Three-Dimensional Chromatin Structure of the EBV Genome: A Crucial Factor in Viral Infection
Abstract
1. Introduction
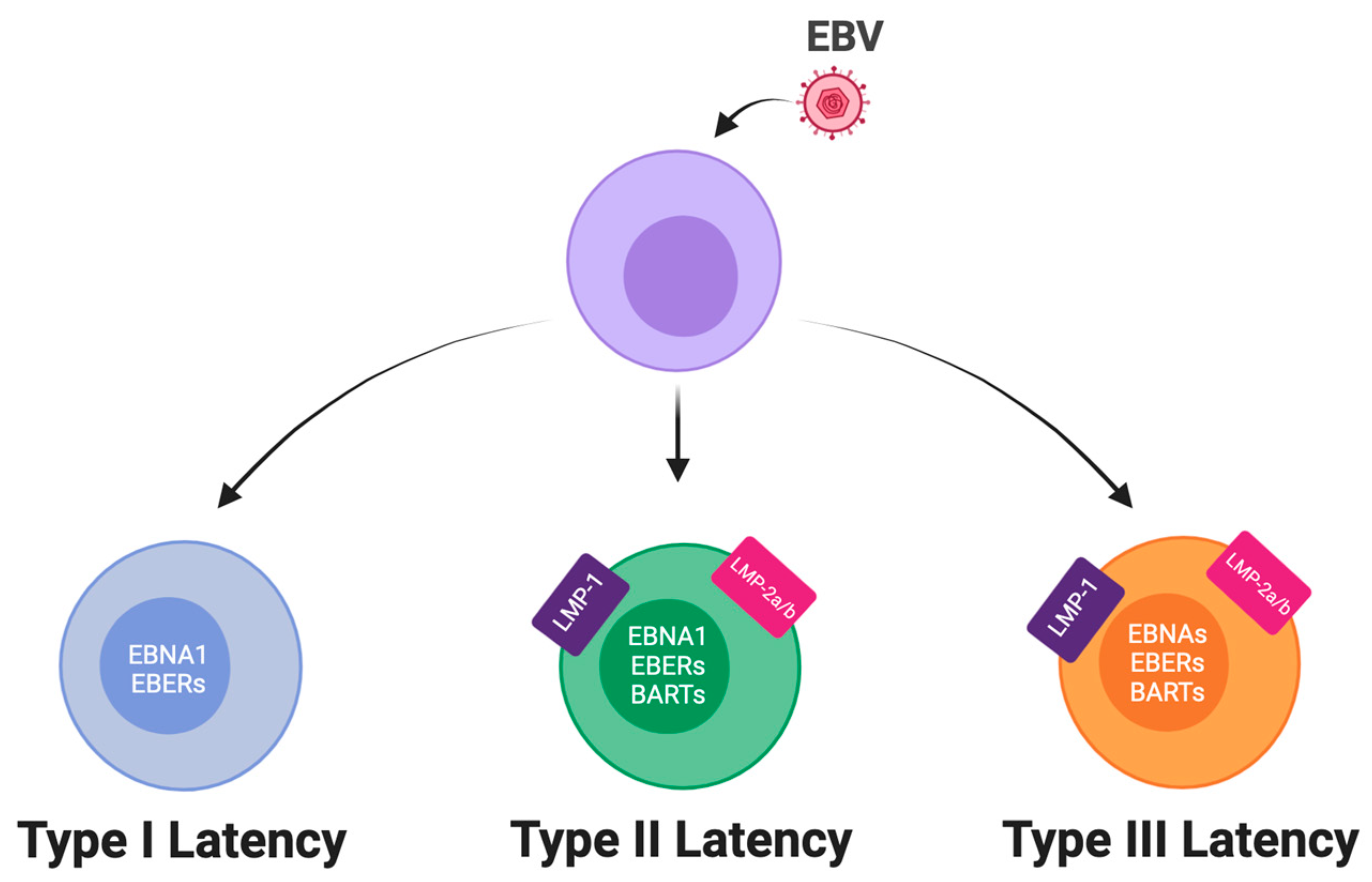
2. EBV Latency
3. EBV and CTCF
4. CTCF and Cohesin
5. Regulation of EBV 3D Structure
6. EBV and Nuclear Lamina
7. EBV and MYC
8. EBV-Associated Enhancers
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964, 1, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein-Barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Dunmire, S.K.; Verghese, P.S.; Balfour, H.H. Primary Epstein-Barr virus infection. J. Clin. Virol. 2018, 102, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Gewurz, B.; Longnecker, R.; Cohen, J.I. Epstein-Barr Virus. In Fields Virology, 7th ed.; Howley, P., Knipe, D.M., Cohen, J.I., Damania, B., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2021; pp. 324–388. [Google Scholar]
- Khan, G.; Hashim, M.J. Global burden of deaths from Epstein-Barr virus attributable malignancies 1990–2010. Infect. Agents Cancer 2014, 9, 38. [Google Scholar] [CrossRef]
- Wong, Y.; Meehan, M.T.; Burrows, S.R.; Doolan, D.L.; Miles, J.J. Estimating the global burden of Epstein-Barr virus-related cancers. J. Cancer Res. Clin. Oncol. 2022, 148, 31–46. [Google Scholar] [CrossRef]
- Hsu, J.L.; Glaser, S.L. Epstein-barr virus-associated malignancies: Epidemiologic patterns and etiologic implications. Crit. Rev. Oncol. Hematol. 2000, 34, 27–53. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A. The Global Landscape of EBV-Associated Tumors. Front. Oncol. 2019, 9, 713. [Google Scholar] [CrossRef]
- Farrell, P.J. Epstein-Barr Virus and Cancer. Annu. Rev. Pathol. 2019, 14, 29–53. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Hawkins, J.B.; Tracy, S.I.; Shapiro, M. The pathogenesis of Epstein-Barr virus persistent infection. Curr. Opin. Virol. 2013, 3, 227–232. [Google Scholar] [CrossRef]
- Cohen, J.I. Epstein-Barr virus infection. N. Engl. J. Med. 2000, 343, 481–492. [Google Scholar] [CrossRef]
- Cohen, J.I. Herpesvirus latency. J. Clin. Investig. 2020, 130, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Babcock, G.J.; Decker, L.L.; Volk, M.; Thorley-Lawson, D.A. EBV persistence in memory B cells in vivo. Immunity 1998, 9, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Rosemarie, Q.; Sugden, B. Epstein-Barr Virus: How Its Lytic Phase Contributes to Oncogenesis. Microorganisms 2020, 8, 1824. [Google Scholar] [CrossRef]
- Ma, S.D.; Hegde, S.; Young, K.H.; Sullivan, R.; Rajesh, D.; Zhou, Y.; Jankowska-Gan, E.; Burlingham, W.J.; Sun, X.; Gulley, M.L.; et al. A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J. Virol. 2011, 85, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.K.; Gulley, M.L.; Feng, W.H.; Delecluse, H.J.; Holley-Guthrie, E.; Kenney, S.C. Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J. Virol. 2005, 79, 13993–14003. [Google Scholar] [CrossRef] [PubMed]
- Okuno, Y.; Murata, T.; Sato, Y.; Muramatsu, H.; Ito, Y.; Watanabe, T.; Okuno, T.; Murakami, N.; Yoshida, K.; Sawada, A.; et al. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat. Microbiol. 2019, 4, 404–413. [Google Scholar] [CrossRef]
- Guo, R.; Gewurz, B.E. Epigenetic control of the Epstein-Barr lifecycle. Curr. Opin. Virol. 2022, 52, 78–88. [Google Scholar] [CrossRef]
- Pei, Y.; Wong, J.H.; Robertson, E.S. Herpesvirus Epigenetic Reprogramming and Oncogenesis. Annu. Rev. Virol. 2020, 7, 309–331. [Google Scholar] [CrossRef]
- Buschle, A.; Hammerschmidt, W. Epigenetic lifestyle of Epstein-Barr virus. Semin. Immunopathol. 2020, 42, 131–142. [Google Scholar] [CrossRef]
- Babcock, G.J.; Hochberg, D.; Thorley-Lawson, A.D. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 2000, 13, 497–506. [Google Scholar] [CrossRef]
- Babcock, G.J.; Thorley-Lawson, D.A. Tonsillar memory B cells, latently infected with Epstein-Barr virus, express the restricted pattern of latent genes previously found only in Epstein-Barr virus-associated tumors. Proc. Natl. Acad. Sci. USA 2000, 97, 12250–12255. [Google Scholar] [CrossRef] [PubMed]
- Rowe, M.; Lear, A.L.; Croom-Carter, D.; Davies, A.H.; Rickinson, A.B. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J. Virol. 1992, 66, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.M.; Tanizawa, H.; Caruso, L.B.; Hulse, M.; Kossenkov, A.; Madzo, J.; Keith, K.; Tan, Y.; Boyle, S.; Lieberman, P.M.; et al. The three-dimensional structure of Epstein-Barr virus genome varies by latency type and is regulated by PARP1 enzymatic activity. Nat. Commun. 2022, 13, 187. [Google Scholar] [CrossRef] [PubMed]
- Tempera, I.; Klichinsky, M.; Lieberman, P.M. EBV latency types adopt alternative chromatin conformations. PLoS Pathog. 2011, 7, e1002180. [Google Scholar] [CrossRef] [PubMed]
- Woisetschlaeger, M.; Yandava, C.N.; Furmanski, L.A.; Strominger, J.L.; Speck, S.H. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. USA 1990, 87, 1725–1729. [Google Scholar] [CrossRef]
- Nonkwelo, C.; Skinner, J.; Bell, A.; Rickinson, A.; Sample, J. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J. Virol. 1996, 70, 623–627. [Google Scholar] [CrossRef]
- Trivedi, P.; Spinsanti, P.; Cuomo, L.; Volpe, M.; Takada, K.; Frati, L.; Faggioni, A. Differential regulation of Epstein-Barr virus (EBV) latent gene expression in Burkitt lymphoma cells infected with a recombinant EBV strain. J. Virol. 2001, 75, 4929–4935. [Google Scholar] [CrossRef]
- Jones, C.H.; Hayward, S.D.; Rawlins, D.R. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J. Virol. 1989, 63, 101–110. [Google Scholar] [CrossRef]
- Sample, J.; Henson, E.B.; Sample, C. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J. Virol. 1992, 66, 4654–4661. [Google Scholar] [CrossRef]
- Chiang, A.K.; Tao, Q.; Srivastava, G.; Ho, F.C. Nasal NK- and T-cell lymphomas share the same type of Epstein-Barr virus latency as nasopharyngeal carcinoma and Hodgkin’s disease. Int. J. Cancer 1996, 68, 285–290. [Google Scholar] [CrossRef]
- Price, A.M.; Luftig, M.A. To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis. PLoS Pathog. 2015, 11, e1004656. [Google Scholar] [CrossRef] [PubMed]
- Thorley-Lawson, D.A.; Gross, A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 2004, 350, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Rooney, C.M.; Brimmell, M.; Buschle, M.; Allan, G.; Farrell, P.J.; Kolman, J.L. Host cell and EBNA-2 regulation of Epstein-Barr virus latent-cycle promoter activity in B lymphocytes. J. Virol. 1992, 66, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gregory, C.D.; Rowe, M.; Rickinson, A.B.; Wang, D.; Birkenbach, M.; Kikutani, H.; Kishimoto, T.; Kieff, E. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc. Natl. Acad. Sci. USA 1987, 84, 3452–3456. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tsang, S.F.; Kurilla, M.G.; Cohen, J.I.; Kieff, E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 1990, 64, 3407–3416. [Google Scholar] [CrossRef]
- Abbot, S.D.; Rowe, M.; Cadwallader, K.; Ricksten, A.; Gordon, J.; Wang, F.; Rymo, L.; Rickinson, A.B. Epstein-Barr virus nuclear antigen 2 induces expression of the virus-encoded latent membrane protein. J. Virol. 1990, 64, 2126–2134. [Google Scholar] [CrossRef]
- Price, A.M.; Tourigny, J.P.; Forte, E.; Salinas, R.E.; Dave, S.S.; Luftig, M.A. Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-kappaB activation. J. Virol. 2012, 86, 11096–11106. [Google Scholar] [CrossRef]
- Brink, A.A.; Dukers, D.F.; Van den Brule, A.J.; Oudejans, J.J.; Middeldorp, J.M.; Meijer, C.J.; Jiwa, M. Presence of Epstein-Barr virus latency type III at the single cell level in post-transplantation lymphoproliferative disorders and AIDS related lymphomas. J. Clin. Pathol. 1997, 50, 911–918. [Google Scholar] [CrossRef]
- Takacs, M.; Banati, F.; Koroknai, A.; Segesdi, J.; Salamon, D.; Wolf, H.; Niller, H.H.; Minarovits, J. Epigenetic regulation of latent Epstein-Barr virus promoters. Biochim. Biophys. Acta 2010, 1799, 228–235. [Google Scholar] [CrossRef]
- Tempera, I.; Lieberman, P.M. Chromatin organization of gammaherpesvirus latent genomes. Biochim. Biophys. Acta 2010, 1799, 236–245. [Google Scholar] [CrossRef]
- Tempera, I.; Lieberman, P.M. Epigenetic regulation of EBV persistence and oncogenesis. Semin. Cancer Biol. 2014, 26, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Masucci, M.G.; Contreras-Salazar, B.E.R.T.H.A.; Ragnar, E.; Falk, K.E.R.S.T.I.N.; Minarovits, J.A.N.O.S.; Ernberg, I.N.G.E.M.A.R.; Klein, G.E.O.R.G.E. 5-Azacytidine up regulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt’s lymphoma line rael. J. Virol. 1989, 63, 3135–3141. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D.; Hayward, S.D.; Ling, P.D.; Samid, D.; Ambinder, R.F. Transcriptional activation of the Epstein-Barr virus latency C promoter after 5-azacytidine treatment: Evidence that demethylation at a single CpG site is crucial. Mol. Cell. Biol. 1995, 15, 6150–6159. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.; Robertson, K.D.; Manns, A.; Hildesheim, A.; Ambinder, R.F. The Epstein-Barr virus major latent promoter Qp is constitutively active, hypomethylated, and methylation sensitive. J. Virol. 1998, 72, 7075–7083. [Google Scholar] [CrossRef] [PubMed]
- Ambinder, R.F.; Robertson, K.D.; Tao, Q. DNA methylation and the Epstein-Barr virus. Semin. Cancer Biol. 1999, 9, 369–375. [Google Scholar] [CrossRef]
- Mentzer, S.J.; Fingeroth, J.; Reilly, J.J.; Perrine, S.P.; Faller, D.V. Arginine butyrate-induced susceptibility to ganciclovir in an Epstein-Barr-virus-associated lymphoma. Blood Cells Mol. Dis. 1998, 24, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Westphal, E.M.; Blackstock, W.; Feng, W.; Israel, B.; Kenney, S.C. Activation of lytic Epstein-Barr virus (EBV) infection by radiation and sodium butyrate in vitro and in vivo: A potential method for treating EBV-positive malignancies. Cancer Res. 2000, 60, 5781–5788. [Google Scholar]
- Bergbauer, M.; Kalla, M.; Schmeinck, A.; Göbel, C.; Rothbauer, U.; Eck, S.; Benet-Pagès, A.; Strom, T.M.; Hammerschmidt, W. CpG-Methylation Regulates a Class of Epstein-Barr Virus Promoters. PLoS Pathog. 2010, 6, e1001114. [Google Scholar] [CrossRef]
- Kalla, M.; Schmeinck, A.; Bergbauer, M.; Pich, D.; Hammerschmidt, W. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc. Natl. Acad. Sci. USA 2010, 107, 850–855. [Google Scholar] [CrossRef]
- Kintner, C.; Sugden, B. Conservation and progressive methylation of Epstein-Barr viral DNA sequences in transformed cells. J. Virol. 1981, 38, 305–316. [Google Scholar] [CrossRef]
- Woellmer, A.; Arteaga-Salas, J.M.; Hammerschmidt, W. BZLF1 Governs CpG-Methylated Chromatin of Epstein-Barr Virus Reversing Epigenetic Repression. PLoS Pathog. 2012, 8, e1002902. [Google Scholar] [CrossRef] [PubMed]
- Tempera, I.; Wiedmer, A.; Dheekollu, J.; Lieberman, P.M. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog. 2010, 6, e1001048. [Google Scholar] [CrossRef] [PubMed]
- Falk, K.I.; Szekely, L.; Aleman, A.; Ernberg, I. Specific methylation patterns in two control regions of Epstein-Barr virus latency: The LMP-1-coding upstream regulatory region and an origin of DNA replication (oriP). J. Virol. 1998, 72, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.J.; Marendy, E.M.; Dickerson, C.A.; Yetming, K.D.; Sample, C.E.; Sample, J.T. Contributions of CTCF and DNA methyltransferases DNMT1 and DNMT3B to Epstein-Barr virus restricted latency. J. Virol. 2012, 86, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Arvey, A.; Tempera, I.; Tsai, K.; Chen, H.S.; Tikhmyanova, N.; Klichinsky, M.; Leslie, C.; Lieberman, P.M. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe 2012, 12, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Arvey, A.; Tempera, I.; Lieberman, P.M. Interpreting the Epstein-Barr Virus (EBV) epigenome using high-throughput data. Viruses 2013, 5, 1042–1054. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Ichikawa, T.; Okuno, Y.; Sato, Y.; Goshima, F.; Yoshiyama, H.; Kanda, T.; Kimura, H.; Murata, T. Regulation of Epstein-Barr Virus Life Cycle and Cell Proliferation by Histone H3K27 Methyltransferase EZH2 in Akata Cells. Msphere 2018, 3, e00478-18. [Google Scholar] [CrossRef]
- Murata, T.; Kondo, Y.; Sugimoto, A.; Kawashima, D.; Saito, S.; Isomura, H.; Kanda, T.; Tsurumi, T. Epigenetic histone modification of Epstein-Barr virus BZLF1 promoter during latency and reactivation in Raji cells. J. Virol. 2012, 86, 4752–4761. [Google Scholar] [CrossRef]
- Murata, T.; Tsurumi, T. Epigenetic modification of the Epstein-Barr virus BZLF1 promoter regulates viral reactivation from latency. Front. Genet. 2013, 4, 53. [Google Scholar] [CrossRef]
- Xu, H.; Li, X.; Rousseau, B.A.; Akinyemi, I.A.; Frey, T.R.; Zhou, K.; Droske, L.E.; Mitchell, J.A.; McIntosh, M.T.; Bhaduri-McIntosh, S. IFI16 Partners with KAP1 to Maintain Epstein-Barr Virus Latency. J. Virol. 2022, 96, e0102822. [Google Scholar] [CrossRef] [PubMed]
- Burton, E.M.; Akinyemi, I.A.; Frey, T.R.; Xu, H.; Li, X.; Su, L.J.; Zhi, J.; McIntosh, M.T.; Bhaduri-McIntosh, S. A heterochromatin inducing protein differentially recognizes self versus foreign genomes. PLoS Pathog. 2021, 17, e1009447. [Google Scholar] [CrossRef] [PubMed]
- Burton, E.M.; Goldbach-Mansky, R.; Bhaduri-McIntosh, S. A promiscuous inflammasome sparks replication of a common tumor virus. Proc. Natl. Acad. Sci. USA 2020, 117, 1722–1730. [Google Scholar] [CrossRef]
- Li, X.; Burton, E.M.; Koganti, S.; Zhi, J.; Doyle, F.; Tenenbaum, S.A.; Horn, B.; Bhaduri-McIntosh, S. KRAB-ZFP Repressors Enforce Quiescence of Oncogenic Human Herpesviruses. J. Virol. 2018, 92, e00298-18. [Google Scholar] [CrossRef] [PubMed]
- Lobanenkov, V.V.; Nicolas, R.H.; Adler, V.V.; Paterson, H.; Klenova, E.M.; Polotskaja, A.V.; Goodwin, G.H. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5’-flanking sequence of the chicken c-myc gene. Oncogene 1990, 5, 1743–1753. [Google Scholar]
- Klenova, E.M.; Nicolas, R.H.; Paterson, H.F.; Carne, A.F.; Heath, C.M.; Goodwin, G.H.; Neiman, P.E.; Lobanenkov, V.V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 1993, 13, 7612–7624. [Google Scholar]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar] [CrossRef]
- Chau, C.M.; Zhang, X.-Y.; McMahon, S.B.; Lieberman, P.M. Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J. Virol. 2006, 80, 5723–5732. [Google Scholar] [CrossRef]
- Day, L.; Chau, C.M.; Nebozhyn, M.; Rennekamp, A.J.; Showe, M.; Lieberman, P.M. Chromatin profiling of Epstein-Barr virus latency control region. J. Virol. 2007, 81, 6389–6401. [Google Scholar] [CrossRef]
- Holdorf, M.M.; Cooper, S.B.; Yamamoto, K.R.; Miranda, J.J.L. Occupancy of chromatin organizers in the Epstein-Barr virus genome. Virology 2011, 415, 1–5. [Google Scholar] [CrossRef]
- Lupey-Green, L.N.; Moquin, S.A.; Martin, K.A.; McDevitt, S.M.; Hulse, M.; Caruso, L.B.; Pomerantz, R.T.; Miranda, J.L.; Tempera, I. PARP1 restricts Epstein Barr Virus lytic reactivation by binding the BZLF1 promoter. Virology 2017, 507, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.S.; Martin, K.A.; Lu, F.; Lupey, L.N.; Mueller, J.M.; Lieberman, P.M.; Tempera, I. Epigenetic deregulation of the LMP1/LMP2 locus of Epstein-Barr virus by mutation of a single CTCF-cohesin binding site. J. Virol. 2014, 88, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Stedman, W.; Kang, H.; Lin, S.; Kissil, J.L.; Bartolomei, M.S.; Lieberman, P.M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008, 27, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Wiedmer, A.; Yuan, Y.; Robertson, E.; Lieberman, P.M. Coordination of KSHV Latent and Lytic Gene Control by CTCF-Cohesin Mediated Chromosome Conformation. PLoS Pathog. 2011, 7, e1002140. [Google Scholar] [CrossRef]
- Chen, H.S.; Wikramasinghe, P.; Showe, L.; Lieberman, P.M. Cohesins repress Kaposi’s sarcoma-associated herpesvirus immediate early gene transcription during latency. J. Virol. 2012, 86, 9454–9464. [Google Scholar] [CrossRef]
- Kang, H.; Cho, H.; Sung, G.H.; Lieberman, P.M. CTCF regulates Kaposi’s sarcoma-associated herpesvirus latency transcription by nucleosome displacement and RNA polymerase programming. J. Virol. 2013, 87, 1789–1799. [Google Scholar] [CrossRef]
- Li, D.J.; Verma, D.; Mosbruger, T.; Swaminathan, S. CTCF and Rad21 act as host cell restriction factors for Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic replication by modulating viral gene transcription. PLoS Pathog. 2014, 10, e1003880. [Google Scholar] [CrossRef]
- Chen, Y.J.; Chen, Y.L.; Chang, Y.; Wu, C.C.; Ko, Y.C.; Tsao, S.W.; Chen, J.Y.; Lin, S.F. Epstein-Barr Virus Rta-Mediated Accumulation of DNA Methylation Interferes with CTCF Binding in both Host and Viral Genomes. J. Virol. 2017, 91, e00736-17. [Google Scholar] [CrossRef]
- Chen, H.S.; De Leo, A.; Wang, Z.; Kerekovic, A.; Hills, R.; Lieberman, P.M. BET-Inhibitors Disrupt Rad21-Dependent Conformational Control of KSHV Latency. PLoS Pathog. 2017, 13, e1006100. [Google Scholar] [CrossRef]
- Li, D.; Mosbruger, T.; Verma, D.; Swaminathan, S. Complex Interactions between Cohesin and CTCF in Regulation of Kaposi’s Sarcoma-Associated Herpesvirus Lytic Transcription. J. Virol. 2020, 94, e01279-19. [Google Scholar] [CrossRef]
- Campbell, M.; Chantarasrivong, C.; Yanagihashi, Y.; Inagaki, T.; Davis, R.R.; Nakano, K.; Kumar, A.; Tepper, C.G.; Izumiya, Y. KSHV Topologically Associating Domains in Latent and Reactivated Viral Chromatin. J. Virol. 2022, 96, e0056522. [Google Scholar] [CrossRef] [PubMed]
- Amelio, A.L.; McAnany, P.K.; Bloom, D.C. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J. Virol. 2006, 80, 2358–2368. [Google Scholar] [CrossRef] [PubMed]
- Ertel, M.K.; Cammarata, A.L.; Hron, R.J.; Neumann, D.M. CTCF occupation of the herpes simplex virus 1 genome is disrupted at early times postreactivation in a transcription-dependent manner. J. Virol. 2012, 86, 12741–12759. [Google Scholar] [CrossRef] [PubMed]
- Lang, F.; Li, X.; Vladimirova, O.; Hu, B.; Chen, G.; Xiao, Y.; Singh, V.; Lu, D.; Li, L.; Han, H.; et al. CTCF interacts with the lytic HSV-1 genome to promote viral transcription. Sci. Rep. 2017, 7, 39861. [Google Scholar] [CrossRef] [PubMed]
- Washington, S.D.; Edenfield, S.I.; Lieux, C.; Watson, Z.L.; Taasan, S.M.; Dhummakupt, A.; Bloom, D.C.; Neumann, D.M. Depletion of the Insulator Protein CTCF Results in Herpes Simplex Virus 1 Reactivation In Vivo. J. Virol. 2018, 92, e00173-18. [Google Scholar] [CrossRef]
- Washington, S.D.; Musarrat, F.; Ertel, M.K.; Backes, G.L.; Neumann, D.M. CTCF Binding Sites in the Herpes Simplex Virus 1 Genome Display Site-Specific CTCF Occupation, Protein Recruitment, and Insulator Function. J. Virol. 2018, 92, e00156-18. [Google Scholar] [CrossRef]
- Singh, P.; Collins, M.F.; Johns, R.N.; Manuel, K.A.; Ye, Z.A.; Bloom, D.C.; Neumann, D.M. Deletion of the CTRL2 Insulator in HSV-1 Results in the Decreased Expression of Genes Involved in Axonal Transport and Attenuates Reactivation In Vivo. Viruses 2022, 14, 909. [Google Scholar] [CrossRef]
- Singh, P.; Neumann, D.M. Cohesin subunit Rad21 binds to the HSV-1 genome near CTCF insulator sites during latency in vivo. J. Virol. 2021, 95, e00364-21. [Google Scholar] [CrossRef]
- Martínez, F.P.; Cruz, R.; Lu, F.; Plasschaert, R.; Deng, Z.; Rivera-Molina, Y.A.; Bartolomei, M.S.; Lieberman, P.M.; Tang, Q. CTCF binding to the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication. J. Virol. 2014, 88, 7389–7401. [Google Scholar] [CrossRef]
- Mehta, K.; Gunasekharan, V.; Satsuka, A.; Laimins, L.A. Human papillomaviruses activate and recruit SMC1 cohesin proteins for the differentiation-dependent life cycle through association with CTCF insulators. PLoS Pathog. 2015, 11, e1004763. [Google Scholar] [CrossRef]
- Paris, C.; Pentland, I.; Groves, I.; Roberts, D.C.; Powis, S.J.; Coleman, N.; Roberts, S.; Parish, J.L. CCCTC-binding factor recruitment to the early region of the human papillomavirus 18 genome regulates viral oncogene expression. J. Virol. 2015, 89, 4770–4785. [Google Scholar] [CrossRef]
- Pentland, I.; Campos-León, K.; Cotic, M.; Davies, K.J.; Wood, C.D.; Groves, I.J.; Burley, M.; Coleman, N.; Stockton, J.D.; Noyvert, B.; et al. Disruption of CTCF-YY1-dependent looping of the human papillomavirus genome activates differentiation-induced viral oncogene transcription. PLoS Biol. 2018, 16, e2005752. [Google Scholar] [CrossRef] [PubMed]
- Degner, S.C.; Verma-Gaur, J.; Wong, T.P.; Bossen, C.; Iverson, G.M.; Torkamani, A.; Vettermann, C.; Lin, Y.C.; Ju, Z.; Schulz, D.; et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc. Natl. Acad. Sci. USA 2011, 108, 9566–9571. [Google Scholar] [CrossRef] [PubMed]
- Millau, J.-F.; Gaudreau, L. CTCF, cohesin, and histone variants: Connecting the genome. Biochem. Cell Biol. 2011, 89, 505–513. [Google Scholar] [CrossRef]
- Degner, S.C.; Wong, T.P.; Jankevicius, G.; Feeney, A.J. Cutting edge: Developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J. Immunol. 2009, 182, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.; Peters, J.M.; Uhlmann, F. Splitting the chromosome: Cutting the ties that bind sister chromatids. Science 2000, 288, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Losada, A.; Hirano, T. Shaping the metaphase chromosome: Coordination of cohesion and condensation. Bioessays 2001, 23, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Kojic, A.; Cuadrado, A.; De Koninck, M.; Giménez-Llorente, D.; Rodríguez-Corsino, M.; Gómez-López, G.; Le Dily, F.; Marti-Renom, M.A.; Losada, A. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat. Struct. Mol. Biol. 2018, 25, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Mach, P.; Kos, P.I.; Zhan, Y.; Cramard, J.; Gaudin, S.; Tünnermann, J.; Marchi, E.; Eglinger, J.; Zuin, J.; Kryzhanovska, M.; et al. Cohesin and CTCF control the dynamics of chromosome folding. Nat. Genet. 2022, 54, 1907–1918. [Google Scholar] [CrossRef]
- Reisman, D.; Sugden, B. Trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol. Cell. Biol. 1986, 6, 3838–3846. [Google Scholar]
- Guo, R.; Jiang, C.; Zhang, Y.; Govande, A.; Trudeau, S.J.; Chen, F.; Fry, C.J.; Puri, R.; Wolinsky, E.; Schineller, M.; et al. MYC Controls the Epstein-Barr Virus Lytic Switch. Mol. Cell 2020, 78, 653–669. [Google Scholar] [CrossRef]
- Lupey-Green, L.N.; Caruso, L.B.; Madzo, J.; Martin, K.A.; Tan, Y.; Hulse, M.; Tempera, I. PARP1 Stabilizes CTCF Binding and Chromatin Structure To Maintain Epstein-Barr Virus Latency Type. J. Virol. 2018, 92, e00755-18. [Google Scholar] [CrossRef] [PubMed]
- Messner, S.; Altmeyer, M.; Zhao, H.; Pozivil, A.; Roschitzki, B.; Gehrig, P.; Rutishauser, D.; Huang, D.; Caflisch, A.; Hottiger, M.O. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010, 38, 6350–6362. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Ginjala, V.; Pant, V.; Chernukhin, I.; Whitehead, J.; Docquier, F.; Farrar, D.; Tavoosidana, G.; Mukhopadhyay, R.; Kanduri, C.; et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 2004, 36, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Farrar, D.; Rai, S.; Chernukhin, I.; Jagodic, M.; Ito, Y.; Yammine, S.; Ohlsson, R.; Murrell, A.; Klenova, E. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol. Cell. Biol. 2010, 30, 1199–1216. [Google Scholar] [CrossRef]
- Martin, K.A.; Lupey, L.N.; Tempera, I. Epstein-Barr Virus Oncoprotein LMP1 Mediates Epigenetic Changes in Host Gene Expression through PARP1. J. Virol. 2016, 90, 8520–8530. [Google Scholar] [CrossRef]
- Grady, S.L.; Hwang, J.; Vastag, L.; Rabinowitz, J.D.; Shenk, T. Herpes simplex virus 1 infection activates poly(ADP-ribose) polymerase and triggers the degradation of poly(ADP-ribose) glycohydrolase. J. Virol. 2012, 86, 8259–8268. [Google Scholar] [CrossRef]
- Li, Z.; Yamauchi, Y.; Kamakura, M.; Murayama, T.; Goshima, F.; Kimura, H.; Nishiyama, Y. Herpes simplex virus requires poly(ADP-ribose) polymerase activity for efficient replication and induces extracellular signal-related kinase-dependent phosphorylation and ICP0-dependent nuclear localization of tankyrase 1. J. Virol. 2012, 86, 492–503. [Google Scholar] [CrossRef]
- Ohsaki, E.; Ueda, K.; Sakakibara, S.; Do, E.; Yada, K.; Yamanishi, K. Poly(ADP-ribose) polymerase 1 binds to Kaposi’s sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol. 2004, 78, 9936–9946. [Google Scholar] [CrossRef]
- Chung, W.C.; Park, J.H.; Kang, H.R.; Song, M.J. Downregulation of Poly(ADP-Ribose) Polymerase 1 by a Viral Processivity Factor Facilitates Lytic Replication of Gammaherpesvirus. J. Virol. 2015, 89, 9676–9682. [Google Scholar] [CrossRef]
- Gwack, Y.; Nakamura, H.; Lee, S.H.; Souvlis, J.; Yustein, J.T.; Gygi, S.; Kung, H.J.; Jung, J.U. Poly(ADP-ribose) polymerase 1 and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol. Cell. Biol. 2003, 23, 8282–8294. [Google Scholar] [CrossRef] [PubMed]
- Peric-Hupkes, D.; van Steensel, B. Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Oldenburg, A.R.; Delbarre, E.; Freberg, C.T.; Duband-Goulet, I.; Eskeland, R.; Buendia, B.; Collas, P. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome. Res. 2013, 23, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar] [CrossRef]
- Worman, H.J.; Lazaridis, I.; Georgatos, S.D. Nuclear lamina heterogeneity in mammalian cells. Differential expression of the major lamins and variations in lamin B phosphorylation. J. Biol. Chem. 1988, 263, 12135–12141. [Google Scholar] [CrossRef]
- Hutchison, C.J.; Bridger, J.M.; Cox, L.S.; Kill, I.R. Weaving a pattern from disparate threads: Lamin function in nuclear assembly and DNA replication. J. Cell Sci. 1994, 107 Pt 12, 3259–3269. [Google Scholar] [CrossRef]
- de Noronha, C.M.; Sherman, M.P.; Lin, H.W.; Cavrois, M.V.; Moir, R.D.; Goldman, R.D.; Greene, W.C. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science 2001, 294, 1105–1108. [Google Scholar] [CrossRef]
- Gonnella, R.; Farina, A.; Santarelli, R.; Raffa, S.; Feederle, R.; Bei, R.; Granato, M.; Modesti, A.; Frati, L.; Delecluse, H.J.; et al. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: Interactions with BFRF1 and with the nuclear lamina. J. Virol. 2005, 79, 3713–3727. [Google Scholar] [CrossRef]
- Scott, E.S.; O’Hare, P. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 2001, 75, 8818–8830. [Google Scholar] [CrossRef]
- Muranyi, W.; Haas, J.; Wagner, M.; Krohne, G.; Koszinowski, U.H. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 2002, 297, 854–857. [Google Scholar] [CrossRef]
- Caruso, L.B.; Guo, R.; Keith, K.; Madzo, J.; Maestri, D.; Boyle, S.; Wasserman, J.; Kossenkov, A.; Gewurz, B.E.; Tempera, I. The nuclear lamina binds the EBV genome during latency and regulates viral gene expression. PLoS Pathog. 2022, 18, e1010400. [Google Scholar] [CrossRef] [PubMed]
- Okabe, A.; Huang, K.K.; Matsusaka, K.; Fukuyo, M.; Xing, M.; Ong, X.; Hoshii, T.; Usui, G.; Seki, M.; Mano, Y.; et al. Cross-species chromatin interactions drive transcriptional rewiring in Epstein–Barr virus–positive gastric adenocarcinoma. Nat. Genet. 2020, 52, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Moquin, S.A.; Thomas, S.; Whalen, S.; Warburton, A.; Fernandez, S.G.; McBride, A.A.; Pollard, K.S.; Miranda, J.L. The Epstein-Barr Virus Episome Maneuvers between Nuclear Chromatin Compartments during Reactivation. J. Virol. 2018, 92, e01413-17. [Google Scholar] [CrossRef] [PubMed]
- Poleshko, A.; Smith, C.L.; Nguyen, S.C.; Sivaramakrishnan, P.; Wong, K.G.; Murray, J.I.; Lakadamyali, M.; Joyce, E.F.; Jain, R.; Epstein, J.A. H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis. Elife 2019, 8, e49278. [Google Scholar] [CrossRef]
- Holla, S.; Dhakshnamoorthy, J.; Folco, H.D.; Balachandran, V.; Xiao, H.; Sun, L.L.; Wheeler, D.; Zofall, M.; Grewal, S.I. Positioning Heterochromatin at the Nuclear Periphery Suppresses Histone Turnover to Promote Epigenetic Inheritance. Cell 2020, 180, 150–164. [Google Scholar] [CrossRef]
- Zhao, B.; Zou, J.; Wang, H.; Johannsen, E.; Peng, C.W.; Quackenbush, J.; Mar, J.C.; Morton, C.C.; Freedman, M.L.; Blacklow, S.C.; et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc. Natl. Acad. Sci. USA 2011, 8, 14902–14907. [Google Scholar] [CrossRef]
- Ding, W.; Wang, C.; Narita, Y.; Wang, H.; Leong, M.M.L.; Huang, A.; Liao, Y.; Liu, X.; Okuno, Y.; Kimura, H.; et al. The Epstein-Barr Virus Enhancer Interaction Landscapes in Virus-Associated Cancer Cell Lines. J. Virol. 2022, 96, e0073922. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caruso, L.B.; Maestri, D.; Tempera, I. Three-Dimensional Chromatin Structure of the EBV Genome: A Crucial Factor in Viral Infection. Viruses 2023, 15, 1088. https://doi.org/10.3390/v15051088
Caruso LB, Maestri D, Tempera I. Three-Dimensional Chromatin Structure of the EBV Genome: A Crucial Factor in Viral Infection. Viruses. 2023; 15(5):1088. https://doi.org/10.3390/v15051088
Chicago/Turabian StyleCaruso, Lisa Beatrice, Davide Maestri, and Italo Tempera. 2023. "Three-Dimensional Chromatin Structure of the EBV Genome: A Crucial Factor in Viral Infection" Viruses 15, no. 5: 1088. https://doi.org/10.3390/v15051088
APA StyleCaruso, L. B., Maestri, D., & Tempera, I. (2023). Three-Dimensional Chromatin Structure of the EBV Genome: A Crucial Factor in Viral Infection. Viruses, 15(5), 1088. https://doi.org/10.3390/v15051088






