The Three-Cornered Alfalfa Hopper, Spissistilus festinus, Is a Vector of Grapevine Red Blotch Virus in Vineyards
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Spissistilus festinus Population
2.3. GRBV Acquisition by S. festinus in the Vineyards
2.4. Transmission Assays of GRBV by S. festinus from Infected Grapevines to Healthy Grapevines in the Greenhouse
2.5. Inoculation of P. vulgaris with GRBV in the Greenhouse and Virus Acquisition by S. festinus
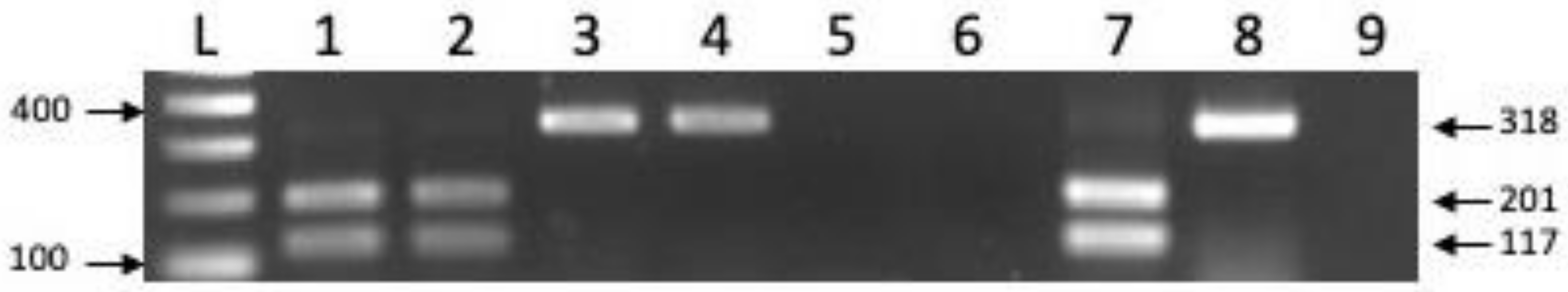
2.6. Agrobacterium tumefaciens Detection via PCR
2.7. Transmission of Grapevine Red Blotch Virus by S. festinus from Infected P. vulgaris to Grapevines in the Vineyard
2.8. Nucleic Acid Extraction from Plant and S. festinus Tissues
2.9. GRBV Detection via PCR and qPCR
2.10. GRBV Phylogenetic Clade Determination via Restriction Digestion of Rep PCR Amplicons
2.11. Statistics
3. Results
3.1. GRBV Titer Is Consistent in the Middle and Lower Canopy of Infected ‘Cabernet franc’ Vines Selected for Acquisition Experiments in the Vineyard in June
3.2. Spissistilus festinus Acquires GRBV from Infected Grapevines in the Vineyards
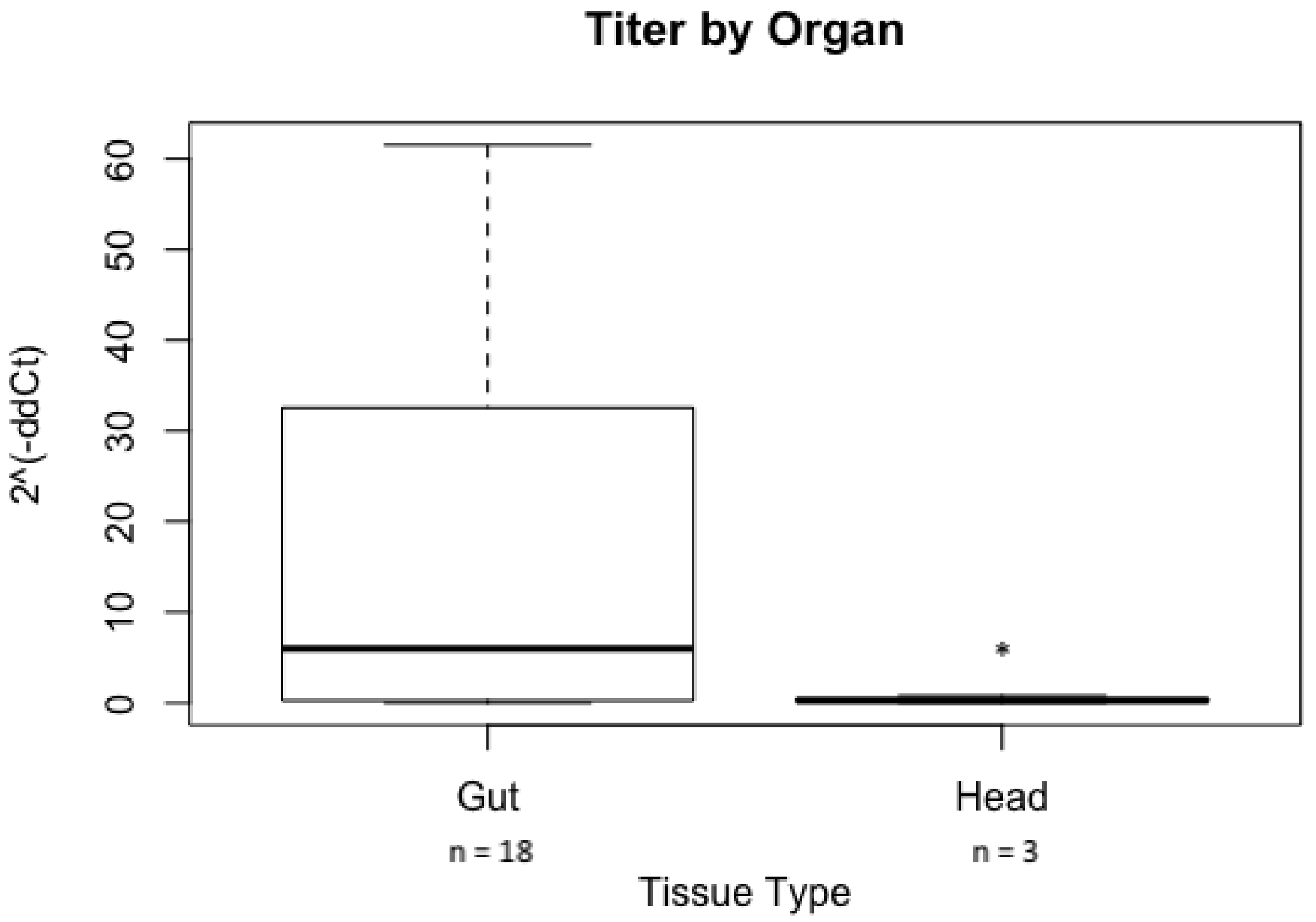
3.3. GRBV Titer in S. festinus after Acquisition from Infected Grapevines in the Vineyard Vary by Insect Tissue Type
3.4. The Rate of GRBV Ingestion by Spissistilus festinus from Infected Grapevines in the Vineyard Is Similar between Phylogenetic Clades and Canopy Position
3.5. Spissistilus festinus Transmit GRBV from Infected Grapevines to Healthy Grapevines in the Greenhouse
3.6. Restricting the Feeding Area of S. festinus on Grapevine Tissue and Limiting the Number of Insects Increase the Ability to Document GRBV Transmission in the Vineyard
3.7. GRBV Is Detected in S. festinus-Inoculated Grapevines after One Dormancy, but No Disease Symptoms Are Apparent
3.8. Co-Infection of GRBV Isolates from Both Phylogenetic Clades Can Be Detected in a Single Vine in the Vineyard
3.9. Girdling and Foliar Reddening Due to S. festinus Feeding in the Vineyard Is Not Indicative of GRBV Transmission
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cieniewicz, E.J.; Pethybridge, S.J.; Gorny, A.; Madden, L.V.; McLane, H.; Perry, K.L.; Fuchs, M. Spatiotemporal Spread of Grapevine Red Blotch-Associated Virus in a California Vineyard. Virus Res. 2017, 241, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Sudarshana, M.R.; Perry, K.L.; Fuchs, M.F. Grapevine Red Blotch-Associated Virus, an Emerging Threat to the Grapevine Industry. Phytopathology 2015, 105, 1026–1032. [Google Scholar] [CrossRef]
- Cieniewicz, E.J.; Pethybridge, S.J.; Loeb, G.; Perry, K.; Fuchs, M. Insights into the Ecology of Grapevine red blotch virus in a Diseased Vineyard. Phytopathology 2017, 108, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Rumbaugh, A.C.; Girardello, R.C.; Cooper, M.L.; Plank, C.; Kurtural, S.K.; Oberholster, A. Impact of Rootstock and Season on Red Blotch Disease Expression in Cabernet Sauvignon (V. Vinifera). Plants 2021, 10, 1583. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Ulate, B.; Hopfer, H.; Figueroa-Balderas, R.; Ye, Z.; Rivero, R.M.; Albacete, A.; Pérez-Alfocea, F.; Koyama, R.; Anderson, M.M.; Smith, R.J.; et al. Red Blotch Disease Alters Grape Berry Development and Metabolism by Interfering with the Transcriptional and Hormonal Regulation of Ripening. J. Exp. Bot. 2017, 68, 1225–1238. [Google Scholar] [CrossRef]
- Cauduro Girardello, R.; Rich, V.; Smith, R.J.; Brenneman, C.; Heymann, H.; Oberholster, A. The Impact of Grapevine Red Blotch Disease on Vitis vinifera L. Chardonnay Grape and Wine Composition and Sensory Attributes over Three Seasons. J. Sci. Food Agric. 2020, 100, 1436–1447. [Google Scholar] [CrossRef]
- Martínez-Lüscher, J.; Plank, C.M.; Brillante, L.; Cooper, M.L.; Smith, R.J.; Al-Rwahnih, M.; Yu, R.; Oberholster, A.; Girardello, R.; Kurtural, S.K. Grapevine Red Blotch Virus May Reduce Carbon Translocation Leading to Impaired Grape Berry Ripening. J. Agric. Food Chem. 2019, 67, 2437–2448. [Google Scholar] [CrossRef]
- Ricketts, K.D.; Gómez, M.I.; Fuchs, M.F.; Martinson, T.E.; Smith, R.J.; Cooper, M.L.; Moyer, M.M.; Wise, A. Mitigating the Economic Impact of Grapevine Red Blotch: Optimizing Disease Management Strategies in US Vineyards. Am. J. Enol. Vitic. 2017, 68, 127–135. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Dave, A.; Anderson, M.M.; Rowhani, A.; Uyemoto, J.K.; Sudarshana, M.R. Association of a DNA Virus with Grapevines Affected by Red Blotch Disease in California. Phytopathology 2013, 103, 1069–1076. [Google Scholar] [CrossRef]
- Krenz, B.; Thompson, J.R.; Fuchs, M.; Perry, K.L. Complete Genome Sequence of a New Circular DNA Virus from Grapevine. J. Virol. 2012, 86, 7715. [Google Scholar] [CrossRef]
- Yepes, L.M.; Cieniewicz, E.; Krenz, B.; McLane, H.; Thompson, J.R.; Perry, K.L.; Fuchs, M. Causative Role of Grapevine Red Blotch Virus in Red Blotch Disease. Phytopathology 2018, 108, 902–909. [Google Scholar] [CrossRef]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Rivera Bustamante, R.F.; Zerbini, F.M.; Adkins, S.; et al. World Management of Geminiviruses. Annu. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.W.; Rivera-Bustamante, R.; Murilo Zerbini, F.; Martin, D.P. Capulavirus and Grablovirus: Two New Genera in the Family Geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.R. Analysis of the Genome of Grapevine Red Blotch Virus and Related Grabloviruses Indicates Diversification Prior to the Arrival of Vitis Vinifera in North America. J. Gen. Virol. 2022, 103, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Krenz, B.; Thompson, J.R.; McLane, H.L.; Fuchs, M.; Perry, K.L. Grapevine Red Blotch-Associated Virus Is Widespread in the United States. Phytopathology 2014, 104, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Bahder, B.W.; Zalom, F.G.; Sudarshana, M.R. An Evaluation of the Flora Adjacent to Wine Grape Vineyards for the Presence of Alternative Host Plants of Grapevine Red Blotch-Associated Virus. Plant Dis. 2016, 100, 1571–1574. [Google Scholar] [CrossRef]
- Wilson, H.; Hogg, B.N.; Blaisdell, G.K.; Andersen, J.C.; Yazdani, A.S.; Billings, A.C.; Ooi, K.M.; Soltani, N.; Almeida, R.P.P.; Cooper, M.L.; et al. Survey of Vineyard Insects and Plants to Identify Potential Insect Vectors and Noncrop Reservoirs of Grapevine Red Blotch Virus. PhytoFrontiers 2022, 2, 66–73. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Rowhani, A.; Golino, D. First Report of Grapevine Red Blotch-Associated Virus in Archival Grapevine Material From Sonoma County, California. Plant Dis. 2015, 99, 895. [Google Scholar] [CrossRef]
- Schoelz, J.; Volenberg, D.; Adhab, M.; Fang, Z.; Klassen, V.; Spinka, C.; Rwahnih, M.A. A Survey of Viruses Found in Grapevine Cultivars Grown in Missouri. Am. J. Enol. Vitic. 2021, 72, 73–84. [Google Scholar] [CrossRef]
- Soltani, N.; Hu, R.; Hensley, D.D.; Lockwood, D.L.; Perry, K.L.; Hajimorad, M.R. A Survey for Nine Major Viruses of Grapevines in Tennessee Vineyards. Plant Health Prog. 2020, 21, 157–161. [Google Scholar] [CrossRef]
- Cieniewicz, E.; Thompson, J.R.; McLane, H.; Perry, K.L.; Dangl, G.S.; Corbett, Q.; Martinson, T.; Wise, A.; Wallis, A.; O’Connell, J.; et al. Prevalence and Genetic Diversity of Grabloviruses in Free-Living Vitis spp. Plant Dis. 2018, 102, 2308–2316. [Google Scholar] [CrossRef] [PubMed]
- Perry, K.; Mclane, H.; Hyder, M.; Dangl, G.; Thompson, J.; Fuchs, M. Grapevine Red Blotch-Associated Virus Is Present in Free-Living Vitis spp. Proximal to Cultivated Grapevines. Phytopathology 2016, 106, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Achala, N.; DeShields, J.B.; Levin, A.D.; Hilton, R.; Rijal, J. Epidemiology of Grapevine Red Blotch Disease Progression in Southern Oregon Vineyards. Am. J. Enol. Vitic. 2022, 73, 116–124. [Google Scholar] [CrossRef]
- Cieniewicz, E.J.; Qiu, W.; Saldarelli, P.; Fuchs, M. Believing Is Seeing: Lessons from Emerging Viruses in Grapevine. J. Plant Pathol. 2020, 102, 619–632. [Google Scholar] [CrossRef]
- Fall, M.L.; Xu, D.; Lemoyne, P.; Moussa, I.E.B.; Beaulieu, C.; Carisse, O. A Diverse Virome of Leafroll-Infected Grapevine Unveiled by DsRNA Sequencing. Viruses 2020, 12, 1142. [Google Scholar] [CrossRef]
- Poojari, S.; Lowery, D.; Rott, M.; Schmidt, A.; Úrbez-Torres, J.R. Incidence, Distribution and Genetic Diversity of Grapevine Red Blotch Virus in British Columbia. Can. J. Plant Pathol. 2017, in press. [CrossRef]
- Xiao, H.; Kim, W.-S.; Meng, B. A Highly Effective and Versatile Technology for the Isolation of RNAs from Grapevines and Other Woody Perennials for Use in Virus Diagnostics. Virol. J. 2015, 12, 171. [Google Scholar] [CrossRef]
- Lim, S.; Igori, D.; Zhao, F.; Moon, J.S.; Cho, I.-S.; Choi, G.-S. First Report of Grapevine Red Blotch-Associated Virus on Grapevine in Korea. Plant Dis. 2016, 100, 1957. [Google Scholar] [CrossRef]
- Reynard, J.-S.; Brodard, J.; Dubuis, N.; Zufferey, V.; Schumpp, O.; Schaerer, S.; Gugerli, P. Grapevine Red Blotch Virus: Absence in Swiss Vineyards and Analysis of Potential Detrimental Effect on Viticultural Performance. Plant Dis. 2017, 102, 651–655. [Google Scholar] [CrossRef]
- Gasperin-Bulbarela, J.; Licea-Navarro, A.F.; Pino-Villar, C.; Hernández-Martínez, R.; Carrillo-Tripp, J. First Report of Grapevine Red Blotch Virus in Mexico. Plant Dis. 2018, 103, 381. [Google Scholar] [CrossRef]
- Luna, F.; Debat, H.; Moyano, S.; Zavallo, D.; Asurmendi, S.; Gomez-Talquenca, S. First Report of Grapevine Red Blotch Virus Infecting Grapevine in Argentina. J. Plant Pathol. 2019, 101, 1239. [Google Scholar] [CrossRef]
- Marwal, A.; Kumar, R.; Paul Khurana, S.M.; Gaur, R.K. Complete Nucleotide Sequence of a New Geminivirus Isolated from Vitis Vinifera in India: A Symptomless Host of Grapevine Red Blotch Virus. Virusdisease 2019, 30, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Bertazzon, N.; Migliaro, D.; Rossa, A.; Filippin, L.; Casarin, S.; Giust, M.; Brancadoro, L.; Crespan, M.; Angelini, E. Grapevine Red Blotch Virus Is Sporadically Present in a Germplasm Collection in Northern Italy. J. Plant Dis. Prot. 2021, 128, 1115–1119. [Google Scholar] [CrossRef]
- Reynard, J.-S.; Brodard, J.; Dubuis, N.; Kellenberger, I.; Spilmont, A.-S.; Roquis, D.; Maliogka, V.; Marchal, C.; Dedet, S.; Gning, O.; et al. Screening of Grapevine Red Blotch Virus in Two European Ampelographic Collections. J. Plant Pathol. 2022, 104, 9–15. [Google Scholar] [CrossRef]
- Grapevine Red Blotch Detections—Agriculture. Available online: https://agriculture.vic.gov.au/biosecurity/moving-plants-and-plant-products/biosecurity-updates/grapevine-red-blotch-virus-detections (accessed on 14 November 2022).
- Cieniewicz, E.; Flasco, M.; Brunelli, M.; Onwumelu, A.; Wise, A.; Fuchs, M.F. Differential Spread of Grapevine Red Blotch Virus in California and New York Vineyards. Phytobiomes J. 2019, 3, 203–211. [Google Scholar] [CrossRef]
- Dalton, D.T.; Hilton, R.J.; Kaiser, C.; Daane, K.M.; Sudarshana, M.R.; Vo, J.; Zalom, F.G.; Buser, J.Z.; Walton, V.M. Spatial Associations of Vines Infected with Grapevine Red Blotch Virus in Oregon Vineyards. Plant Dis. 2019, 103, 1507–1514. [Google Scholar] [CrossRef]
- Bahder, B.W.; Zalom, F.G.; Jayanth, M.; Sudarshana, M.R. Phylogeny of Geminivirus Coat Protein Sequences and Digital PCR Aid in Identifying Spissistilus festinus as a Vector of Grapevine Red Blotch-Associated Virus. Phytopathology 2016, 106, 1223–1230. [Google Scholar] [CrossRef]
- Flasco, M.; Hoyle, V.; Cieniewicz, E.; Roy, B.; McLane, H.; Perry, K.L.; Loeb, G.M.; Nault, B.; Cilia, M.; Fuchs, M. Grapevine Red Blotch Virus Is Transmitted by the Three-Cornered Alfalfa Hopper in a Circulative, Nonpropagative Mode with Unique Attributes. Phytopathology 2021, 111, 1851–1861. [Google Scholar] [CrossRef]
- Hoyle, V.; Flasco, M.T.; Choi, J.; Cieniewicz, E.J.; McLane, H.; Perry, K.; Dangl, G.; Rwahnih, M.A.; Heck, M.; Loeb, G.; et al. Transmission of Grapevine Red Blotch Virus by Spissistilus festinus [Say, 1830] (Hemiptera: Membracidae) between Free-Living Vines and Vitis Vinifera ‘Cabernet Franc’. Viruses 2022, 14, 1156. [Google Scholar] [CrossRef]
- Cieniewicz, E.; Poplaski, V.; Brunelli, M.; Dombroskie, J.; Fuchs, M. Two Distinct Genotypes of Spissistilus festinus (Say, 1830) (Hemiptera, Membracidae) in the United States Revealed by Phylogenetic and Morphological Analyses. Insects 2020, 11, 80. [Google Scholar] [CrossRef]
- Deeba, F.; Hyder, M.Z.; Shah, S.H.; Naqvi, S.M.S. Multiplex PCR Assay for Identification of Commonly Used Disarmed Agrobacterium Tumefaciens Strains. SpringerPlus 2014, 3, 358. [Google Scholar] [CrossRef]
- Setiono, F.J.; Chatterjee, D.; Fuchs, M.; Perry, K.L.; Thompson, J.R. The Distribution and Detection of Grapevine Red Blotch Virus in Its Host Depend on Time of Sampling and Tissue Type. Plant Dis. 2018, 102, 2187–2193. [Google Scholar] [CrossRef]
- Czosnek, H. Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Springer Science & Business Media: Dordrecht, The Netherlands, 2007; ISBN 978-1-4020-4769-5. [Google Scholar]
- Morra, M.R.; Petty, I.T.D. Tissue Specificity of Geminivirus Infection Is Genetically Determined. Plant Cell 2000, 12, 2259–2270. [Google Scholar] [CrossRef]
- Preto, C.R.; Sudarshana, M.R.; Zalom, F.G. Feeding and Reproductive Hosts of Spissistilus festinus (Say) (Hemiptera: Membracidae) Found in Californian Vineyards. J. Econ. Entomol. 2018, 111, 2531–2535. [Google Scholar] [CrossRef] [PubMed]
- Preto, C.R.; Sudarshana, M.R.; Bollinger, M.L.; Zalom, F.G. Vitis vinifera (Vitales: Vitaceae) as a Reproductive Host of Spissistilus festinus (Hemiptera: Membracidae). J. Insect Sci. 2018, 18, 20. [Google Scholar] [CrossRef] [PubMed]
- Preto, C.R.; Bahder, B.W.; Bick, E.N.; Sudarshana, M.R.; Zalom, F.G. Seasonal Dynamics of Spissistilus Festinus (Hemiptera: Membracidae) in a Californian Vineyard. J. Econ. Entomol. 2019, 112, 1138–1144. [Google Scholar] [CrossRef]
- Wilson, H.; Yazdani, A.S.; Daane, K.M. Influence of Riparian Habitat and Ground Covers on Threecornered Alfalfa Hopper (Hemiptera: Membracidae) Populations in Vineyards. J. Econ. Entomol. 2020, 113, 2354–2361. [Google Scholar] [CrossRef] [PubMed]
- Blaisdell, G.K.; Cooper, M.L.; Kuhn, E.J.; Taylor, K.A.; Daane, K.M.; Almeida, R.P.P. Disease Progression of Vector-Mediated Grapevine Leafroll-Associated Virus 3 Infection of Mature Plants under Commercial Vineyard Conditions. Eur. J. Plant Pathol. 2016, 146, 105–116. [Google Scholar] [CrossRef]
- Feil, H.; Feil, W.S.; Purcell, A.H. Effects of Date of Inoculation on the Within-Plant Movement of Xylella fastidiosa and Persistence of Pierce’s Disease Within Field Grapevines. Phytopathology 2003, 93, 244–251. [Google Scholar] [CrossRef]
- Purcell, A.H. Vector Preference and Inoculation Efficiency as Components of Resistance to Pierce’s Disease in European Grape Cultivars. Phytopathology 1981, 71, 429. [Google Scholar] [CrossRef]
- Hooks, C.R.R.; Manandhar, R.; Perez, E.P.; Wang, K.-H.; Almeida, R.P.P. Comparative Susceptibility of Two Banana Cultivars to Banana Bunchy Top Virus Under Laboratory and Field Environments. J. Econ. Entomol. 2009, 102, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Fiore, N.; Prodan, S.; Montealegre, J.; Aballay, E.; Pino, A.M.; Zamorano, A. Survey of Grapevine Viruses in Chile. J. Plant Pathol. 2008, 90, 125–130. [Google Scholar]
- Fuchs, M.; Martinson, T.E.; Loeb, G.M.; Hoch, H.C. Survey for the Three Major Leafroll Disease-Associated Viruses in Finger Lakes Vineyards in New York. Plant Dis. 2009, 93, 395–401. [Google Scholar] [CrossRef]
- Sharma, A.M.; Baraff, B.; Hutchins, J.T.; Wong, M.K.; Blaisdell, G.K.; Cooper, M.L.; Daane, K.M.; Almeida, R.P.P. Relative Prevalence of Grapevine Leafroll-Associated Virus Species in Wine Grape-Growing Regions of California. PLoS ONE 2015, 10, e0142120. [Google Scholar] [CrossRef]
- LaFond, H.F.; Volenberg, D.S.; Schoelz, J.E.; Finke, D.L. Identification of Potential Grapevine Red Blotch Virus Vector in Missouri Vineyards. Am. J. Enol. Vitic. 2022, 73, 247–255. [Google Scholar] [CrossRef]
- Kahl, D.; Úrbez-Torres, J.R.; Kits, J.; Hart, M.; Nyirfa, A.; Lowery, D.T. Identification of Candidate Insect Vectors of Grapevine Red Blotch Virus by Means of an Artificial Feeding Diet. Can. J. Plant Pathol. 2021, 43, 905–913. [Google Scholar] [CrossRef]


| Sleeve Configuration | 2020 a | 2021 b | 2022 c | Total d | Total f | ||||
|---|---|---|---|---|---|---|---|---|---|
| NY e | CA | NY | CA | NY | CA | NY | CA | ||
| Tootsie roll g | 0/10 | n/a | 0/4 | 0/6 | n/a | n/a | 0/14 | 0/6 | 0/20 |
| Peppermint candy h | n/a | n/a | 0/11 | 0/15 | n/a | n/a | 0/11 | 0/15 | 0/26 |
| Lollipop i | n/a | n/a | 0/5 | n/a | 5/11 | 1/20 | 5/16 | 1/20 | 6/36 |
| Total j | 0/10 | 0/41 | 6/31 | 6/82 | |||||
| Sleeve Configuration | Vine Number | Girdles a | Red Leaves b | GRBV c |
|---|---|---|---|---|
| Lollipop d | 1 | 3 | 0 | - |
| 2 | 3 | 0 | - | |
| 3 | 2 | 0 | - | |
| 4 | 1 | 0 | - | |
| 5 | 2 | 0 | - | |
| Peppermint candy e | 1 | 15 | 1 | - |
| 2 | 23 | 1 | - | |
| 3 | 10 | 3 | - | |
| 4 | 19 | 2 | - | |
| 5 | 16 | 2 | - | |
| 6 | 16 | 2 | - | |
| 7 | 9 | 0 | - | |
| 8 | 12 | 2 | - | |
| 9 | 14 | 5 | - | |
| 10 | 13 | 3 | - | |
| 11 | 10 | 2 | - | |
| 12 * | 15 | 3 | - | |
| Tootsie roll f | 1 | 17 | 5 | - |
| 2 | 30 | 4 | - | |
| 3 | 21 | 6 | - | |
| 4 | 20 | 9 | - | |
| 5 * | 25 | 4 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flasco, M.T.; Hoyle, V.; Cieniewicz, E.J.; Loeb, G.; McLane, H.; Perry, K.; Fuchs, M.F. The Three-Cornered Alfalfa Hopper, Spissistilus festinus, Is a Vector of Grapevine Red Blotch Virus in Vineyards. Viruses 2023, 15, 927. https://doi.org/10.3390/v15040927
Flasco MT, Hoyle V, Cieniewicz EJ, Loeb G, McLane H, Perry K, Fuchs MF. The Three-Cornered Alfalfa Hopper, Spissistilus festinus, Is a Vector of Grapevine Red Blotch Virus in Vineyards. Viruses. 2023; 15(4):927. https://doi.org/10.3390/v15040927
Chicago/Turabian StyleFlasco, Madison T., Victoria Hoyle, Elizabeth J. Cieniewicz, Greg Loeb, Heather McLane, Keith Perry, and Marc F. Fuchs. 2023. "The Three-Cornered Alfalfa Hopper, Spissistilus festinus, Is a Vector of Grapevine Red Blotch Virus in Vineyards" Viruses 15, no. 4: 927. https://doi.org/10.3390/v15040927
APA StyleFlasco, M. T., Hoyle, V., Cieniewicz, E. J., Loeb, G., McLane, H., Perry, K., & Fuchs, M. F. (2023). The Three-Cornered Alfalfa Hopper, Spissistilus festinus, Is a Vector of Grapevine Red Blotch Virus in Vineyards. Viruses, 15(4), 927. https://doi.org/10.3390/v15040927








