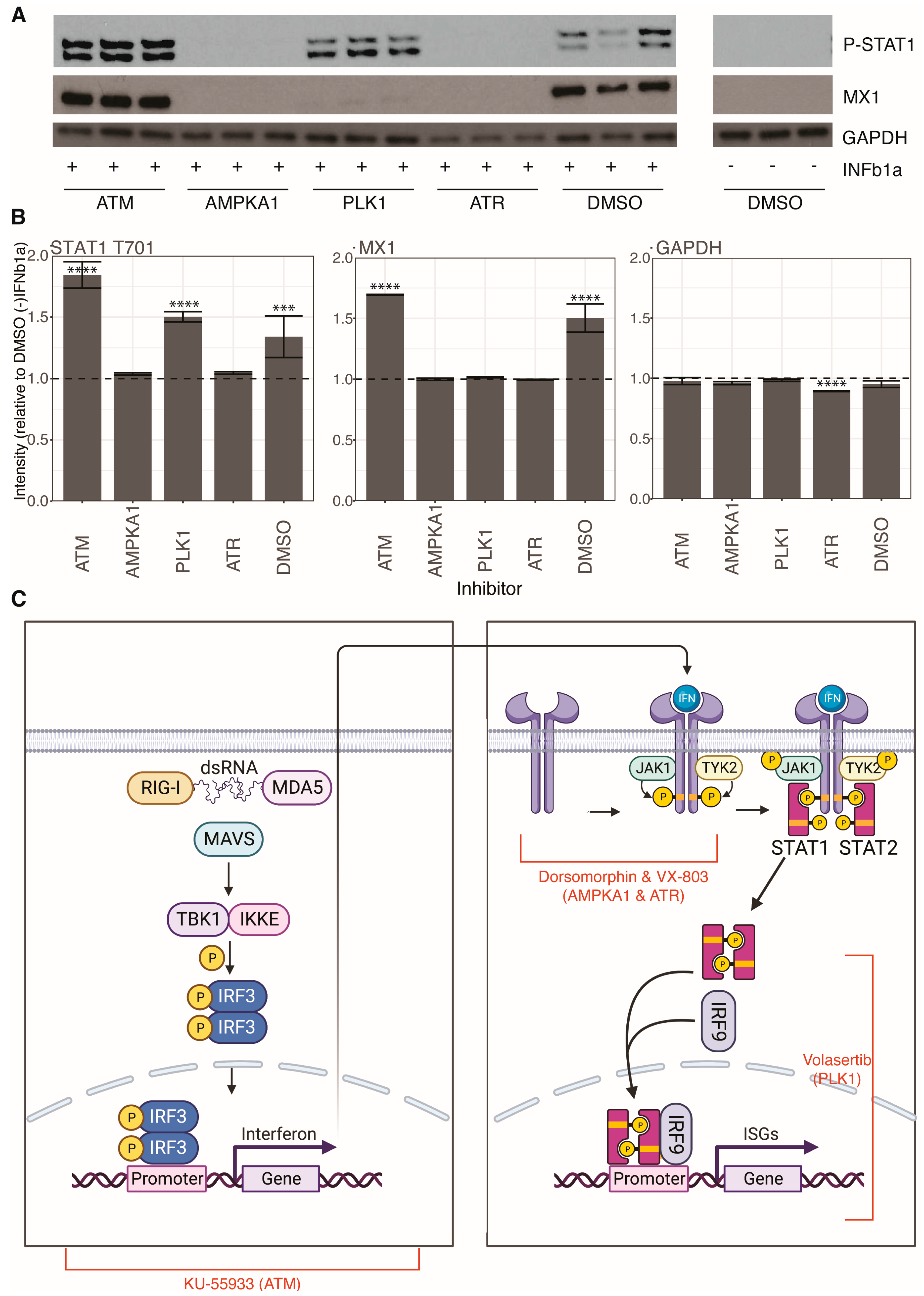
3.5. Cellular Kinase Inhibitors Block Innate Immune Signaling by Different Mechanisms
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goubau, D.; Deddouche, S.; Reis e Sousa, C. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Michael Lavigne, G.; Russell, H.; Sherry, B.; Ke, R. Autocrine and paracrine interferon signalling as ‘ring vaccination’ and ‘contact tracing’ strategies to suppress virus infection in a host. Proc. Biol. Sci. 2021, 288, 20203002. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Ashour, J.; Laurent-Rolle, M.; Shi, P.Y.; Garcia-Sastre, A. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 2009, 83, 5408–5418. [Google Scholar] [CrossRef]
- Grant, A.; Ponia, S.S.; Tripathi, S.; Balasubramaniam, V.; Miorin, L.; Sourisseau, M.; Schwarz, M.C.; Sanchez-Seco, M.P.; Evans, M.J.; Best, S.M.; et al. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe 2016, 19, 882–890. [Google Scholar] [CrossRef]
- Morrison, J.; Laurent-Rolle, M.; Maestre, A.M.; Rajsbaum, R.; Pisanelli, G.; Simon, V.; Mulder, L.C.; Fernandez-Sesma, A.; Garcia-Sastre, A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013, 9, e1003265. [Google Scholar] [CrossRef]
- Hale, B.G.; Randall, R.E.; Ortin, J.; Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Börold, J.; Eletto, D.; Busnadiego, I.; Mair, N.K.; Moritz, E.; Schiefer, S.; Schmidt, N.; Petric, P.P.; Wong, W.W.-L.; Schwemmle, M.; et al. BRD9 is a druggable component of interferon-stimulated gene expression and antiviral activity. EMBO Rep. 2021, 22, e52823. [Google Scholar] [CrossRef]
- Higgins, C.A.; Nilsson-Payant, B.E.; Kurland, A.P.; Adhikary, P.; Golynker, I.; Danziger, O.; Panis, M.; Rosenberg, B.R.; tenOever, B.; Johnson, J.R. SARS-CoV-2 hijacks p38ß/MAPK11 to promote viral protein translation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Johnson, J.R.; Crosby, D.C.; Hultquist, J.F.; Kurland, A.P.; Adhikary, P.; Li, D.; Marlett, J.; Swann, J.; Huttenhain, R.; Verschueren, E.; et al. Global post-translational modification profiling of HIV-1-infected cells reveals mechanisms of host cellular pathway remodeling. Cell Rep. 2022, 39, 110690. [Google Scholar] [CrossRef]
- Bruderer, R.; Bernhardt, O.M.; Gandhi, T.; Xuan, Y.; Sondermann, J.; Schmidt, M.; Gomez-Varela, D.; Reiter, L. Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Mol. Cell. Proteom. 2017, 16, 2296–2309. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef]
- Choi, M.; Chang, C.Y.; Clough, T.; Broudy, D.; Killeen, T.; MacLean, B.; Vitek, O. MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014, 30, 2524–2526. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Gene Ontology, C. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Hernandez-Armenta, C.; Ochoa, D.; Goncalves, E.; Saez-Rodriguez, J.; Beltrao, P. Benchmarking substrate-based kinase activity inference using phosphoproteomic data. Bioinformatics 2017, 33, 1845–1851. [Google Scholar] [CrossRef]
- Bachman, J.A.; Sorger, P.K.; Gyori, B.M. Assembling a corpus of phosphoproteomic annotations using ProtMapper to normalize site information from databases and text mining. bioRxiv 2022, 822668. [Google Scholar] [CrossRef]
- Cheng, A.; Grant, C.E.; Noble, W.S.; Bailey, T.L. MoMo: Discovery of statistically significant post-translational modification motifs. Bioinformatics 2019, 35, 2774–2782. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.F.; Schwartz, D. Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinform. 2011, 35, 13.15.1–13.15.24. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, M.; Hoffmann, H.H.; Scull, M.A.; Gilmore, R.H.; Bell, K.L.; Ciancanelli, M.; Wilson, S.J.; Crotta, S.; Yu, Y.; Flatley, B.; et al. A serpin shapes the extracellular environment to prevent influenza A virus maturation. Cell 2015, 160, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; Yaron, T.M.; Huntsman, E.M.; Kerelsky, A.; Song, J.; Regev, A.; Lin, T.Y.; Liberatore, K.; Cizin, D.M.; Cohen, B.M.; et al. An atlas of substrate specificities for the human serine/threonine kinome. Nature 2023, 613, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e619. [Google Scholar] [CrossRef]
- Chevrier, N.; Mertins, P.; Artyomov, M.N.; Shalek, A.K.; Iannacone, M.; Ciaccio, M.F.; Gat-Viks, I.; Tonti, E.; DeGrace, M.M.; Clauser, K.R.; et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell 2011, 147, 853–867. [Google Scholar] [CrossRef]
- Hartlova, A.; Erttmann, S.F.; Raffi, F.A.; Schmalz, A.M.; Resch, U.; Anugula, S.; Lienenklaus, S.; Nilsson, L.M.; Kroger, A.; Nilsson, J.A.; et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 2015, 42, 332–343. [Google Scholar] [CrossRef]
- Feng, X.; Tubbs, A.; Zhang, C.; Tang, M.; Sridharan, S.; Wang, C.; Jiang, D.; Su, D.; Zhang, H.; Chen, Z.; et al. ATR inhibition potentiates ionizing radiation-induced interferon response via cytosolic nucleic acid-sensing pathways. EMBO J. 2020, 39, e104036. [Google Scholar] [CrossRef]
- Zhang, Q.; Green, M.D.; Lang, X.; Lazarus, J.; Parsels, J.D.; Wei, S.; Parsels, L.A.; Shi, J.; Ramnath, N.; Wahl, D.R.; et al. Inhibition of ATM Increases Interferon Signaling and Sensitizes Pancreatic Cancer to Immune Checkpoint Blockade Therapy. Cancer Res. 2019, 79, 3940–3951. [Google Scholar] [CrossRef]
- Prantner, D.; Perkins, D.J.; Vogel, S.N. AMP-activated Kinase (AMPK) Promotes Innate Immunity and Antiviral Defense through Modulation of Stimulator of Interferon Genes (STING) Signaling. J. Biol. Chem. 2017, 292, 292–304. [Google Scholar] [CrossRef]
- Bergholz, J.S.; Zhao, J.J. How Compensatory Mechanisms and Adaptive Rewiring Have Shaped Our Understanding of Therapeutic Resistance in Cancer. Cancer Res. 2021, 81, 6074–6077. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Perez-Riverol, Y.; Sharma, V.; Carver, J.J.; Mendoza, L.; Kundu, D.J.; Wang, S.; Bandla, C.; Kamatchinathan, S.; et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023, 51, D1539–D1548. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Garcia-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurland, A.P.; Bonaventure, B.; Johnson, J.R. A Chemical Proteomics Approach to Discover Regulators of Innate Immune Signaling. Viruses 2023, 15, 1112. https://doi.org/10.3390/v15051112
Kurland AP, Bonaventure B, Johnson JR. A Chemical Proteomics Approach to Discover Regulators of Innate Immune Signaling. Viruses. 2023; 15(5):1112. https://doi.org/10.3390/v15051112
Chicago/Turabian StyleKurland, Andrew P., Boris Bonaventure, and Jeffrey R. Johnson. 2023. "A Chemical Proteomics Approach to Discover Regulators of Innate Immune Signaling" Viruses 15, no. 5: 1112. https://doi.org/10.3390/v15051112
APA StyleKurland, A. P., Bonaventure, B., & Johnson, J. R. (2023). A Chemical Proteomics Approach to Discover Regulators of Innate Immune Signaling. Viruses, 15(5), 1112. https://doi.org/10.3390/v15051112





