The Burden of Epstein–Barr Virus (EBV) and Its Determinants among Adult HIV-Positive Individuals in Ethiopia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Time Period, and Setting
2.2. Source and Study Population
2.3. Inclusion and Exclusion Criteria
2.4. Study Variables
2.5. Sample Collection
2.6. Sample Preparation
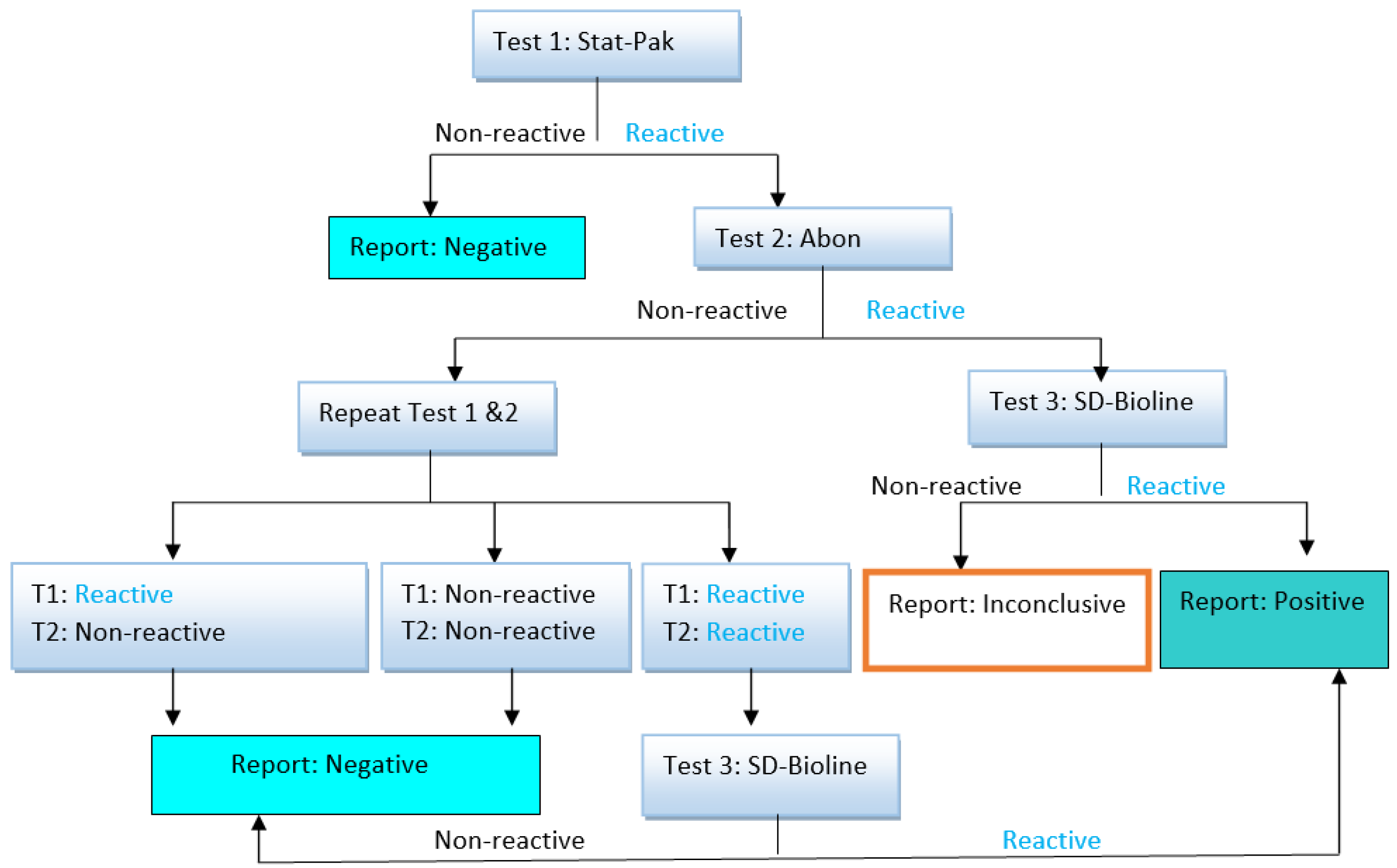
2.7. HIV Serology Test
2.8. EBV Serology Test
2.9. DNA Extraction
2.10. Real-Time Quantification PCR from Genomic DNA for EBV Load/Copy Number Determination
2.11. Data Collection and Quality Assurance
2.12. Statistical Analysis
3. Results
3.1. Socio-Demographic Characteristics
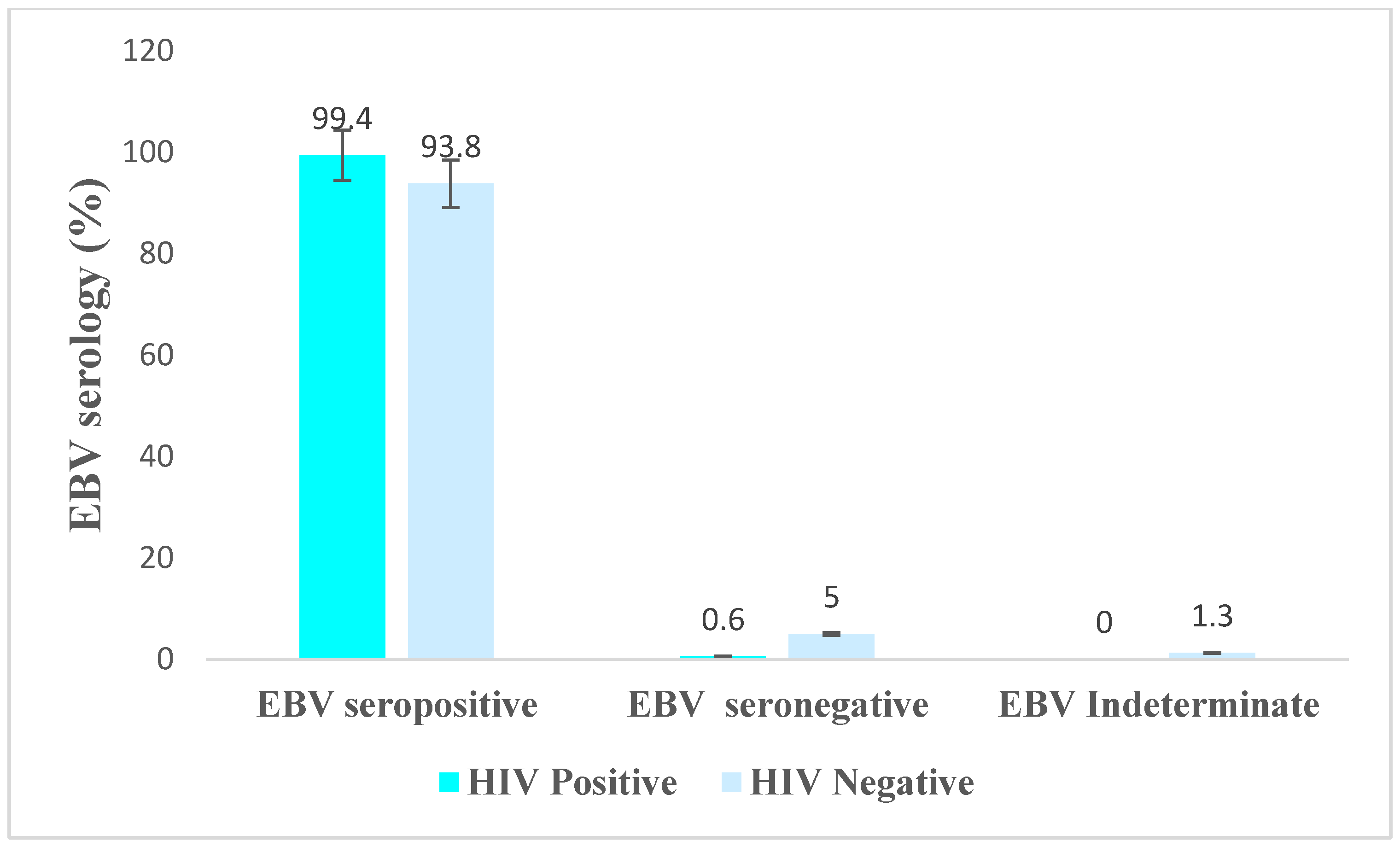
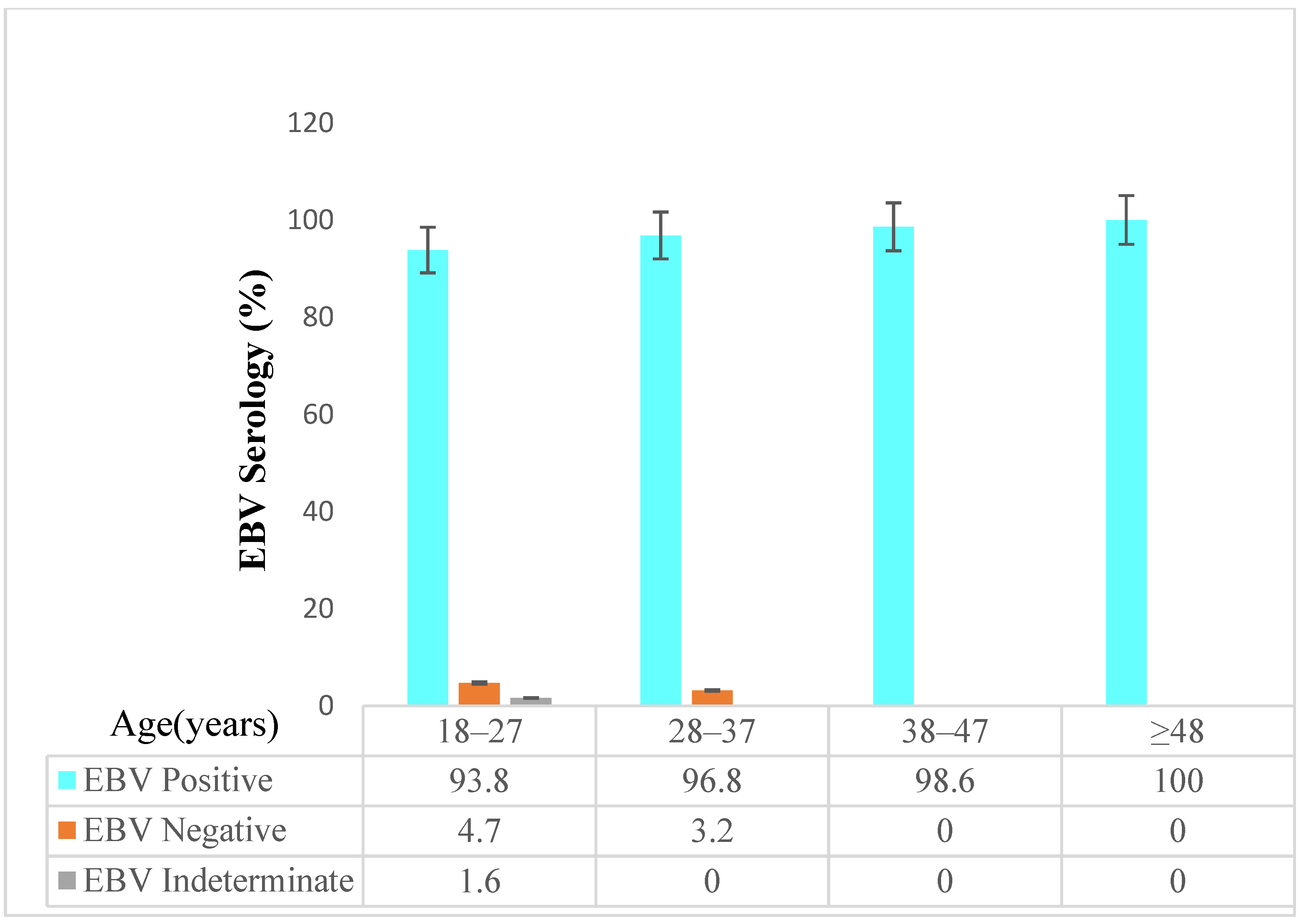
3.2. Detection of EBV Antibody
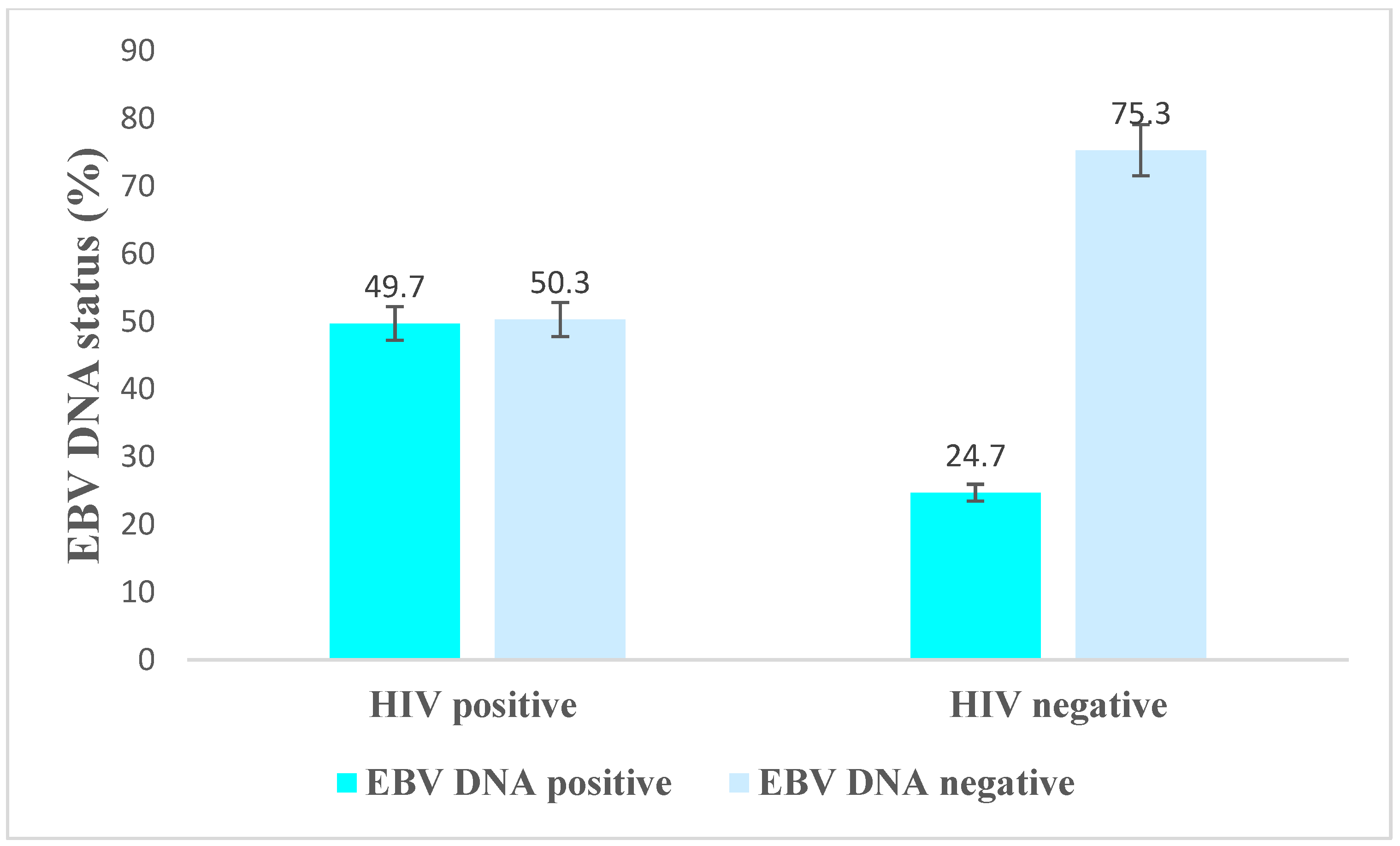
3.3. EBV DNA Positivity by Demographic and Clinical Characteristics among HIV-Positive Individuals
3.4. Associated Factors of EBV DNA Positivity among HIV-Positive and HIV-Negative Individuals
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADC | AIDS-defining cancer |
| AIDS | Acquired immune deficiency syndrome |
| ART | Antiretroviral therapy |
| cART | Combined antiretroviral therapy |
| DNA | Deoxyribonucleic acid |
| DLBCL | Diffuse large B cell lymphoma |
| DMSO | Dimethyl sulfoxide |
| EBER | Epstein–Barr virus (EBV)-encoded small RNAs |
| EBNA 1 | Epstein–Barr nuclear antigen 1 |
| EBV | Epstein–Barr virus |
| HBV | Hepatitis B virus |
| HCV | Hepatitis C virus |
| HHV-8 | Human herpesvirus-8 |
| HIV | Human immunodeficiency viruses |
| HL | Hodgkin lymphoma |
| HPV | Human papillomavirus |
| IgG | Immunoglobulin G |
| KSHV | Kaposi’s sarcoma-associated herpesvirus |
| NHL | Non-Hodgkin lymphoma |
| PBMCs | Peripheral blood mononuclear cells |
| PBS | Phosphate buffer saline |
| PCR | Polymerase chain reaction |
| PWH | People living with HIV |
| qRT PCR | Quantitative real-time polymerase chain reaction |
| RDT | Rapid diagnostic test |
| RNA | Ribonucleic acid |
| SSA | Sub-Saharan Africa |
| TASH | Tikur Anbessa Specialised Hospital |
| VCA | Viral capsid antigens |
References
- Shahriar, S.; Araf, Y.; Ahmad, R.; Kattel, P.; Sah, G.S.; Rahaman, T.I.; Sadiea, R.Z.; Sultana, S.; Islam, M.S.; Zheng, C.; et al. Insights Into the Coinfections of Human Immunodeficiency Virus-Hepatitis B Virus, Human Immunodeficiency Virus-Hepatitis C Virus, and Hepatitis B Virus-Hepatitis C Virus: Prevalence, Risk Factors, Pathogenesis, Diagnosis, and Treatment. Front. Microbiol. 2022, 12, 780887. [Google Scholar] [CrossRef]
- Riedel, D.J.; Tang, L.S.; Rositch, A.F. The role of viral co-infection in HIV-associated non-AIDS-related cancers. Curr. HIV/AIDS Rep. 2015, 12, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Calabresi, A.; Ferraresi, A.; Festa, A.; Scarcella, C.; Donato, F.; Vassallo, F.; Limina, R.M.; Castelli, F.; Quiros-Roldan, E.; Brescia HIV Cancer Study Group. Incidence of AIDS-defining cancers and virus-related and non-virus-related non-AIDS-defining cancers among HIV-infected patients compared with the general population in a large health district of Northern Italy, 1999–2009. HIV Med. 2013, 14, 481–490. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin-Drubin, M.E.; Munger, K. Viruses associated with human cancer. Biochim. Biophys. Acta 2008, 1782, 127–150. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A. The Global Landscape of EBV-Associated Tumors. Front. Oncol. 2019, 9, 713. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A. EBV persistence—Introducing the virus. In Epstein Barr Virus Volume 1; Springer: Berlin/Heidelberg, Germany, 2015; pp. 151–209. [Google Scholar]
- Bibas, M.; Antinori, A. EBV and HIV-related lymphoma. Mediterr. J. Hematol. Infect. Dis. 2009, 1, e2009032. [Google Scholar] [CrossRef] [PubMed]
- Ling, P.D.; Vilchez, R.A.; Keitel, W.A.; Poston, D.G.; Peng, R.S.; White, Z.S.; Visnegarwala, F.; Lewis, D.E.; Butel, J.S. Epstein-Barr virus DNA loads in adult human immunodeficiency virus type 1-infected patients receiving highly active antiretroviral therapy. Clin. Infect. Dis. 2003, 37, 1244–1249. [Google Scholar] [CrossRef] [PubMed]
- Merlo, A.; Turrini, R.; Dolcetti, R.; Martorelli, D.; Muraro, E.; Comoli, P.; Rosato, A. The interplay between Epstein-Barr virus and the immune system: A rationale for adoptive cell therapy of EBV-related disorders. Haematologica 2010, 95, 1769. [Google Scholar] [CrossRef] [PubMed]
- Shindiapina, P.; Ahmed, E.H.; Mozhenkova, A.; Abebe, T.; Baiocchi, R.A. Immunology of EBV-Related Lymphoproliferative Disease in HIV-Positive Individuals. Front. Oncol. 2020, 10, 1723. [Google Scholar] [CrossRef]
- Mandal, D.; Desai, D.; Sinha, S. High prevalence of plasma EBV DNA among the HIV positive individuals, with or without malignancies, attending the clinic at AIIMS, New Delhi. Virusdisease 2021, 32, 137–139. [Google Scholar] [CrossRef]
- Kharsany, A.B.; Karim, Q.A. HIV infection and AIDS in sub-Saharan Africa: Current status, challenges and opportunities. Open AIDS J. 2016, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- The Ethiopian Public Health Institute, Ministry of Health. HIV Related Estimates and Projections in Ethiopia for the Year 2021–2022; The Ethiopian Public Health Institute: Addis Ababa, Ethiopia, 2022. [Google Scholar]
- Central Statistical Agency. Ethiopia Demographic and Health Survey; Central Statistical Agency: Addis Ababa, Ethiopia, 2016. [Google Scholar]
- Ethiopian Public Health Institute. Ethiopia Population-Based HIV Impact Assessment 2017–2018; Ethiopian Public Health Institute: Addis Ababa, Ethiopia, 2020. [Google Scholar]
- Memirie, S.T.; Habtemariam, M.K.; Asefa, M.; Deressa, B.T.; Abayneh, G.; Tsegaye, B.; Abraha, M.W.; Ababi, G.; Jemal, A.; Rebbeck, T.R. Estimates of cancer incidence in Ethiopia in 2015 using population-based registry data. J. Glob. Oncol. 2018, 4, 1–11. [Google Scholar] [CrossRef]
- Palella, F.J., Jr.; Delaney, K.M.; Moorman, A.C.; Loveless, M.O.; Fuhrer, J.; Satten, G.A.; Aschman, D.J.; Holmberg, S.D.; Investigators, H.O.S. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 1998, 338, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Simard, E.P.; Pfeiffer, R.M.; Engels, E.A. Cumulative incidence of cancer among individuals with acquired immunodeficiency syndrome in the United States. Cancer 2011, 117, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Anselmi, A.; Vendrame, D.; Rampon, O.; Giaquinto, C.; Zanchetta, M.; De Rossi, A. Immune reconstitution in human immunodeficiency virus type 1-infected children with different virological responses to anti-retroviral therapy. Clin. Exp. Immunol. 2007, 150, 442–450. [Google Scholar] [CrossRef]
- Sachithanandham, J.; Kannangai, R.; Pulimood, S.; Desai, A.; Abraham, A.; Abraham, O.; Ravi, V.; Samuel, P.; Sridharan, G. Significance of Epstein-Barr virus (HHV-4) and CMV (HHV-5) infection among subtype-C human immunodeficiency virus-infected individuals. Indian J. Med. Microbiol. 2014, 32, 261–269. [Google Scholar] [CrossRef]
- Ministry of Health. National Guidelines for Comprehensive HIV Prevention, Care and Treatment; Ministry of Health: Addis Ababa, Ethiopia, 2018. [Google Scholar]
- Ahmed, E.H.; Lustberg, M.; Hale, C.; Sloan, S.; Mao, C.; Zhang, X.; Ozer, H.G.; Schlotter, S.; Smith, P.L.; Jeney, F. Follicular Helper and Regulatory T Cells Drive the Development of Spontaneous Epstein–Barr Virus Lymphoproliferative Disorder. Cancers 2023, 15, 3046. [Google Scholar] [CrossRef]
- Cesarman, E. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett. 2011, 305, 163–174. [Google Scholar] [CrossRef]
- Luzuriaga, K.; Sullivan, J.L. Infectious Mononucleosis. N. Engl. J. Med. 2010, 362, 1993–2000. [Google Scholar] [CrossRef]
- Xu, G.J.; Kula, T.; Xu, Q.; Li, M.Z.; Vernon, S.D.; Ndung’u, T.; Ruxrungtham, K.; Sanchez, J.; Brander, C.; Chung, R.T. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015, 348, aaa0698. [Google Scholar] [CrossRef]
- Keramat, F.; Basir, H.R.G.; Ahmadpour, S.; Moradi, A. Seroprevalence of Epstein-Barr Virus in HIV-Infected Patients, A Case Control Study. J. Kermanshah Univ. Med. Sci. 2019, 23, e84967. [Google Scholar] [CrossRef]
- McClain, K.L.; Leach, C.T.; Jenson, H.B.; Joshi, V.V.; Pollock, B.H.; Parmley, R.T.; DiCarlo, F.J.; Chadwick, E.G.; Murphy, S.B. Association of Epstein–Barr virus with leiomyosarcomas in young people with AIDS. N. Engl. J. Med. 1995, 332, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Kyaw, M.T.; Hurren, L.; Evans, L.; Moss, D.J.; Cooper, D.A.; Benson, E.; Esmore, D.; Sculley, T.B. Expression of B-type Epstein-Barr virus in HIV-infected patients and cardiac transplant recipients. AIDS Res. Hum. Retroviruses 1992, 8, 1869–1874. [Google Scholar] [CrossRef] [PubMed]
- Schaftenaar, E.; Verjans, G.M.; Getu, S.; McIntyre, J.A.; Struthers, H.E.; Osterhaus, A.D.; Peters, R.P. High seroprevalence of human herpesviruses in HIV-infected individuals attending primary healthcare facilities in rural South Africa. PLoS ONE 2014, 9, e99243. [Google Scholar] [CrossRef] [PubMed]
- Adjei, A.A.; Armah, H.B.; Gbagbo, F.; Boamah, I.; Adu-Gyamfi, C.; Asare, I. Seroprevalence of HHV-8, CMV, and EBV among the general population in Ghana, West Africa. BMC Infect. Dis. 2008, 8, 111. [Google Scholar] [CrossRef]
- Sharifipour, S.; Rad, K.D. Seroprevalence of Epstein–Barr virus among children and adults in Tehran, Iran. New Microbes New Infect. 2020, 34, 100641. [Google Scholar] [CrossRef]
- Smatti, M.K.; Yassine, H.M.; AbuOdeh, R.; AlMarawani, A.; Taleb, S.A.; Althani, A.A.; Nasrallah, G.K. Prevalence and molecular profiling of Epstein Barr virus (EBV) among healthy blood donors from different nationalities in Qatar. PLoS ONE 2017, 12, e0189033. [Google Scholar] [CrossRef]
- Corcoran, C.; Rebe, K.; van der Plas, H.; Myer, L.; Hardie, D.R. The predictive value of cerebrospinal fluid Epstein-Barr viral load as a marker of primary central nervous system lymphoma in HIV-infected persons. J. Clin. Virol. 2008, 42, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Opintan, J.A.; Awadzi, B.K.; Biney, I.J.; Ganu, V.; Doe, R.; Kenu, E.; Adu, R.F.; Osei, M.M.; Akumwena, A.; Grigg, M.E. High rates of cerebral toxoplasmosis in HIV patients presenting with meningitis in Accra, Ghana. Trans. R. Soc. Trop. Med. Hyg. 2017, 111, 464–471. [Google Scholar] [CrossRef]
- Rajasingham, R.; Rhein, J.; Klammer, K.; Musubire, A.; Nabeta, H.; Akampurira, A.; Mossel, E.C.; Williams, D.A.; Boxrud, D.J.; Crabtree, M.B. Epidemiology of meningitis in an HIV-infected Ugandan cohort. Am. J. Trop. Med. Hyg. 2015, 92, 274. [Google Scholar] [CrossRef]
- Kelly, M.J.; Benjamin, L.A.; Cartwright, K.; Ajdukiewicz, K.M.; Cohen, D.B.; Menyere, M.; Galbraith, S.; Guiver, M.; Neuhann, F.; Solomon, T. Epstein-barr virus coinfection in cerebrospinal fluid is associated with increased mortality in Malawian adults with bacterial meningitis. J. Infect. Dis. 2012, 205, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Liu, X.; Zhou, S.; Ning, Z.; Zheng, H.; Gao, M.; Ding, Y.; Yao, W.; Liao, X.; He, N. Differential prevalence and correlates of whole blood Epstein–Barr virus DNA between HIV-positive and HIV-negative men who have sex with men in Shanghai, China. Epidemiol. Infect. 2017, 145, 2330–2340. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ammatuna, P.; Campisi, G.; Giovannelli, L.; Giambelluca, D.; Alaimo, C.; Mancuso, S.; Margiotta, V. Presence of Epstein–Barr virus, cytomegalovirus and human papillomavirus in normal oral mucosa of HIV-infected and renal transplant patients. Oral Dis. 2001, 7, 34–40. [Google Scholar] [PubMed]
- Santos, L.; Azevedo, K.; Silva, L.; Oliveira, L. Epstein-Barr virus in oral mucosa from human immunodeficiency virus positive patients. Rev. Assoc. Médica Bras. 2014, 60, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Nishiwaki, M.; Fujimuro, M.; Teishikata, Y.; Inoue, H.; Sasajima, H.; Nakaso, K.; Nakashima, K.; Sadanari, H.; Yamamoto, T.; Fujiwara, Y. Epidemiology of Epstein–Barr virus, cytomegalovirus, and kaposi’s sarcoma-associated herpesvirus infections in peripheral blood leukocytes revealed by a multiplex PCR assay. J. Med. Virol. 2006, 78, 1635–1642. [Google Scholar] [CrossRef]
- Sousa, H.; Silva, J.; Azevedo, L.; Pinto-Correia, A.L.; Catarino, R.; Pinto, D.; Lopes, C.; Medeiros, R. Epstein-Barr virus in healthy individuals from Portugal. Acta Médica Port. 2011, 24, 707–712. [Google Scholar]
- Abusalah, M.A.H.; Gan, S.H.; Al-Hatamleh, M.A.; Irekeola, A.A.; Shueb, R.H.; Yean Yean, C. Recent advances in diagnostic approaches for epstein–barr virus. Pathogens 2020, 9, 226. [Google Scholar] [CrossRef]
- Liao, H.M.; Liu, H.; Chin, P.J.; Li, B.; Hung, G.C.; Tsai, S.; Otim, I.; Legason, I.D.; Ogwang, M.D.; Reynolds, S.J.; et al. Epstein-Barr Virus in Burkitt Lymphoma in Africa Reveals a Limited Set of Whole Genome and LMP-1 Sequence Patterns: Analysis of Archival Datasets and Field Samples From Uganda, Tanzania, and Kenya. Front. Oncol. 2022, 12, 812224. [Google Scholar] [CrossRef]
- Lupo, J.; Germi, R.; Lancar, R.; Algarte-Genin, M.; Hendel-Chavez, H.; Taoufik, Y.; Mounier, N.; Partisani, M.; Bonnet, F.; Meyohas, M.-C. Prospective evaluation of blood Epstein–Barr virus DNA load and antibody profile in HIV-related non-Hodgkin lymphomas. AIDS 2021, 35, 861–868. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Wen, Y. Lessons from Epstein-Barr virus DNA detection in cerebrospinal fluid as a diagnostic tool for EBV-induced central nervous system dysfunction among HIV-positive patients. Biomed. Pharmacother. 2022, 145, 112392. [Google Scholar] [CrossRef]
- Mbopi-Kéou, F.-X.; Bélec, L.; Teo, C.G.; Scully, C.; Porter, S.R. Synergism between HIV and other viruses in the mouth. Lancet Infect. Dis. 2002, 2, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Timotewos, G.; Solomon, A.; Mathewos, A.; Addissie, A.; Bogale, S.; Wondemagegnehu, T.; Aynalem, A.; Ayalnesh, B.; Dagnechew, H.; Bireda, W. First data from a population based cancer registry in Ethiopia. Cancer Epidemiol. 2018, 53, 93–98. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | HIV Status | Total (260) | % | ||||
|---|---|---|---|---|---|---|---|
| Positive (n = 179) | % | Negative (n = 81) | % | ||||
| Sex | Male | 66 | 36.9 | 31 | 38.3 | 97 | 37.3 |
| Female | 113 | 63.1 | 50 | 61.7 | 163 | 62.7 | |
| Age | 18–28 | 25 | 14.0 | 39 | 48.1 | 64 | 24.6 |
| 29–38 | 38 | 21.2 | 25 | 30.9 | 63 | 24.2 | |
| 39–48 | 59 | 33.0 | 12 | 14.8 | 71 | 27.3 | |
| >49 | 57 | 31.8 | 5 | 6.2 | 62 | 23.8 | |
| Marital status | Unmarried | 51 | 28.5 | 46 | 56.8 | 97 | 37.3 |
| Married | 74 | 41.3 | 31 | 38.3 | 105 | 40.4 | |
| Divorced | 23 | 12.8 | 4 | 4.9 | 27 | 10.4 | |
| Widowed | 31 | 17.3 | 0 | 0.0 | 31 | 11.9 | |
| Educational status | Illiterate | 16 | 8.9 | 0 | 0.0 | 16 | 6.2 |
| Primary School | 57 | 31.8 | 9 | 11.1 | 66 | 25.4 | |
| Secondary School | 76 | 42.5 | 28 | 34.6 | 104 | 40.0 | |
| College diploma and above | 30 | 16.8 | 44 | 54.3 | 74 | 28.5 | |
| Source of income | Government employee | 26 | 14.5 | 62 | 76.5 | 88 | 33.8 |
| Stay-at-home spouse | 27 | 15.1 | 0 | 0.0 | 27 | 10.4 | |
| Private sector employee | 53 | 29.6 | 0 | 0.0 | 53 | 20.4 | |
| Self-employed | 43 | 24.0 | 0 | 0.0 | 43 | 16.5 | |
| Student | 18 | 10.1 | 1 | 1.2 | 19 | 7.3 | |
| Unemployed | 12 | 6.7 | 18 | 22.2 | 30 | 11.5 | |
| Average monthly income in ETB | <1000 | 52 | 29.1 | 1 | 1.2 | 53 | 20.4 |
| 1001–2000 | 37 | 20.7 | 13 | 16.0 | 50 | 19.2 | |
| 2001–3000 | 25 | 14.0 | 12 | 14.8 | 37 | 14.2 | |
| 3001–4000 | 15 | 8.4 | 26 | 32.1 | 41 | 15.8 | |
| >4000 | 19 | 10.6 | 11 | 13.6 | 30 | 11.5 | |
| NA | 31 | 17.3 | 18 | 22.2 | 49 | 18.8 | |
| Characteristics | EBV DNA Status | Total | % | X2 | p-Value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | % | Negative | % | ||||||
| Sex | Male | 30 | 45.5% | 36 | 54.5% | 66 | 37% | 0.76 | 0.383 |
| Female | 59 | 52.2% | 54 | 47.8% | 113 | 63% | |||
| Age | <35 | 20 | 36.4% | 35 | 63.6% | 55 | 31% | 0.57 | 0.017 |
| ≥35 | 69 | 55.6% | 55 | 44.4% | 124 | 69% | |||
| Marital status | Unmarried | 23 | 45.1% | 28 | 54.9% | 51 | 28% | 6.85 | 0.077 |
| Married | 33 | 44.6% | 41 | 55.4% | 74 | 41% | |||
| Divorced | 11 | 47.8% | 12 | 52.2% | 23 | 13% | |||
| Widowed | 22 | 71.0% | 9 | 29.0% | 31 | 17% | |||
| Educational status | Illiterate | 9 | 56.3% | 7 | 43.8% | 16 | 9% | 1.53 | 0.676 |
| Primary school | 31 | 54.4% | 26 | 45.6% | 57 | 32% | |||
| Secondary school | 34 | 44.7% | 42 | 55.3% | 76 | 42% | |||
| College diploma and above | 15 | 50.0% | 15 | 50.0% | 30 | 17% | |||
| Source of income | Government employee | 15 | 57.7% | 11 | 42.3% | 26 | 15% | 12.62 | 0.027 |
| Stay-at-home spouse | 17 | 63.0% | 10 | 37.0% | 27 | 15% | |||
| Private sector employee | 25 | 47.2% | 28 | 52.8% | 53 | 30% | |||
| Self-employed | 5 | 27.8% | 13 | 72.2% | 18 | 10% | |||
| Student | 2 | 16.7% | 10 | 83.3% | 12 | 7% | |||
| Unemployed | 25 | 58.1% | 18 | 41.9% | 43 | 24% | |||
| Average monthly income in ETB | <1000 | 29 | 55.8% | 23 | 44.2% | 52 | 29% | 2.67 | 0.751 |
| 1001–2000 | 20 | 54.1% | 17 | 45.9% | 37 | 21% | |||
| 2001–3000 | 12 | 48.0% | 13 | 52.0% | 25 | 14% | |||
| 3001–4000 | 7 | 46.7% | 8 | 53.3% | 15 | 8% | |||
| >4000 | 9 | 47.4% | 10 | 52.6% | 19 | 11% | |||
| NA | 12 | 38.7% | 19 | 61.3% | 31 | 17% | |||
| Most recent VL | <1000 | 74 | 46.5% | 85 | 53.5% | 159 | 89% | 5.76 | 0.016 |
| >1000 | 15 | 75.0% | 5 | 25.0% | 20 | 11% | |||
| Most recent CD4count | <200 | 14 | 51.9% | 13 | 48.1% | 27 | 16% | 0.96 | 0.757 |
| ≥200 | 70 | 46.7% | 74 | 51.4% | 144 | 84% | |||
| ART | 1st line | 64 | 50.0% | 64 | 50.0% | 128 | 72% | 2.57 | 0.277 |
| 2nd line | 19 | 43.2% | 25 | 56.8% | 44 | 25% | |||
| 3rd line | 4 | 80.0% | 1 | 20.0% | 5 | 3% | |||
| Regimen type | ATV/r (PI based) | 19 | 43.2% | 25 | 56.8% | 44 | 25% | 4.70 | 0.195 |
| Darunavir (PI based) | 4 | 80.0% | 1 | 20.0% | 5 | 3% | |||
| DTG (INSTI based) | 62 | 51.7% | 58 | 48.3% | 120 | 68% | |||
| EFV/NVP (NNRTI based) | 2 | 25.0% | 6 | 75.0% | 8 | 5% | |||
| Characteristics | COR (95% CI) | AOR (95% CI) | |
|---|---|---|---|
| Age | <35 | 1 | 1 |
| ≥35 | 1.75 (1.06–2.89) | 1.34 (0.65–2.72) | |
| Marital status | Unmarried | 1 | 1 |
| Married | 1.04 (0.59–1.84) | 0.78 (0.37–1.64) | |
| Divorced | 1.16 (0.49–2.78) | 0.77 (0.28–2.12) | |
| Widowed | 4.14 (1.72–9.97) | 2.71 (0.95–7.71) | |
| Educational status | Illiterate | 1.43 (4.83–4.25) | 0.39 (0.11–1.50) |
| Primary school | 0.99 (0.51–1.92) | 0.49 (0.20–1.19) | |
| Secondary school | 0.54 (0.29–1.00) | 0.32 (0.14–0.69) | |
| College diploma and above | 1 | 1 | |
| Average monthly income | <1000 | 1.75 (0.80–3.84) | 1.28 (0.503–3.24) |
| 1001–2000 | 0.967 (0.43–2.16) | 1.03 (0.404–2.62) | |
| 2001–3000 | 0.79 (0.33–1.90) | 0.76 (0.285–2.03) | |
| 3001–4000 | 0.41 (0.16–1.04) | 0.49 (0.17–1.44) | |
| >4000 | 2.17 (0.86–5.40) | 2.08 (0.72–6.01) | |
| NA | 1 | 1 | |
| HIV status | Negative | 1 | 1 |
| Positive | 3.02 (1.68–5.41) | 3.05 (1.4–6.67) | |
| Characteristics | COR (95% CI) | AOR (95% CI) | |
|---|---|---|---|
| Age | <35 | 1 | 1 |
| ≥35 | 2.20 (1.14–4.22) | 2.16 (0.89–5.30) | |
| Marital status | Unmarried | 1 | 1 |
| Married | 0.98 (0.48–2.01) | 0.49 (0.172–1.40) | |
| Divorced | 1.12 (0.42–3.00) | 0.63 (1.90–2.11) | |
| Widowed | 2.98 (1.15–7.71) | 1.37 (0.40–4.71) | |
| Source of income | Government employee | 1 | 1 |
| Stay-at-home spouse | 1.25 (0.41–3.75) | 1.94 (0.504–7.50) | |
| Private sector employee | 0.66(0.25–1.69) | 0.92(3.15–2.72) | |
| Self-employed | 0.28 (0.08–1.03) | 0.35 (0.078–1.56) | |
| Student | 0.147 (0.03–0.81) | 0.154 (0.014–1.64) | |
| Unemployed | 1.02 (0.38–2.73) | 0.97 (0.27–3.50) | |
| Average monthly income | <1000 | 2.0 (0.81–4.94) | 1.31 (0.37–4.69) |
| 1001–2000 | 1.86 (0.71–4.91) | 1.11 (0.28–4.47) | |
| 2001–3000 | 1.46 (0.503–4.25) | 0.94 (0.21–4.15) | |
| 3001–4000 | 1.38 (0.40–4.81) | 1.25 (0.21–7.48) | |
| >4000 | 1.43 (0.45–4.52) | 1.11 (0.21–5.76) | |
| NA | 1 | 1 | |
| Most recent VL | <1000 | 1 | 1 |
| >1000 | 3.45(1.20–9.94) | 5.81 (1.4–24.13) | |
| Regimen type | ATV/r based (PI based) | 1 | 1 |
| Darunavir based (PI based) | 2.30 (0.41–12.60) | 2.4 (0.185–31.1) | |
| DTG based (INSTI based) | 12.00 (0.80–180.9) | 1.5 (0.66–3.38) | |
| EFV/NVP based (NNRTI based) | 3.21 (0.62–16.53) | 0.297 (0.039–2.28) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zealiyas, K.; Teshome, S.; Berhe, N.; Amogne, W.; Haile, A.F.; Abate, E.; Yimer, G.; Weigel, C.; Ahmed, E.H.; Abebe, T.; et al. The Burden of Epstein–Barr Virus (EBV) and Its Determinants among Adult HIV-Positive Individuals in Ethiopia. Viruses 2023, 15, 1743. https://doi.org/10.3390/v15081743
Zealiyas K, Teshome S, Berhe N, Amogne W, Haile AF, Abate E, Yimer G, Weigel C, Ahmed EH, Abebe T, et al. The Burden of Epstein–Barr Virus (EBV) and Its Determinants among Adult HIV-Positive Individuals in Ethiopia. Viruses. 2023; 15(8):1743. https://doi.org/10.3390/v15081743
Chicago/Turabian StyleZealiyas, Kidist, Seifegebriel Teshome, Nega Berhe, Wondwossen Amogne, Aklilu Feleke Haile, Ebba Abate, Getnet Yimer, Christoph Weigel, Elshafa Hassan Ahmed, Tamrat Abebe, and et al. 2023. "The Burden of Epstein–Barr Virus (EBV) and Its Determinants among Adult HIV-Positive Individuals in Ethiopia" Viruses 15, no. 8: 1743. https://doi.org/10.3390/v15081743
APA StyleZealiyas, K., Teshome, S., Berhe, N., Amogne, W., Haile, A. F., Abate, E., Yimer, G., Weigel, C., Ahmed, E. H., Abebe, T., & Baiocchi, R. (2023). The Burden of Epstein–Barr Virus (EBV) and Its Determinants among Adult HIV-Positive Individuals in Ethiopia. Viruses, 15(8), 1743. https://doi.org/10.3390/v15081743







