Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Antiviral Prophylaxis, Treatment and Routine Screening
2.3. Virological Testing
2.4. Nucleic Acid Extraction and r(RT-)PCR Screening
2.5. Statistical Methods
3. Results
3.1. Patient Characteristics
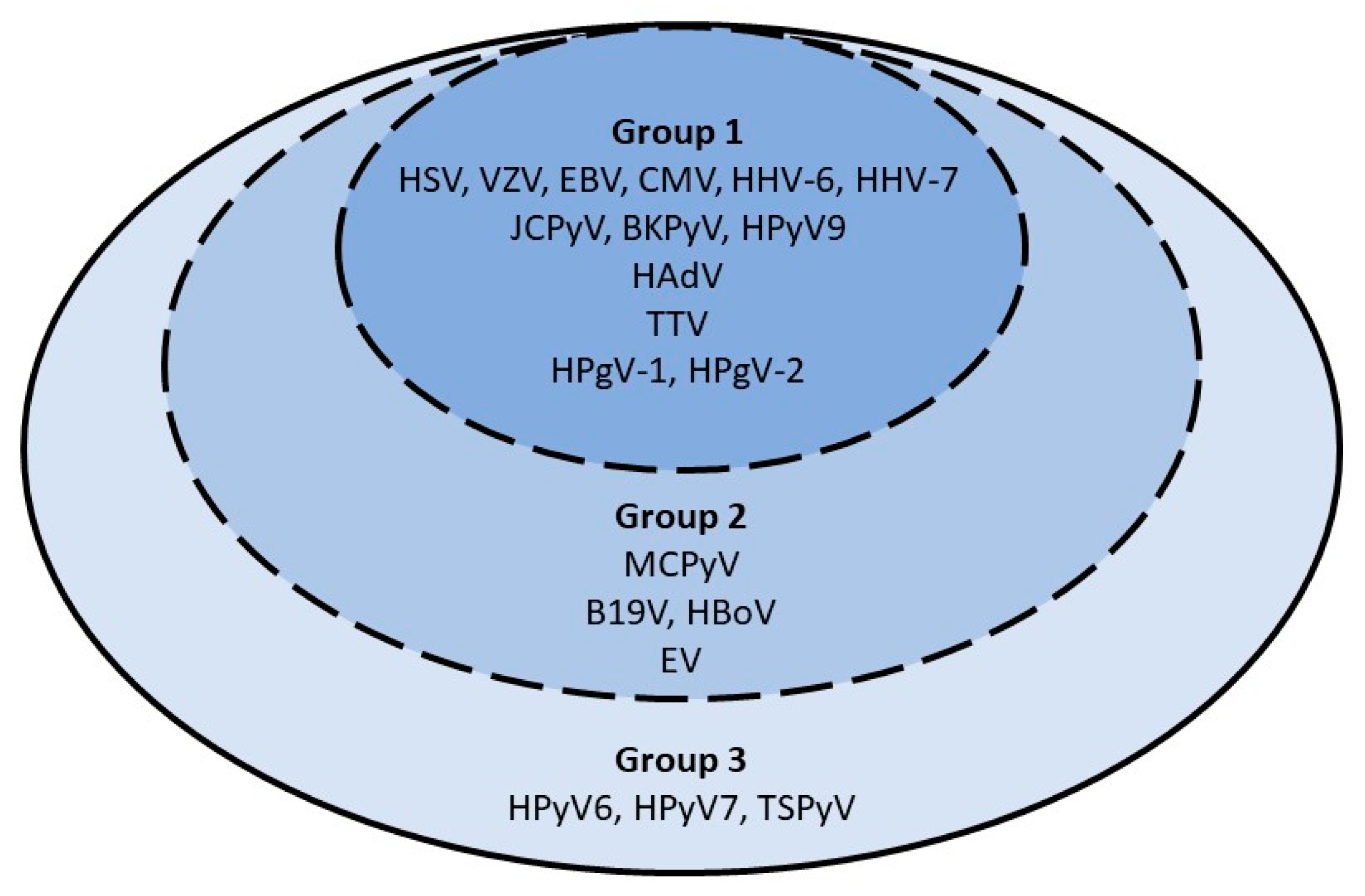
3.2. Virological Testing
3.2.1. Period Prevalence
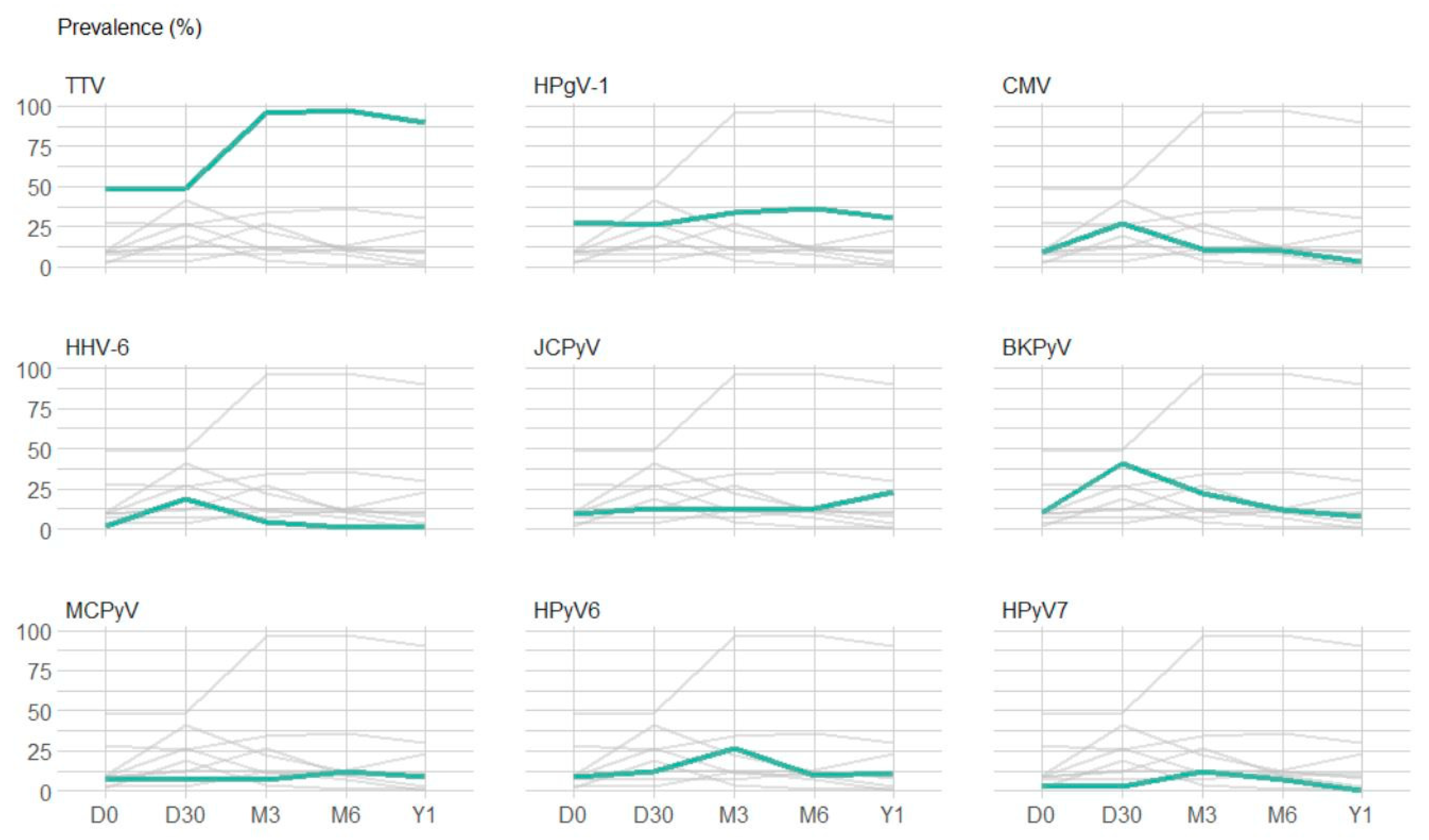
3.2.2. Point Prevalence and Dynamic Evolution of Prevalence
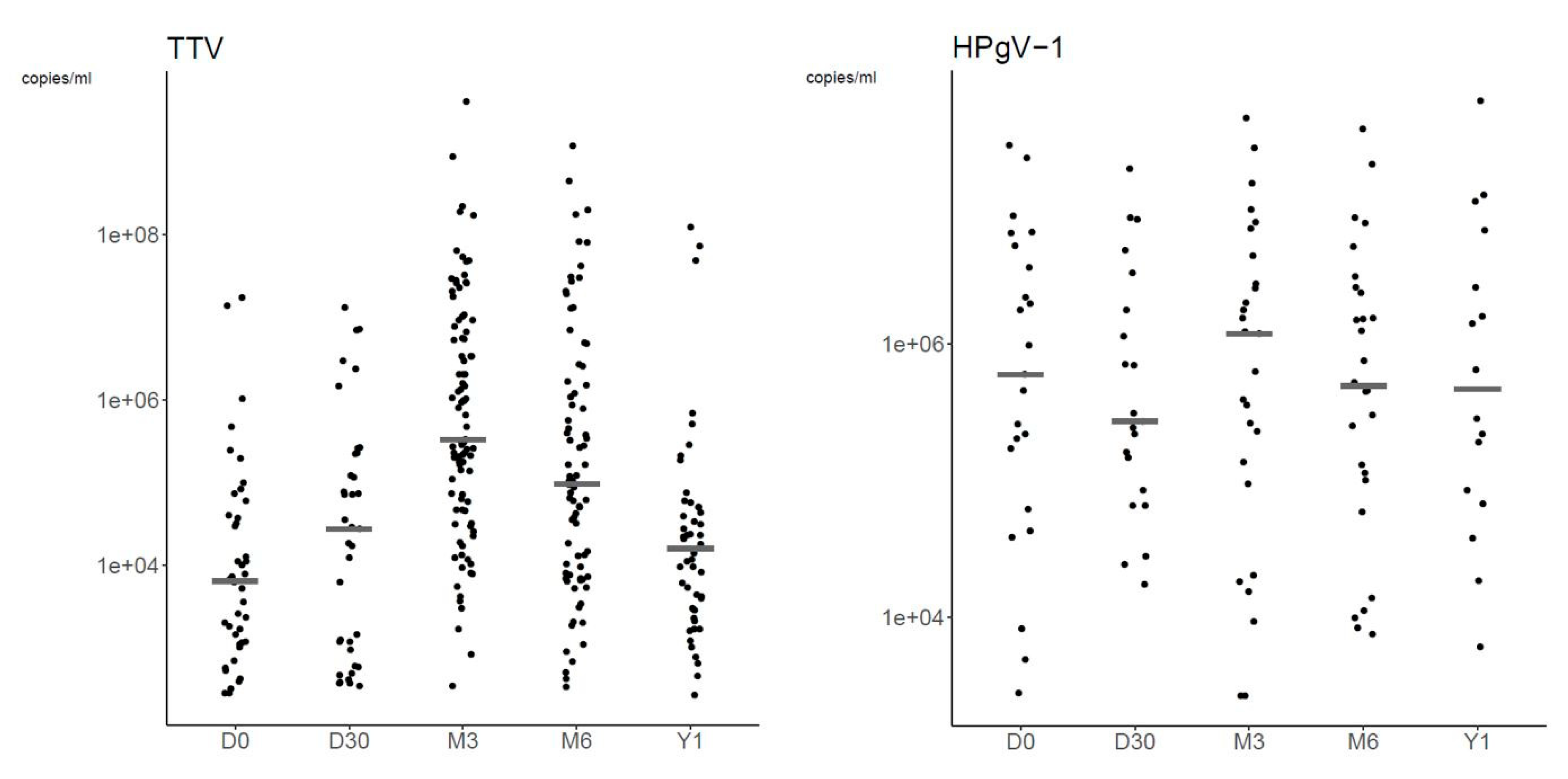
3.2.3. Plasma Viral Loads
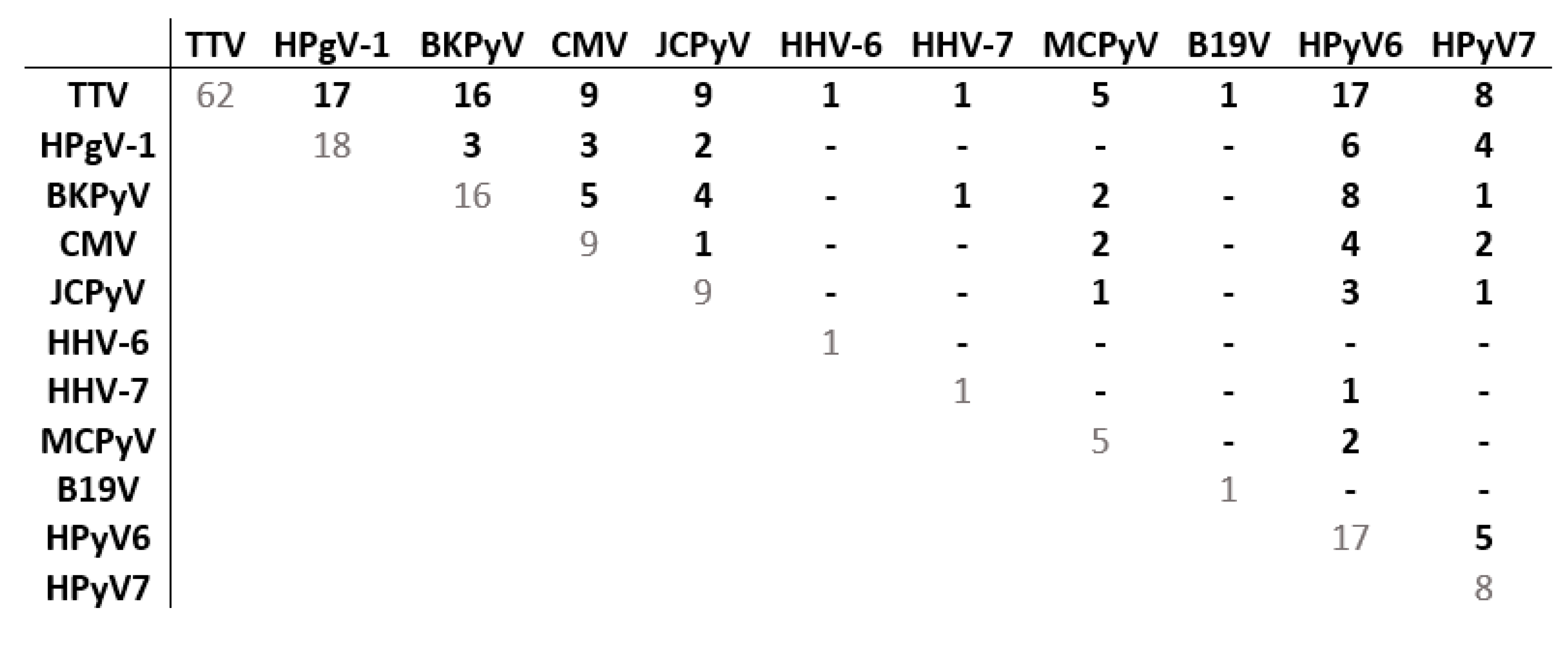
3.2.4. Co-Detections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin-Pena, A.; Aguilar-Guisado, M.; Espigado, I.; Parody, R.; Miguel Cisneros, J. Prospective study of infectious complications in allogeneic hematopoietic stem cell transplant recipients. Clin. Transplant. 2011, 25, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.L.; Dayer, J.A.; Masouridi-Levrat, S.; Combescure, C.; Boely, E.; Khanna, N.; Mueller, N.J.; Kleber, M.; Medinger, M.; Halter, J.; et al. Microbiologically documented infections after adult allogeneic hematopoietic cell transplantation: A 5-year analysis within the Swiss Transplant Cohort study. Transpl. Infect. Dis. 2020, 22, e13289. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.L.; Cordey, S.; Simonetta, F.; Brito, F.; Docquier, M.; Turin, L.; van Delden, C.; Boely, E.; Dantin, C.; Pradier, A.; et al. Human pegivirus persistence in human blood virome after allogeneic haematopoietic stem-cell transplantation. Clin. Microbiol. Infect. 2019, 25, 225–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanella, M.C.; Cordey, S.; Kaiser, L. Beyond cytomegalovirus and Epstein-Barr virus: A review of viruses composing the blood virome of solid organ transplant and hematopoietic stem cell transplant recipients. Clin. Microbiol. Rev. 2020, 33, e00027-20. [Google Scholar] [CrossRef]
- Zanella, M.C.; Cordey, S.; Laubscher, F.; Docquier, M.; Vieille, G.; Van Delden, C.; Braunersreuther, V.; Ta, M.K.; Lobrinus, J.A.; Masouridi-Levrat, S.; et al. Unmasking viral sequences by metagenomic next-generation sequencing in adult human blood samples during steroid-refractory/dependent graft-versus-host disease. Microbiome 2021, 9, 28. [Google Scholar] [CrossRef]
- Hill, J.A.; Mayer, B.T.; Xie, H.; Leisenring, W.M.; Huang, M.L.; Stevens-Ayers, T.; Milano, F.; Delaney, C.; Sorror, M.L.; Sandmaier, B.M.; et al. The cumulative burden of double-stranded DNA virus detection after allogeneic HCT is associated with increased mortality. Blood 2017, 129, 2316–2325. [Google Scholar] [CrossRef] [Green Version]
- Hill, J.A.; Mayer, B.T.; Xie, H.; Leisenring, W.M.; Huang, M.L.; Stevens-Ayers, T.; Milano, F.; Delaney, C.; Jerome, K.R.; Zerr, D.M.; et al. Kinetics of Double-Stranded DNA Viremia After Allogeneic Hematopoietic Cell Transplantation. Clin. Infect. Dis. 2018, 66, 368–375. [Google Scholar] [CrossRef] [Green Version]
- Royston, L.; Royston, E.; Masouridi-Levrat, S.; Vernaz, N.; Chalandon, Y.; Van Delden, C.; Neofytos, D. Letermovir Primary Prophylaxis in High-Risk Hematopoietic Cell Transplant Recipients: A Matched Cohort Study. Vaccines 2021, 9, 372. [Google Scholar] [CrossRef]
- Schibler, M.; Yerly, S.; Vieille, G.; Docquier, M.; Turin, L.; Kaiser, L.; Tapparel, C. Critical analysis of rhinovirus RNA load quantification by real-time reverse transcription-PCR. J. Clin. Microbiol. 2012, 50, 2868–2872. [Google Scholar] [CrossRef] [Green Version]
- Armand, P.; Gibson, C.J.; Cutler, C.; Ho, V.T.; Koreth, J.; Alyea, E.P.; Ritz, J.; Sorror, M.L.; Lee, S.J.; Deeg, H.J.; et al. A disease risk index for patients undergoing allogeneic stem cell transplantation. Blood 2012, 120, 905–913. [Google Scholar] [CrossRef] [Green Version]
- Mouton, W.; Conrad, A.; Bal, A.; Boccard, M.; Malcus, C.; Ducastelle-Lepretre, S.; Balsat, M.; Barraco, F.; Larcher, M.V.; Fossard, G.; et al. Torque Teno Virus Viral Load as a Marker of Immune Function in Allogeneic Haematopoietic Stem Cell Transplantation Recipients. Viruses 2020, 12, 1292. [Google Scholar] [CrossRef]
- Wohlfarth, P.; Leiner, M.; Schoergenhofer, C.; Hopfinger, G.; Goerzer, I.; Puchhammer-Stoeckl, E.; Rabitsch, W. Torquetenovirus Dynamics and Immune Marker Properties in Patients Following Allogeneic Hematopoietic Stem Cell Transplantation: A Prospective Longitudinal Study. Biol. Blood Marrow Transplant. 2018, 24, 194–199. [Google Scholar] [CrossRef] [Green Version]
- Albert, E.; Solano, C.; Gimenez, E.; Focosi, D.; Perez, A.; Macera, L.; Pinana, J.L.; Mateo, E.M.; Boluda, J.C.H.; Maggi, F.; et al. Kinetics of Alphatorquevirus plasma DNAemia at late times after allogeneic hematopoietic stem cell transplantation. Med. Microbiol. Immunol. 2019, 208, 253–258. [Google Scholar] [CrossRef]
- Gilles, R.; Herling, M.; Holtick, U.; Heger, E.; Awerkiew, S.; Fish, I.; Holler, K.; Sierra, S.; Knops, E.; Kaiser, R.; et al. Dynamics of Torque Teno virus viremia could predict risk of complications after allogeneic hematopoietic stem cell transplantation. Med. Microbiol. Immunol. 2017, 206, 355–362. [Google Scholar] [CrossRef]
- Kaczorowska, J.; van der Hoek, L. Human anelloviruses: Diverse, omnipresent and commensal members of the virome. FEMS Microbiol. Rev. 2020, 44, 305–313. [Google Scholar] [CrossRef] [Green Version]
- Arze, C.A.; Springer, S.; Dudas, G.; Patel, S.; Bhattacharyya, A.; Swaminathan, H.; Brugnara, C.; Delagrave, S.; Ong, T.; Kahvejian, A.; et al. Global genome analysis reveals a vast and dynamic anellovirus landscape within the human virome. Cell Host Microbe 2021, 29, 1305–1315. [Google Scholar] [CrossRef]
- Cebria-Mendoza, M.; Arbona, C.; Larrea, L.; Diaz, W.; Arnau, V.; Pena, C.; Bou, J.V.; Sanjuan, R.; Cuevas, J.M. Deep viral blood metagenomics reveals extensive anellovirus diversity in healthy humans. Sci. Rep. 2021, 11, 6921. [Google Scholar] [CrossRef]
- Pradier, A.; Masouridi-Levrat, S.; Bosshard, C.; Dantin, C.; Vu, D.L.; Zanella, M.C.; Boely, E.; Tapparel, C.; Kaiser, L.; Chalandon, Y.; et al. Torque Teno Virus as a Potential Biomarker for Complications and Survival After Allogeneic Hematopoietic Stem Cell Transplantation. Front. Immunol. 2020, 11, 998. [Google Scholar] [CrossRef]
- Focosi, D.; Maggi, F.; Albani, M.; Macera, L.; Ricci, V.; Gragnani, S.; Di Beo, S.; Ghimenti, M.; Antonelli, G.; Bendinelli, M.; et al. Torquetenovirus viremia kinetics after autologous stem cell transplantation are predictable and may serve as a surrogate marker of functional immune reconstitution. J. Clin. Virol. 2010, 47, 189–192. [Google Scholar] [CrossRef]
- Albert, E.; Solano, C.; Pascual, T.; Torres, I.; Macera, L.; Focosi, D.; Maggi, F.; Gimenez, E.; Amat, P.; Navarro, D. Dynamics of Torque Teno virus plasma DNAemia in allogeneic stem cell transplant recipients. J. Clin. Virol. 2017, 94, 22–28. [Google Scholar] [CrossRef]
- Schmitz, J.; Kobbe, G.; Kondakci, M.; Schuler, E.; Magorsch, M.; Adams, O. The Value of Torque Teno Virus (TTV) as a Marker for the Degree of Immunosuppression in Adult Patients after Hematopoietic Stem Cell Transplantation (HSCT). Biol. Blood Marrow Transplant. 2020, 26, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ma, H.H.; Lau, G.K.; Leung, Y.K.; Yao, C.L.; Chong, Y.T.; Tang, W.H.; Yao, J.L. Prevalence of hepatitis G virus infection and homology of different viral strains in Southern China. World J. Gastroenterol. 2002, 8, 1081–1087. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Liang, Y.; Hu, L.; Chen, S. Prevalence and risk factors of human pegivirus type 1 infection in hematopoietic stem cell transplantation patients. Int. J. Infect. Dis. 2019, 85, 111–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chivero, E.T.; Stapleton, J.T. Tropism of human pegivirus (formerly known as GB virus C/hepatitis G virus) and host immunomodulation: Insights into a highly successful viral infection. J. Gen. Virol. 2015, 96, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Cebria-Mendoza, M.; Bracho, M.A.; Arbona, C.; Larrea, L.; Diaz, W.; Sanjuan, R.; Cuevas, J.M. Exploring the Diversity of the Human Blood Virome. Viruses 2021, 13, 2322. [Google Scholar] [CrossRef]
- Stapleton, J.T.; Chaloner, K.; Martenson, J.A.; Zhang, J.; Klinzman, D.; Xiang, J.; Sauter, W.; Desai, S.N.; Landay, A. GB virus C infection is associated with altered lymphocyte subset distribution and reduced T cell activation and proliferation in HIV-infected individuals. PLoS ONE 2012, 7, e50563. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Ruiz, M.; Forque, L.; Albert, E.; Redondo, N.; Gimenez, E.; Lopez-Medrano, F.; Gonzalez, E.; Polanco, N.; Ruiz-Merlo, T.; Parra, P.; et al. Human pegivirus type 1 infection in kidney transplant recipients: Replication kinetics and clinical correlates. Transplant. Infect. Dis. 2022, 24, e13771. [Google Scholar] [CrossRef]
- Graninger, M.; Aberle, S.; Gorzer, I.; Jaksch, P.; Puchhammer-Stockl, E. Human pegivirus 1 infection in lung transplant recipients: Prevalence, clinical relevance and kinetics of viral replication under immunosuppressive therapy. J. Clin. Virol. 2021, 143, 104937. [Google Scholar] [CrossRef]
- Fama, A.; Larson, M.C.; Link, B.K.; Habermann, T.M.; Feldman, A.L.; Call, T.G.; Ansell, S.M.; Liebow, M.; Xiang, J.; Maurer, M.J.; et al. Human Pegivirus Infection and Lymphoma Risk: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2020, 71, 1221–1228. [Google Scholar] [CrossRef]
- Laskin, B.L.; Denburg, M.R.; Furth, S.L.; Moatz, T.; Altrich, M.; Kleiboeker, S.; Lutzko, C.; Zhu, X.; Blackard, J.T.; Jodele, S.; et al. The Natural History of BK Polyomavirus and the Host Immune Response After Stem Cell Transplantation. Clin. Infect. Dis. 2020, 71, 3044–3054. [Google Scholar] [CrossRef]
- Tan, C.S.; Broge, T.A., Jr.; Ngo, L.; Gheuens, S.; Viscidi, R.; Bord, E.; Rosenblatt, J.; Wong, M.; Avigan, D.; Koralnik, I.J. Immune reconstitution after allogeneic hematopoietic stem cell transplantation is associated with selective control of JC virus reactivation. Biol. Blood Marrow Transplant. 2014, 20, 992–999. [Google Scholar] [CrossRef] [Green Version]
- Hill, J.A. Human herpesvirus 6 in transplant recipients: An update on diagnostic and treatment strategies. Curr. Opin. Infect. Dis. 2019, 32, 584–590. [Google Scholar] [CrossRef]
- Dulery, R.; Salleron, J.; Dewilde, A.; Rossignol, J.; Boyle, E.M.; Gay, J.; de Berranger, E.; Coiteux, V.; Jouet, J.P.; Duhamel, A.; et al. Early human herpesvirus type 6 reactivation after allogeneic stem cell transplantation: A large-scale clinical study. Biol. Blood Marrow Transplant. 2012, 18, 1080–1089. [Google Scholar] [CrossRef] [Green Version]
- Boutolleau, D.; Fernandez, C.; Andre, E.; Imbert-Marcille, B.M.; Milpied, N.; Agut, H.; Gautheret-Dejean, A. Human herpesvirus (HHV)-6 and HHV-7: Two closely related viruses with different infection profiles in stem cell transplantation recipients. J. Infect. Dis. 2003, 187, 179–186. [Google Scholar] [CrossRef]
- de Pagter, P.J.; Schuurman, R.; Visscher, H.; de Vos, M.; Bierings, M.; van Loon, A.M.; Uiterwaal, C.S.; van Baarle, D.; Sanders, E.A.; Boelens, J. Human herpes virus 6 plasma DNA positivity after hematopoietic stem cell transplantation in children: An important risk factor for clinical outcome. Biol. Blood Marrow Transplant. 2008, 14, 831–839. [Google Scholar] [CrossRef] [Green Version]
- Inazawa, N.; Hori, T.; Hatakeyama, N.; Yamamoto, M.; Yoto, Y.; Nojima, M.; Suzuki, N.; Shimizu, N.; Tsutsumi, H. Large-scale multiplex polymerase chain reaction assay for diagnosis of viral reactivations after allogeneic hematopoietic stem cell transplantation. J. Med. Virol. 2015, 87, 1427–1435. [Google Scholar] [CrossRef]
- Styczynski, J.; van der Velden, W.; Fox, C.P.; Engelhard, D.; de la Camara, R.; Cordonnier, C.; Ljungman, P. Management of Epstein-Barr Virus infections and post-transplant lymphoproliferative disorders in patients after allogeneic hematopoietic stem cell transplantation: Sixth European Conference on Infections in Leukemia (ECIL-6) guidelines. Haematologica 2016, 101, 803–811. [Google Scholar] [CrossRef]
- Zhou, J.R.; Shi, D.Y.; Wei, R.; Wang, Y.; Yan, C.H.; Zhang, X.H.; Xu, L.P.; Liu, K.Y.; Huang, X.J.; Sun, Y.Q. Co-Reactivation of Cytomegalovirus and Epstein-Barr Virus Was Associated With Poor Prognosis After Allogeneic Stem Cell Transplantation. Front. Immunol. 2020, 11, 620891. [Google Scholar] [CrossRef]
- Rahiala, J.; Koskenvuo, M.; Sadeghi, M.; Waris, M.; Vuorinen, T.; Lappalainen, M.; Saarinen-Pihkala, U.; Allander, T.; Soderlund-Venermo, M.; Hedman, K.; et al. Polyomaviruses BK, JC, KI, WU, MC, and TS in children with allogeneic hematopoietic stem cell transplantation. Pediatr. Transplant. 2016, 20, 424–431. [Google Scholar] [CrossRef]
- Kapoor, A.; Simmonds, P.; Slikas, E.; Li, L.; Bodhidatta, L.; Sethabutr, O.; Triki, H.; Bahri, O.; Oderinde, B.S.; Baba, M.M.; et al. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 2010, 201, 1633–1643. [Google Scholar] [CrossRef]
- Arthur, J.L.; Higgins, G.D.; Davidson, G.P.; Givney, R.C.; Ratcliff, R.M. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog 2009, 5, e1000391. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Slikas, E.; Simmonds, P.; Chieochansin, T.; Naeem, A.; Shaukat, S.; Alam, M.M.; Sharif, S.; Angez, M.; Zaidi, S.; et al. A newly identified bocavirus species in human stool. J. Infect. Dis. 2009, 199, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Pinana, J.L.; Madrid, S.; Perez, A.; Hernandez-Boluda, J.C.; Gimenez, E.; Terol, M.J.; Calabuig, M.; Navarro, D.; Solano, C. Epidemiologic and Clinical Characteristics of Coronavirus and Bocavirus Respiratory Infections after Allogeneic Stem Cell Transplantation: A Prospective Single-Center Study. Biol. Blood Marrow Transplant. 2018, 24, 563–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, B.C.L.; Dabilla, N.A.S.; Almeida, T.N.; Fiaccadori, F.S.; de Souza, T.T.; Cardoso, D.; Arantes, A.M.; Souza, M. Human bocavirus detection and quantification in fecal and serum specimens from recipients of allogeneic hematopoietic stem cell transplantation: A longitudinal study. J. Med. Virol. 2022, 94, 594–600. [Google Scholar] [CrossRef]
- Ogimi, C.; Martin, E.T.; Xie, H.; Campbell, A.P.; Waghmare, A.; Jerome, K.R.; Leisenring, W.M.; Milano, F.; Englund, J.A.; Boeckh, M. Role of Human Bocavirus Respiratory Tract Infection in Hematopoietic Cell Transplant Recipients. Clin. Infect. Dis. 2021, 73, e4392–e4399. [Google Scholar] [CrossRef]
- Berg, M.G.; Lee, D.; Coller, K.; Frankel, M.; Aronsohn, A.; Cheng, K.; Forberg, K.; Marcinkus, M.; Naccache, S.N.; Dawson, G.; et al. Discovery of a Novel Human Pegivirus in Blood Associated with Hepatitis C Virus Co-Infection. PLoS Pathog 2015, 11, e1005325. [Google Scholar] [CrossRef]
- Coller, K.E.; Bruce, V.; Cassidy, M.; Gersch, J.; Frankel, M.B.; Vallari, A.; Cloherty, G.; Hackett, J., Jr.; Evans, J.L.; Page, K.; et al. Chronic Human Pegivirus 2 without Hepatitis C Virus Co-infection. Emerg. Infect. Dis. 2020, 26, 265–272. [Google Scholar] [CrossRef]
- Strenger, V.; Kessler, H.H.; Stelzl, E.; Aberle, S.W.; Keldorfer, M.; Zach, K.; Karastaneva, A.; Sperl, D.; Lackner, H.; Benesch, M.; et al. Enterovirus infections in pediatric hematologic/oncologic patients. Pediatr. Blood Cancer 2019, 66, e27448. [Google Scholar] [CrossRef]
- Matsumura-Kimoto, Y.; Inamoto, Y.; Tajima, K.; Kawajiri, A.; Tanaka, T.; Hirakawa, T.; Ino, K.; Asao, Y.; Tamogami, H.; Kono, C.; et al. Association of Cumulative Steroid Dose with Risk of Infection after Treatment for Severe Acute Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2016, 22, 1102–1107. [Google Scholar] [CrossRef] [Green Version]
- Miller, H.K.; Braun, T.M.; Stillwell, T.; Harris, A.C.; Choi, S.; Connelly, J.; Couriel, D.; Goldstein, S.; Kitko, C.L.; Magenau, J.; et al. Infectious Risk after Allogeneic Hematopoietic Cell Transplantation Complicated by Acute Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2017, 23, 522–528. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, A.; Wang, C.; Srivastava, D.K.; Burnette, K.; Shenep, J.L.; Leung, W.; Hayden, R.T. Timeline, epidemiology, and risk factors for bacterial, fungal, and viral infections in children and adolescents after allogeneic hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2013, 19, 94–101. [Google Scholar] [CrossRef] [Green Version]
- Eriguchi, Y.; Takashima, S.; Oka, H.; Shimoji, S.; Nakamura, K.; Uryu, H.; Shimoda, S.; Iwasaki, H.; Shimono, N.; Ayabe, T.; et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood 2012, 120, 223–231. [Google Scholar] [CrossRef] [Green Version]
- Levinson, A.; Pinkney, K.; Jin, Z.; Bhatia, M.; Kung, A.L.; Foca, M.D.; George, D.; Garvin, J.H.; Sosna, J.; Karamehmet, E.; et al. Acute gastrointestinal graft-vs-host disease is associated with increased enteric bacterial bloodstream infection density in pediatric allogeneic hematopoietic cell transplant recipients. Clin. Infect. Dis. 2015, 61, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Sayer, H.G.; Longton, G.; Bowden, R.; Pepe, M.; Storb, R. Increased risk of infection in marrow transplant patients receiving methylprednisolone for graft-versus-host disease prevention. Blood 1994, 84, 1328–1332. [Google Scholar] [CrossRef] [Green Version]
- Martin, P.J.; Rizzo, J.D.; Wingard, J.R.; Ballen, K.; Curtin, P.T.; Cutler, C.; Litzow, M.R.; Nieto, Y.; Savani, B.N.; Schriber, J.R.; et al. First- and second-line systemic treatment of acute graft-versus-host disease: Recommendations of the American Society of Blood and Marrow Transplantation. Biol. Blood Marrow Transplant. 2012, 18, 1150–1163. [Google Scholar] [CrossRef] [Green Version]
- Vu, D.L.; Bosch, A.; Pinto, R.M.; Guix, S. Epidemiology of Classic and Novel Human Astrovirus: Gastroenteritis and Beyond. Viruses 2017, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Cordey, S.; Vu, D.L.; Schibler, M.; L’Huillier, A.G.; Brito, F.; Docquier, M.; Posfay-Barbe, K.M.; Petty, T.J.; Turin, L.; Zdobnov, E.M.; et al. Astrovirus MLB2, a New Gastroenteric Virus Associated with Meningitis and Disseminated Infection. Emerg Infect Dis 2016, 22, 846–853. [Google Scholar] [CrossRef] [Green Version]
- Cordey, S.; Vu, D.L.; Zanella, M.C.; Turin, L.; Mamin, A.; Kaiser, L. Novel and classical human astroviruses in stool and cerebrospinal fluid: Comprehensive screening in a tertiary care hospital, Switzerland. Emerg. Microbes Infect. 2017, 6, e84. [Google Scholar] [CrossRef]
- Piret, J.; Boivin, G. Antiviral Drugs Against Herpesviruses. In Antiviral Drug Discovery and Development; Springer: Singapore, 2021; Volume 1322, pp. 1–30. [Google Scholar] [CrossRef]
- Hatakeyama, N.; Suzuki, N.; Kudoh, T.; Hori, T.; Mizue, N.; Tsutsumi, H. Successful cidofovir treatment of adenovirus-associated hemorrhagic cystitis and renal dysfunction after allogenic bone marrow transplant. Pediatr. Infect. Dis. J. 2003, 22, 928–929. [Google Scholar] [CrossRef]
- Kadambi, P.V.; Josephson, M.A.; Williams, J.; Corey, L.; Jerome, K.R.; Meehan, S.M.; Limaye, A.P. Treatment of refractory BK virus-associated nephropathy with cidofovir. Am. J. Transplant. 2003, 3, 186–191. [Google Scholar] [CrossRef]
- Vats, A.; Shapiro, R.; Singh Randhawa, P.; Scantlebury, V.; Tuzuner, A.; Saxena, M.; Moritz, M.L.; Beattie, T.J.; Gonwa, T.; Green, M.D.; et al. Quantitative viral load monitoring and cidofovir therapy for the management of BK virus-associated nephropathy in children and adults. Transplantation 2003, 75, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Canavan, T.N.; Baddley, J.W.; Pavlidakey, P.; Tallaj, J.A.; Elewski, B.E. Human polyomavirus-7-associated eruption successfully treated with acitretin. Am. J. Transplant. 2018, 18, 1278–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubert, M.; Boyle, N.M.; Stone, D.; Stensland, L.; Huang, M.-L.; Magaret, A.S.; Galetto, R.; Rawlings, D.J.; Scharenberg, A.M.; Jerome, K.R. In vitro Inactivation of Latent HSV by Targeted Mutagenesis Using an HSV-specific Homing Endonuclease. Mol. Ther. Nucleic Acids 2014, 3, e146. [Google Scholar] [CrossRef] [PubMed]
- Corey, L.; Huang, M.L.; Selke, S.; Wald, A. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time taqman PCR assay. J. Med. Virol. 2005, 76, 350–355. [Google Scholar] [CrossRef]
- Weidmann, M.; Meyer-Konig, U.; Hufert, F.T. Rapid detection of herpes simplex virus and varicella-zoster virus infections by real-time PCR. J. Clin. Microbiol. 2003, 41, 1565–1568. [Google Scholar] [CrossRef] [Green Version]
- Rockett, R.J.; Sloots, T.P.; Bowes, S.; O’Neill, N.; Ye, S.; Robson, J.; Whiley, D.M.; Lambert, S.; Wang, D.; Nissen, M.; et al. Detection of novel polyomaviruses, TSPyV, HPyV6, HPyV7, HPyV9 and MWPyV in feces, urine, blood, respiratory swabs and cerebrospinal fluid. PLoS ONE 2013, 8, e62764. [Google Scholar] [CrossRef] [Green Version]
- Verheyen, J.; Timmen-Wego, M.; Laudien, R.; Boussaad, I.; Sen, S.; Koc, A.; Uesbeck, A.; Mazou, F.; Pfister, H. Detection of adenoviruses and rotaviruses in drinking water sources used in rural areas of Benin, West Africa. Appl. Environ. Microbiol. 2009, 75, 2798–2801. [Google Scholar] [CrossRef] [Green Version]
- Masouridi-Levrat, S.; Pradier, A.; Simonetta, F.; Kaiser, L.; Chalandon, Y.; Roosnek, E. Torque teno virus in patients undergoing allogeneic hematopoietic stem cell transplantation for hematological malignancies. Bone Marrow Transplant. 2016, 51, 440–442. [Google Scholar] [CrossRef] [Green Version]
- Chivero, E.T.; Bhattarai, N.; Rydze, R.T.; Winters, M.A.; Holodniy, M.; Stapleton, J.T. Human pegivirus RNA is found in multiple blood mononuclear cells in vivo and serum-derived viral RNA-containing particles are infectious in vitro. J. Gen. Virol. 2014, 95, 1307–1319. [Google Scholar] [CrossRef]
- Frankel, M.; Forberg, K.; Coller, K.E.; Berg, M.G.; Hackett, J.; Cloherty, G.; Dawson, G.J. Development of a high-throughput multiplexed real time RT-PCR assay for detection of human pegivirus 1 and 2. J. Virol. Methods. 2017, 241, 34–40. [Google Scholar] [CrossRef]
- Cordey, S.; Junier, T.; Gerlach, D.; Gobbini, F.; Farinelli, L.; Zdobnov, E.M.; Winther, B.; Tapparel, C.; Kaiser, L. Rhinovirus genome evolution during experimental human infection. PLoS ONE 2010, 5, e10588. [Google Scholar] [CrossRef] [Green Version]
- Kantola, K.; Sadeghi, M.; Antikainen, J.; Kirveskari, J.; Delwart, E.; Hedman, K.; Söderlund-Venermo, M. Real-time quantitative PCR detection of four human bocaviruses. J. Clin. Microbiol. 2010, 48, 4044–4050. [Google Scholar] [CrossRef] [Green Version]
- Arvia, R.; Sollai, M.; Pierucci, F.; Urso, C.; Massi, D.; Zakrzewska, K. Droplet digital PCR (ddPCR) vs quantitative real-time PCR (qPCR) approach for detection and quantification of Merkel cell polyomavirus (MCPyV) DNA in formalin fixed paraffin embedded (FFPE) cutaneous biopsies. J. Virol. Methods. 2017, 246, 15–20. [Google Scholar] [CrossRef]
- Tapparel, C.; Cordey, S.; Van Belle, S.; Turin, L.; Lee, W.-M.; Regamey, N.; Meylan, P.; Mühlemann, K.; Gobbini, F.; Kaiser, L. New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J. Clin. Microbiol. 2009, 47, 1742–1749. [Google Scholar] [CrossRef] [Green Version]
- Antonsson, A.; Bialasiewicz, S.; Rockett, R.J.; Jacob, K.; Bennett, I.C.; Sloots, T.P. Exploring the prevalence of ten polyomaviruses and two herpes viruses in breast cancer. PLoS ONE 2012, 7, e39842. [Google Scholar] [CrossRef] [Green Version]
- Urbano, P.R.; Nali, L.H.; Bicalho, C.S.; Pierrotti, L.C.; David-Neto, E.; Pannuti, C.S.; Romano, C.M. New findings about trichodysplasia spinulosa-associated polyomavirus (TSPyV)--novel qPCR detects TSPyV-DNA in blood samples. Diagn. Microbiol. Infect. Dis. 2016, 84, 123–124. [Google Scholar] [CrossRef]


| Total N = 109 | |
|---|---|
| Demographics Sex (male), n (%) | 72 (66) |
| Age, median (IQR) | 56 (18) |
| Transplant source, n (%) | |
| Peripheral blood cells | 96 (88) |
| Bone marrow | 13 (12) |
| Underlying disease, n (%) | |
| Acute myeloid leukemia | 59 (54) |
| MDS/MDPS | 27 (25) |
| Acute lymphoid leukemia | 10 (9) |
| Myeloproliferative syndrome | 4 (4) |
| Lymphoma | 4 (4) |
| Chronic lymphocytic leukemia | 2 (2) |
| Myeloma | 2 (2) |
| Chronic myelogenous leukemia | 1 (1) |
| Disease risk index *, n (%) | |
| Low | 5 (5) |
| Intermediate | 71 (65) |
| High | 29 (27) |
| Very high | 4 (4) |
| EBMT risk score, n (%) | |
| 1 | 5 (5) |
| 2 | 10 (9) |
| 3 | 46 (42) |
| 4 | 19 (17) |
| 5 | 19 (17) |
| 6 | 10 (9) |
| EBV donor/recipient constellation, n (%) | |
| +/+ | 96 (88) |
| −/+ | 8 (7) |
| +/− | 3 (3) |
| −/− | 2 (2) |
| CMV donor/recipient constellation, n (%) | |
| +/+ | 46 (42) |
| +/− | 10 (9) |
| −/− | 40 (37) |
| −/+ | 13 (12) |
| Conditioning regimen, n (%) | |
| Reduced intensity conditioning | 74 (68) |
| Myeloablative conditioning | 43 (39) |
| Ex vivo T-cell depletion, n (%) | 19 (17) |
| Donor type, n (%) | |
| HLA identical sibling donor | 28 (26) |
| HLA matched unrelated donor | 47 (43) |
| Haploidentical donor | 24 (22) |
| HLA mismatched unrelated donor | 10 (9) |
| Patients, N | |||||
|---|---|---|---|---|---|
| Day 0 | Day 30 | Month 3 | Month 6 | Year 1 | |
| Group 1 (13 virus) | 103 | 102 | 101 | 89 | 64 |
| Group 2 (17 virus) | 74 | 72 | 73 | 66 | 50 |
| Group 3 (20 virus) | 65 | 65 | 64 | 58 | 44 |
| Missing data * | 6 | 7 | 8 | 20 | 45 |
| Viral Specie | Viral Species Detected N (%) | Viral Species Undetectedor Not Tested N (%) |
|---|---|---|
| TTV | 104 (95) | 5 (5) |
| BKPyV | 57 (52) | 52 (48) |
| HPgV-1 | 51 (47) | 58 (53) |
| JCPyV | 41 (38) | 68 (62) |
| CMV | 36 (33) | 73 (67) |
| HHV-6 | 26 (24) | 83 (76) |
| HPyV6 | 26 (24) | 83 (76) |
| MCPyV | 23 (21) | 86 (79) |
| HPyV7 | 15 (14) | 94 (86) |
| EBV | 12 (11) | 97 (89) |
| HSV | 5 (5) | 104 (95) |
| HAdV | 5 (5) | 104 (95) |
| HHV-7 | 4 (4) | 105 (96) |
| B19V | 3 (3) | 106 (97) |
| VZV | 2 (2) | 107 (98) |
| HPyV9 | - | 109 (100) |
| HPgV-2 | - | 109 (100) |
| HBoV | - | 109 (100) |
| EV | - | 109 (100) |
| TSPyV | - | 109 (100) |
| Viral Species | r(RT-)PCR Result | Time-Point | ||||
|---|---|---|---|---|---|---|
| Day 0 | Day 30 | Month 3 | Month 6 | Year 1 | ||
| TTV | detected | 49 (48%) | 49 (49%) | 97 (96%) | 86 (97%) | 55 (90%) |
| not detected | 54 (52%) | 52 (51%) | 4 (4%) | 3 (3%) | 6 (10%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| HPgV-1 | detected | 29 (28%) | 26 (26%) | 34 (34%) | 32 (36%) | 18 (30%) |
| not detected | 74 (72%) | 75 (74%) | 67 (66%) | 57 (64%) | 43 (70%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| BKPyV | detected | 10 (10%) | 41 (41%) | 22 (22%) | 11 (12%) | 5 (8%) |
| not detected | 93 (90%) | 60 (59%) | 79 (78%) | 78 (88%) | 56 (92%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| JCPyV | detected | 9 (9%) | 13 (13%) | 12 (12%) | 12 (13%) | 14 (23%) |
| not detected | 94 (91%) | 88 (87%) | 89 (88%) | 77 (87%) | 47 (77%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| MCPyV | detected | 5 (7%) | 6 (8%) | 5 (7%) | 8 (12%) | 4 (9%) |
| not detected | 69 (93%) | 65 (92%) | 68 (93%) | 58 (88%) | 39 (91%) | |
| NA | 35 | 38 | 36 | 43 | 66 | |
| HPyV6 | detected | 6 (9%) | 8 (12%) | 17 (27%) | 6 (10%) | 4 (11%) |
| not detected | 59 (91%) | 56 (88%) | 47 (73%) | 52 (90%) | 34 (89%) | |
| NA | 44 | 45 | 45 | 51 | 71 | |
| HPyV7 | detected | 2 (3%) | 2 (3%) | 8 (12%) | 4 (7%) | 0 (0%) |
| not detected | 63 (97%) | 62 (97%) | 56 (88%) | 54 (93%) | 38 (100%) | |
| NA | 44 | 45 | 45 | 51 | 71 | |
| CMV | detected | 9 (9%) | 27 (27%) | 11 (11%) | 9 (10%) | 2 (3%) |
| not detected | 92 (91%) | 74 (73%) | 90 (89%) | 80 (90%) | 59 (97%) | |
| NA | 8 | 8 | 8 | 20 | 48 | |
| HHV-6 | detected | 2 (2%) | 19 (19%) | 4 (4%) | 1 (1%) | 1 (2%) |
| not detected | 101 (98%) | 82 (81%) | 97 (96%) | 88 (99%) | 60 (98%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| HSV-1/2 | detected | 2 (2%) | 2 (2%) | 0 (0%) | 0 (0%) | 1 (2%) |
| not detected | 101 (98%) | 99 (98%) | 101 (100%) | 89 (100%) | 60 (98%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| VZV | detected | 0 (0%) | 0 (0%) | 0 (0%) | 2 (2%) | 0 (0%) |
| not detected | 103 (100%) | 101 (100%) | 101 (100%) | 87 (98%) | 61 (100%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| EBV | detected | 2 (2%) | 5 (5%) | 1 (1%) | 4 (4%) | 3 (5%) |
| not detected | 99 (98%) | 96 (95%) | 100 (99%) | 85 (96%) | 58 (95%) | |
| NA | 8 | 8 | 8 | 20 | 48 | |
| HHV-7 | detected | 0 (0%) | 2 (2%) | 1 (1%) | 0 (0%) | 1 (2%) |
| not detected | 103 (100%) | 99 (98%) | 100 (99%) | 89 (100%) | 60 (98%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| HAdV | detected | 1 (1%) | 3 (3%) | 1 (1%) | 1 (1%) | 1 (2%) |
| not detected | 102 (99%) | 98 (97%) | 100 (99%) | 88 (99%) | 60 (98%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| B19V | detected | 1 (1%) | 2 (3%) | 1 (1%) | 1 (2%) | 2 (5%) |
| not detected | 73 (99%) | 69 (97%) | 72 (99%) | 65 (98%) | 41 (95%) | |
| NA | 35 | 38 | 36 | 43 | 66 | |
| TSPyV | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 65 (100%) | 64 (100%) | 64 (100%) | 58 (100%) | 38 (100%) | |
| NA | 44 | 45 | 45 | 51 | 71 | |
| HPyV9 | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 103 (100%) | 101 (100%) | 101 (100%) | 89 (100%) | 61 (100%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| HBoV | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 74 (100%) | 71 (100%) | 73 (100%) | 66 (100%) | 43 (100%) | |
| NA | 35 | 38 | 36 | 43 | 66 | |
| HPgV-2 | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 103 (100%) | 101 (100%) | 101 (100%) | 89 (100%) | 61 (100%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| EV | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 74 (100%) | 71 (100%) | 73 (100%) | 66 (100%) | 43 (100%) | |
| NA | 35 | 38 | 36 | 43 | 66 | |
| Plasma Viral Load * | ||||
|---|---|---|---|---|
| Time-Point | Viral Species | Patients N | Median (IQ) | Range |
| Day 0 | TTV | 42 | 6.35 × 103 (1.15 × 103–3.92 × 104) | 2.76 × 102–1.73 × 107 |
| HPgV-1 | 23 | 5.96 × 105 (1.16 × 105–4.39 × 106) | 2.79 × 103–2.83 × 107 | |
| Day 30 | TTV | 37 | 2.70 × 104 (9.40 × 102–2.19 × 105) | 3.39 × 102–1.31 × 107 |
| HPgV-1 | 21 | 2.71 × 105 (8.39 × 104–1.76 × 106) | 1.73 × 104–1.89 × 107 | |
| CMV | 16 | 2.02 × 102 (1.18 × 102–3.30 × 102) | 5.8 × 101–5.70 × 103 | |
| HHV-6 | 13 | 1.16 × 103 (8.42 × 102–4.03 × 103) | 4.71 × 102–9.88 × 103 | |
| BKPyV | 5 | 1.17 × 103 (1.11 × 103–1.21 × 103) | 1.04 × 103–1.41 × 103 | |
| Month 3 | TTV | 93 | 3.29 × 105 (4.66 × 104–6.65 × 106) | 3.37 × 102–4.06 × 109 |
| HPgV-1 | 27 | 1.18 × 106 (1.15 × 105–3.60 × 106) | 2.61 × 103–4.49 × 107 | |
| HPyV6 | 7 | 1.3 × 103 (4.53 × 102–2.59 × 103) | 2.64 × 102–7.68 × 103 | |
| HPyV7 | 5 | 1.63 × 103 (1.10 × 103–5.39 × 103) | 2.73 × 102–1.71 × 104 | |
| Month 6 | TTV | 79 | 9.58 × 104 (7.74 × 103–1.57 × 106) | 3.33 × 102–1.18 × 109 |
| HPgV-1 | 28 | 4.90 × 105 (1.10 × 105–2.41 × 106) | 7.45 × 103–3.74 × 107 | |
| JCPyV | 8 | 1.17 × 103 (4.21 × 102–1.62 × 103) | 1.34 × 102–2.76 × 103 | |
| Year 1 | TTV | 48 | 1.58 × 104 (2.92 × 103–4.97 × 104) | 2.62 × 102–1.23 × 108 |
| HPgV-1 | 16 | 4.64 × 105 (8.05 × 104–3.61 × 106) | 6.01 × 103–5.98 × 107 | |
| Number of Viral Species Detected | Patients, N (%) | ||||
|---|---|---|---|---|---|
| Day 0 (N = 65) | Day 30 (N = 65) | Month 3 (N = 64) | Month 6 (N = 58) | Year 1 (N = 44) | |
| 0 | 15 (23.1%) | 6 (9.2%) | 1 (1.6%) | 1 (17.2%) | 1 (2.3%) |
| 1 | 23 (35.4%) | 20 (30.8%) | 17 (26.6%) | 20 (34.5%) | 15 (34.1%) |
| 2 | 18 (27.7%) | 12 (18.5%) | 23 (35.4%) | 21 (36.2%) | 13 (29.5%) |
| 3 | 5 (7.7%) | 18 (27.7%) | 14 (21.9%) | 8 (13.8%) | 4 (9.1%) |
| 4 | 1 (1.5%) | 7 (10.8%) | 5 (7.8%) | 6 (10.3%) | 5 (11.4%) |
| 5 | 0 | 0 | 2 (3.1%) | 2 (3.5%) | 0 |
| 6 | 0 | 1 (1.5%) | 2 (3.1%) | 0 | 0 |
| Missing data * | 3 (4.6%) | 1 (1.5%) | 0 | 0 | 6 (13.6%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanella, M.-C.; Vu, D.-L.; Hosszu-Fellous, K.; Neofytos, D.; Van Delden, C.; Turin, L.; Poncet, A.; Simonetta, F.; Masouridi-Levrat, S.; Chalandon, Y.; et al. Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma. Viruses 2023, 15, 928. https://doi.org/10.3390/v15040928
Zanella M-C, Vu D-L, Hosszu-Fellous K, Neofytos D, Van Delden C, Turin L, Poncet A, Simonetta F, Masouridi-Levrat S, Chalandon Y, et al. Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma. Viruses. 2023; 15(4):928. https://doi.org/10.3390/v15040928
Chicago/Turabian StyleZanella, Marie-Céline, Diem-Lan Vu, Krisztina Hosszu-Fellous, Dionysios Neofytos, Chistian Van Delden, Lara Turin, Antoine Poncet, Federico Simonetta, Stavroula Masouridi-Levrat, Yves Chalandon, and et al. 2023. "Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma" Viruses 15, no. 4: 928. https://doi.org/10.3390/v15040928
APA StyleZanella, M.-C., Vu, D.-L., Hosszu-Fellous, K., Neofytos, D., Van Delden, C., Turin, L., Poncet, A., Simonetta, F., Masouridi-Levrat, S., Chalandon, Y., Cordey, S., & Kaiser, L. (2023). Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma. Viruses, 15(4), 928. https://doi.org/10.3390/v15040928







