Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Antiviral Prophylaxis, Treatment and Routine Screening
2.3. Virological Testing
2.4. Nucleic Acid Extraction and r(RT-)PCR Screening
2.5. Statistical Methods
3. Results
3.1. Patient Characteristics
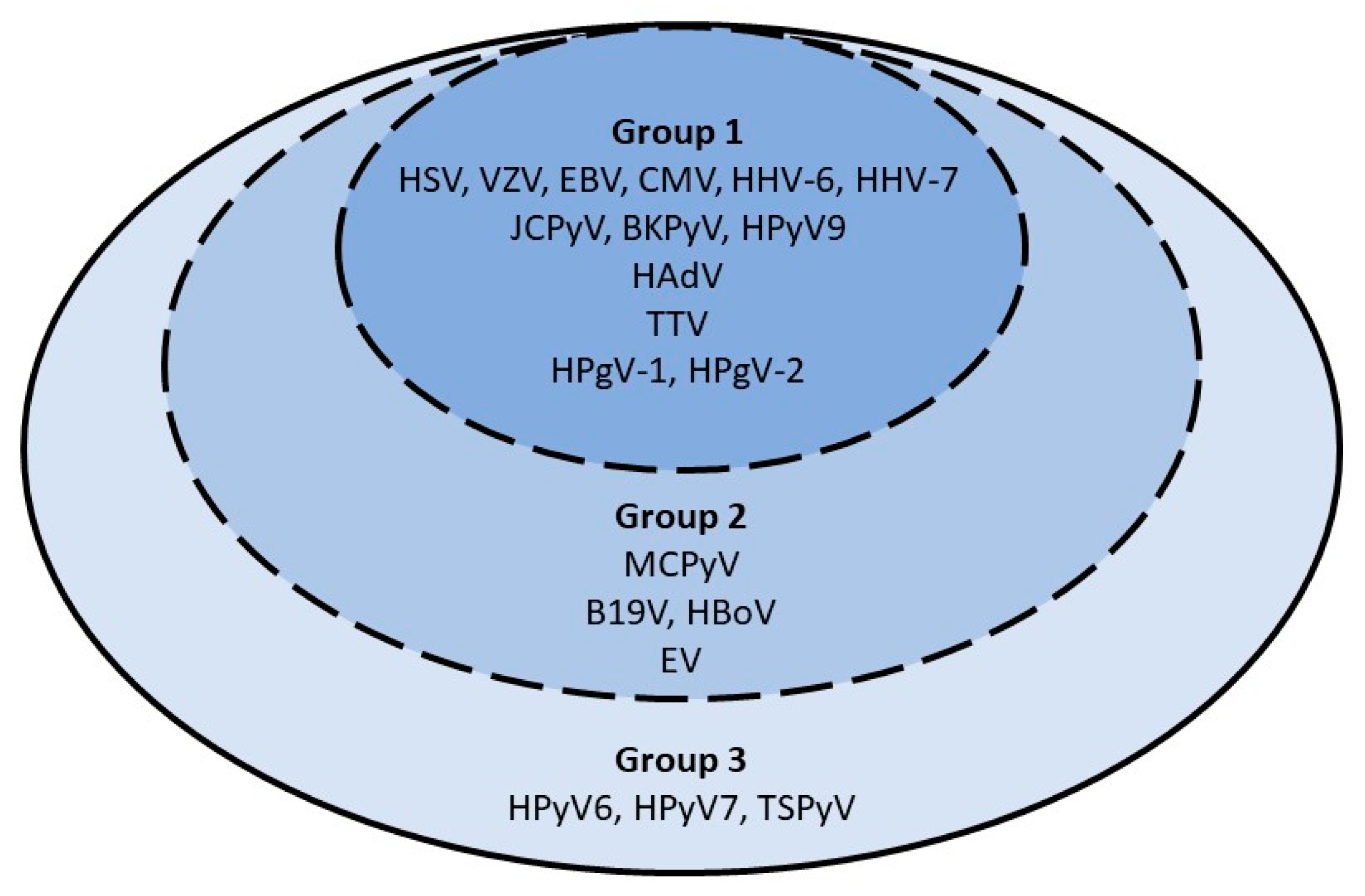
3.2. Virological Testing
3.2.1. Period Prevalence
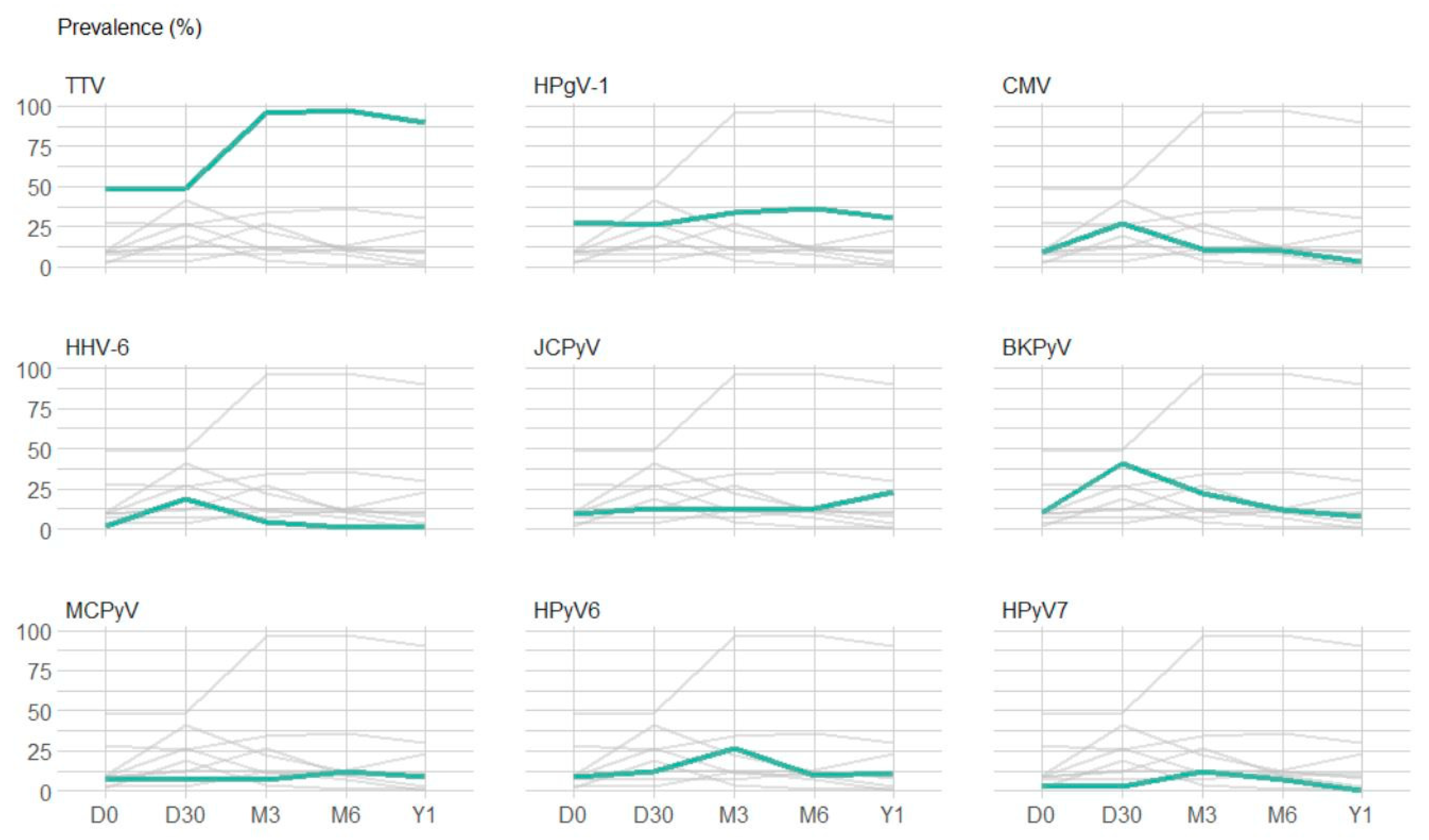
3.2.2. Point Prevalence and Dynamic Evolution of Prevalence
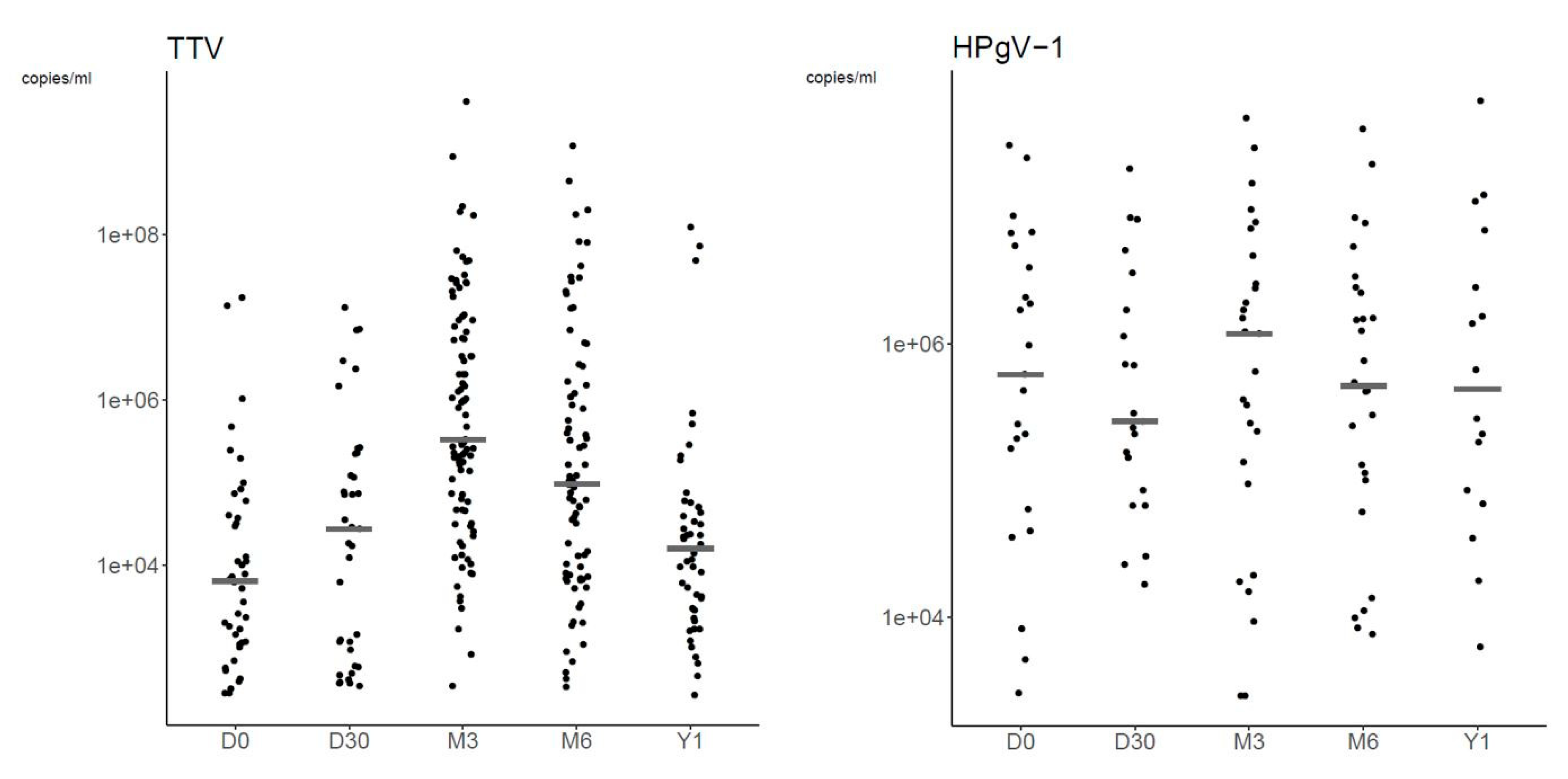
3.2.3. Plasma Viral Loads
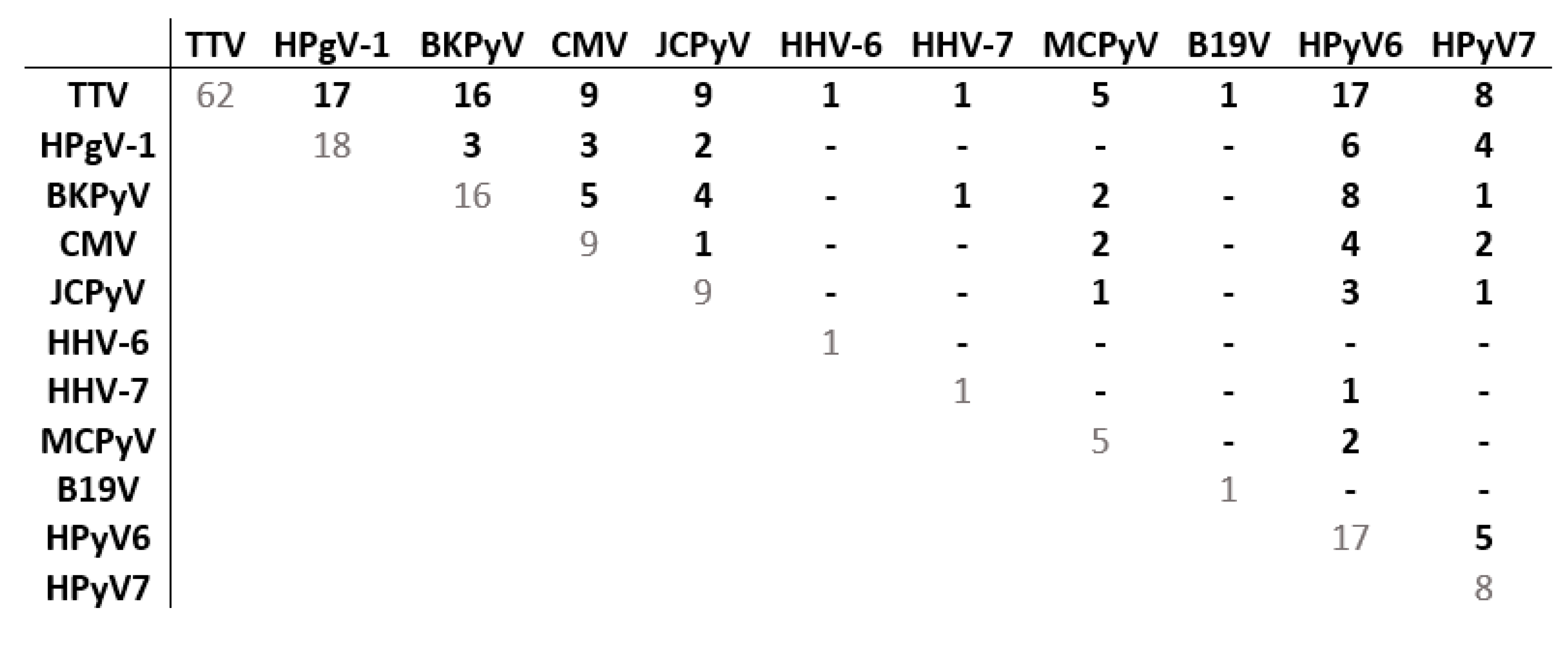
3.2.4. Co-Detections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin-Pena, A.; Aguilar-Guisado, M.; Espigado, I.; Parody, R.; Miguel Cisneros, J. Prospective study of infectious complications in allogeneic hematopoietic stem cell transplant recipients. Clin. Transplant. 2011, 25, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.L.; Dayer, J.A.; Masouridi-Levrat, S.; Combescure, C.; Boely, E.; Khanna, N.; Mueller, N.J.; Kleber, M.; Medinger, M.; Halter, J.; et al. Microbiologically documented infections after adult allogeneic hematopoietic cell transplantation: A 5-year analysis within the Swiss Transplant Cohort study. Transpl. Infect. Dis. 2020, 22, e13289. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.L.; Cordey, S.; Simonetta, F.; Brito, F.; Docquier, M.; Turin, L.; van Delden, C.; Boely, E.; Dantin, C.; Pradier, A.; et al. Human pegivirus persistence in human blood virome after allogeneic haematopoietic stem-cell transplantation. Clin. Microbiol. Infect. 2019, 25, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Zanella, M.C.; Cordey, S.; Kaiser, L. Beyond cytomegalovirus and Epstein-Barr virus: A review of viruses composing the blood virome of solid organ transplant and hematopoietic stem cell transplant recipients. Clin. Microbiol. Rev. 2020, 33, e00027-20. [Google Scholar] [CrossRef]
- Zanella, M.C.; Cordey, S.; Laubscher, F.; Docquier, M.; Vieille, G.; Van Delden, C.; Braunersreuther, V.; Ta, M.K.; Lobrinus, J.A.; Masouridi-Levrat, S.; et al. Unmasking viral sequences by metagenomic next-generation sequencing in adult human blood samples during steroid-refractory/dependent graft-versus-host disease. Microbiome 2021, 9, 28. [Google Scholar] [CrossRef]
- Hill, J.A.; Mayer, B.T.; Xie, H.; Leisenring, W.M.; Huang, M.L.; Stevens-Ayers, T.; Milano, F.; Delaney, C.; Sorror, M.L.; Sandmaier, B.M.; et al. The cumulative burden of double-stranded DNA virus detection after allogeneic HCT is associated with increased mortality. Blood 2017, 129, 2316–2325. [Google Scholar] [CrossRef]
- Hill, J.A.; Mayer, B.T.; Xie, H.; Leisenring, W.M.; Huang, M.L.; Stevens-Ayers, T.; Milano, F.; Delaney, C.; Jerome, K.R.; Zerr, D.M.; et al. Kinetics of Double-Stranded DNA Viremia After Allogeneic Hematopoietic Cell Transplantation. Clin. Infect. Dis. 2018, 66, 368–375. [Google Scholar] [CrossRef]
- Royston, L.; Royston, E.; Masouridi-Levrat, S.; Vernaz, N.; Chalandon, Y.; Van Delden, C.; Neofytos, D. Letermovir Primary Prophylaxis in High-Risk Hematopoietic Cell Transplant Recipients: A Matched Cohort Study. Vaccines 2021, 9, 372. [Google Scholar] [CrossRef]
- Schibler, M.; Yerly, S.; Vieille, G.; Docquier, M.; Turin, L.; Kaiser, L.; Tapparel, C. Critical analysis of rhinovirus RNA load quantification by real-time reverse transcription-PCR. J. Clin. Microbiol. 2012, 50, 2868–2872. [Google Scholar] [CrossRef]
- Armand, P.; Gibson, C.J.; Cutler, C.; Ho, V.T.; Koreth, J.; Alyea, E.P.; Ritz, J.; Sorror, M.L.; Lee, S.J.; Deeg, H.J.; et al. A disease risk index for patients undergoing allogeneic stem cell transplantation. Blood 2012, 120, 905–913. [Google Scholar] [CrossRef]
- Mouton, W.; Conrad, A.; Bal, A.; Boccard, M.; Malcus, C.; Ducastelle-Lepretre, S.; Balsat, M.; Barraco, F.; Larcher, M.V.; Fossard, G.; et al. Torque Teno Virus Viral Load as a Marker of Immune Function in Allogeneic Haematopoietic Stem Cell Transplantation Recipients. Viruses 2020, 12, 1292. [Google Scholar] [CrossRef]
- Wohlfarth, P.; Leiner, M.; Schoergenhofer, C.; Hopfinger, G.; Goerzer, I.; Puchhammer-Stoeckl, E.; Rabitsch, W. Torquetenovirus Dynamics and Immune Marker Properties in Patients Following Allogeneic Hematopoietic Stem Cell Transplantation: A Prospective Longitudinal Study. Biol. Blood Marrow Transplant. 2018, 24, 194–199. [Google Scholar] [CrossRef]
- Albert, E.; Solano, C.; Gimenez, E.; Focosi, D.; Perez, A.; Macera, L.; Pinana, J.L.; Mateo, E.M.; Boluda, J.C.H.; Maggi, F.; et al. Kinetics of Alphatorquevirus plasma DNAemia at late times after allogeneic hematopoietic stem cell transplantation. Med. Microbiol. Immunol. 2019, 208, 253–258. [Google Scholar] [CrossRef]
- Gilles, R.; Herling, M.; Holtick, U.; Heger, E.; Awerkiew, S.; Fish, I.; Holler, K.; Sierra, S.; Knops, E.; Kaiser, R.; et al. Dynamics of Torque Teno virus viremia could predict risk of complications after allogeneic hematopoietic stem cell transplantation. Med. Microbiol. Immunol. 2017, 206, 355–362. [Google Scholar] [CrossRef]
- Kaczorowska, J.; van der Hoek, L. Human anelloviruses: Diverse, omnipresent and commensal members of the virome. FEMS Microbiol. Rev. 2020, 44, 305–313. [Google Scholar] [CrossRef]
- Arze, C.A.; Springer, S.; Dudas, G.; Patel, S.; Bhattacharyya, A.; Swaminathan, H.; Brugnara, C.; Delagrave, S.; Ong, T.; Kahvejian, A.; et al. Global genome analysis reveals a vast and dynamic anellovirus landscape within the human virome. Cell Host Microbe 2021, 29, 1305–1315. [Google Scholar] [CrossRef]
- Cebria-Mendoza, M.; Arbona, C.; Larrea, L.; Diaz, W.; Arnau, V.; Pena, C.; Bou, J.V.; Sanjuan, R.; Cuevas, J.M. Deep viral blood metagenomics reveals extensive anellovirus diversity in healthy humans. Sci. Rep. 2021, 11, 6921. [Google Scholar] [CrossRef]
- Pradier, A.; Masouridi-Levrat, S.; Bosshard, C.; Dantin, C.; Vu, D.L.; Zanella, M.C.; Boely, E.; Tapparel, C.; Kaiser, L.; Chalandon, Y.; et al. Torque Teno Virus as a Potential Biomarker for Complications and Survival After Allogeneic Hematopoietic Stem Cell Transplantation. Front. Immunol. 2020, 11, 998. [Google Scholar] [CrossRef]
- Focosi, D.; Maggi, F.; Albani, M.; Macera, L.; Ricci, V.; Gragnani, S.; Di Beo, S.; Ghimenti, M.; Antonelli, G.; Bendinelli, M.; et al. Torquetenovirus viremia kinetics after autologous stem cell transplantation are predictable and may serve as a surrogate marker of functional immune reconstitution. J. Clin. Virol. 2010, 47, 189–192. [Google Scholar] [CrossRef]
- Albert, E.; Solano, C.; Pascual, T.; Torres, I.; Macera, L.; Focosi, D.; Maggi, F.; Gimenez, E.; Amat, P.; Navarro, D. Dynamics of Torque Teno virus plasma DNAemia in allogeneic stem cell transplant recipients. J. Clin. Virol. 2017, 94, 22–28. [Google Scholar] [CrossRef]
- Schmitz, J.; Kobbe, G.; Kondakci, M.; Schuler, E.; Magorsch, M.; Adams, O. The Value of Torque Teno Virus (TTV) as a Marker for the Degree of Immunosuppression in Adult Patients after Hematopoietic Stem Cell Transplantation (HSCT). Biol. Blood Marrow Transplant. 2020, 26, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ma, H.H.; Lau, G.K.; Leung, Y.K.; Yao, C.L.; Chong, Y.T.; Tang, W.H.; Yao, J.L. Prevalence of hepatitis G virus infection and homology of different viral strains in Southern China. World J. Gastroenterol. 2002, 8, 1081–1087. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Liang, Y.; Hu, L.; Chen, S. Prevalence and risk factors of human pegivirus type 1 infection in hematopoietic stem cell transplantation patients. Int. J. Infect. Dis. 2019, 85, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Chivero, E.T.; Stapleton, J.T. Tropism of human pegivirus (formerly known as GB virus C/hepatitis G virus) and host immunomodulation: Insights into a highly successful viral infection. J. Gen. Virol. 2015, 96, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Cebria-Mendoza, M.; Bracho, M.A.; Arbona, C.; Larrea, L.; Diaz, W.; Sanjuan, R.; Cuevas, J.M. Exploring the Diversity of the Human Blood Virome. Viruses 2021, 13, 2322. [Google Scholar] [CrossRef]
- Stapleton, J.T.; Chaloner, K.; Martenson, J.A.; Zhang, J.; Klinzman, D.; Xiang, J.; Sauter, W.; Desai, S.N.; Landay, A. GB virus C infection is associated with altered lymphocyte subset distribution and reduced T cell activation and proliferation in HIV-infected individuals. PLoS ONE 2012, 7, e50563. [Google Scholar] [CrossRef]
- Fernandez-Ruiz, M.; Forque, L.; Albert, E.; Redondo, N.; Gimenez, E.; Lopez-Medrano, F.; Gonzalez, E.; Polanco, N.; Ruiz-Merlo, T.; Parra, P.; et al. Human pegivirus type 1 infection in kidney transplant recipients: Replication kinetics and clinical correlates. Transplant. Infect. Dis. 2022, 24, e13771. [Google Scholar] [CrossRef]
- Graninger, M.; Aberle, S.; Gorzer, I.; Jaksch, P.; Puchhammer-Stockl, E. Human pegivirus 1 infection in lung transplant recipients: Prevalence, clinical relevance and kinetics of viral replication under immunosuppressive therapy. J. Clin. Virol. 2021, 143, 104937. [Google Scholar] [CrossRef]
- Fama, A.; Larson, M.C.; Link, B.K.; Habermann, T.M.; Feldman, A.L.; Call, T.G.; Ansell, S.M.; Liebow, M.; Xiang, J.; Maurer, M.J.; et al. Human Pegivirus Infection and Lymphoma Risk: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2020, 71, 1221–1228. [Google Scholar] [CrossRef]
- Laskin, B.L.; Denburg, M.R.; Furth, S.L.; Moatz, T.; Altrich, M.; Kleiboeker, S.; Lutzko, C.; Zhu, X.; Blackard, J.T.; Jodele, S.; et al. The Natural History of BK Polyomavirus and the Host Immune Response After Stem Cell Transplantation. Clin. Infect. Dis. 2020, 71, 3044–3054. [Google Scholar] [CrossRef]
- Tan, C.S.; Broge, T.A., Jr.; Ngo, L.; Gheuens, S.; Viscidi, R.; Bord, E.; Rosenblatt, J.; Wong, M.; Avigan, D.; Koralnik, I.J. Immune reconstitution after allogeneic hematopoietic stem cell transplantation is associated with selective control of JC virus reactivation. Biol. Blood Marrow Transplant. 2014, 20, 992–999. [Google Scholar] [CrossRef]
- Hill, J.A. Human herpesvirus 6 in transplant recipients: An update on diagnostic and treatment strategies. Curr. Opin. Infect. Dis. 2019, 32, 584–590. [Google Scholar] [CrossRef]
- Dulery, R.; Salleron, J.; Dewilde, A.; Rossignol, J.; Boyle, E.M.; Gay, J.; de Berranger, E.; Coiteux, V.; Jouet, J.P.; Duhamel, A.; et al. Early human herpesvirus type 6 reactivation after allogeneic stem cell transplantation: A large-scale clinical study. Biol. Blood Marrow Transplant. 2012, 18, 1080–1089. [Google Scholar] [CrossRef]
- Boutolleau, D.; Fernandez, C.; Andre, E.; Imbert-Marcille, B.M.; Milpied, N.; Agut, H.; Gautheret-Dejean, A. Human herpesvirus (HHV)-6 and HHV-7: Two closely related viruses with different infection profiles in stem cell transplantation recipients. J. Infect. Dis. 2003, 187, 179–186. [Google Scholar] [CrossRef]
- de Pagter, P.J.; Schuurman, R.; Visscher, H.; de Vos, M.; Bierings, M.; van Loon, A.M.; Uiterwaal, C.S.; van Baarle, D.; Sanders, E.A.; Boelens, J. Human herpes virus 6 plasma DNA positivity after hematopoietic stem cell transplantation in children: An important risk factor for clinical outcome. Biol. Blood Marrow Transplant. 2008, 14, 831–839. [Google Scholar] [CrossRef]
- Inazawa, N.; Hori, T.; Hatakeyama, N.; Yamamoto, M.; Yoto, Y.; Nojima, M.; Suzuki, N.; Shimizu, N.; Tsutsumi, H. Large-scale multiplex polymerase chain reaction assay for diagnosis of viral reactivations after allogeneic hematopoietic stem cell transplantation. J. Med. Virol. 2015, 87, 1427–1435. [Google Scholar] [CrossRef]
- Styczynski, J.; van der Velden, W.; Fox, C.P.; Engelhard, D.; de la Camara, R.; Cordonnier, C.; Ljungman, P. Management of Epstein-Barr Virus infections and post-transplant lymphoproliferative disorders in patients after allogeneic hematopoietic stem cell transplantation: Sixth European Conference on Infections in Leukemia (ECIL-6) guidelines. Haematologica 2016, 101, 803–811. [Google Scholar] [CrossRef]
- Zhou, J.R.; Shi, D.Y.; Wei, R.; Wang, Y.; Yan, C.H.; Zhang, X.H.; Xu, L.P.; Liu, K.Y.; Huang, X.J.; Sun, Y.Q. Co-Reactivation of Cytomegalovirus and Epstein-Barr Virus Was Associated With Poor Prognosis After Allogeneic Stem Cell Transplantation. Front. Immunol. 2020, 11, 620891. [Google Scholar] [CrossRef]
- Rahiala, J.; Koskenvuo, M.; Sadeghi, M.; Waris, M.; Vuorinen, T.; Lappalainen, M.; Saarinen-Pihkala, U.; Allander, T.; Soderlund-Venermo, M.; Hedman, K.; et al. Polyomaviruses BK, JC, KI, WU, MC, and TS in children with allogeneic hematopoietic stem cell transplantation. Pediatr. Transplant. 2016, 20, 424–431. [Google Scholar] [CrossRef]
- Kapoor, A.; Simmonds, P.; Slikas, E.; Li, L.; Bodhidatta, L.; Sethabutr, O.; Triki, H.; Bahri, O.; Oderinde, B.S.; Baba, M.M.; et al. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 2010, 201, 1633–1643. [Google Scholar] [CrossRef]
- Arthur, J.L.; Higgins, G.D.; Davidson, G.P.; Givney, R.C.; Ratcliff, R.M. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog 2009, 5, e1000391. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Slikas, E.; Simmonds, P.; Chieochansin, T.; Naeem, A.; Shaukat, S.; Alam, M.M.; Sharif, S.; Angez, M.; Zaidi, S.; et al. A newly identified bocavirus species in human stool. J. Infect. Dis. 2009, 199, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Pinana, J.L.; Madrid, S.; Perez, A.; Hernandez-Boluda, J.C.; Gimenez, E.; Terol, M.J.; Calabuig, M.; Navarro, D.; Solano, C. Epidemiologic and Clinical Characteristics of Coronavirus and Bocavirus Respiratory Infections after Allogeneic Stem Cell Transplantation: A Prospective Single-Center Study. Biol. Blood Marrow Transplant. 2018, 24, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.C.L.; Dabilla, N.A.S.; Almeida, T.N.; Fiaccadori, F.S.; de Souza, T.T.; Cardoso, D.; Arantes, A.M.; Souza, M. Human bocavirus detection and quantification in fecal and serum specimens from recipients of allogeneic hematopoietic stem cell transplantation: A longitudinal study. J. Med. Virol. 2022, 94, 594–600. [Google Scholar] [CrossRef]
- Ogimi, C.; Martin, E.T.; Xie, H.; Campbell, A.P.; Waghmare, A.; Jerome, K.R.; Leisenring, W.M.; Milano, F.; Englund, J.A.; Boeckh, M. Role of Human Bocavirus Respiratory Tract Infection in Hematopoietic Cell Transplant Recipients. Clin. Infect. Dis. 2021, 73, e4392–e4399. [Google Scholar] [CrossRef]
- Berg, M.G.; Lee, D.; Coller, K.; Frankel, M.; Aronsohn, A.; Cheng, K.; Forberg, K.; Marcinkus, M.; Naccache, S.N.; Dawson, G.; et al. Discovery of a Novel Human Pegivirus in Blood Associated with Hepatitis C Virus Co-Infection. PLoS Pathog 2015, 11, e1005325. [Google Scholar] [CrossRef]
- Coller, K.E.; Bruce, V.; Cassidy, M.; Gersch, J.; Frankel, M.B.; Vallari, A.; Cloherty, G.; Hackett, J., Jr.; Evans, J.L.; Page, K.; et al. Chronic Human Pegivirus 2 without Hepatitis C Virus Co-infection. Emerg. Infect. Dis. 2020, 26, 265–272. [Google Scholar] [CrossRef]
- Strenger, V.; Kessler, H.H.; Stelzl, E.; Aberle, S.W.; Keldorfer, M.; Zach, K.; Karastaneva, A.; Sperl, D.; Lackner, H.; Benesch, M.; et al. Enterovirus infections in pediatric hematologic/oncologic patients. Pediatr. Blood Cancer 2019, 66, e27448. [Google Scholar] [CrossRef]
- Matsumura-Kimoto, Y.; Inamoto, Y.; Tajima, K.; Kawajiri, A.; Tanaka, T.; Hirakawa, T.; Ino, K.; Asao, Y.; Tamogami, H.; Kono, C.; et al. Association of Cumulative Steroid Dose with Risk of Infection after Treatment for Severe Acute Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2016, 22, 1102–1107. [Google Scholar] [CrossRef]
- Miller, H.K.; Braun, T.M.; Stillwell, T.; Harris, A.C.; Choi, S.; Connelly, J.; Couriel, D.; Goldstein, S.; Kitko, C.L.; Magenau, J.; et al. Infectious Risk after Allogeneic Hematopoietic Cell Transplantation Complicated by Acute Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2017, 23, 522–528. [Google Scholar] [CrossRef]
- Srinivasan, A.; Wang, C.; Srivastava, D.K.; Burnette, K.; Shenep, J.L.; Leung, W.; Hayden, R.T. Timeline, epidemiology, and risk factors for bacterial, fungal, and viral infections in children and adolescents after allogeneic hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2013, 19, 94–101. [Google Scholar] [CrossRef]
- Eriguchi, Y.; Takashima, S.; Oka, H.; Shimoji, S.; Nakamura, K.; Uryu, H.; Shimoda, S.; Iwasaki, H.; Shimono, N.; Ayabe, T.; et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood 2012, 120, 223–231. [Google Scholar] [CrossRef]
- Levinson, A.; Pinkney, K.; Jin, Z.; Bhatia, M.; Kung, A.L.; Foca, M.D.; George, D.; Garvin, J.H.; Sosna, J.; Karamehmet, E.; et al. Acute gastrointestinal graft-vs-host disease is associated with increased enteric bacterial bloodstream infection density in pediatric allogeneic hematopoietic cell transplant recipients. Clin. Infect. Dis. 2015, 61, 350–357. [Google Scholar] [CrossRef]
- Sayer, H.G.; Longton, G.; Bowden, R.; Pepe, M.; Storb, R. Increased risk of infection in marrow transplant patients receiving methylprednisolone for graft-versus-host disease prevention. Blood 1994, 84, 1328–1332. [Google Scholar] [CrossRef]
- Martin, P.J.; Rizzo, J.D.; Wingard, J.R.; Ballen, K.; Curtin, P.T.; Cutler, C.; Litzow, M.R.; Nieto, Y.; Savani, B.N.; Schriber, J.R.; et al. First- and second-line systemic treatment of acute graft-versus-host disease: Recommendations of the American Society of Blood and Marrow Transplantation. Biol. Blood Marrow Transplant. 2012, 18, 1150–1163. [Google Scholar] [CrossRef]
- Vu, D.L.; Bosch, A.; Pinto, R.M.; Guix, S. Epidemiology of Classic and Novel Human Astrovirus: Gastroenteritis and Beyond. Viruses 2017, 9, 33. [Google Scholar] [CrossRef]
- Cordey, S.; Vu, D.L.; Schibler, M.; L’Huillier, A.G.; Brito, F.; Docquier, M.; Posfay-Barbe, K.M.; Petty, T.J.; Turin, L.; Zdobnov, E.M.; et al. Astrovirus MLB2, a New Gastroenteric Virus Associated with Meningitis and Disseminated Infection. Emerg Infect Dis 2016, 22, 846–853. [Google Scholar] [CrossRef]
- Cordey, S.; Vu, D.L.; Zanella, M.C.; Turin, L.; Mamin, A.; Kaiser, L. Novel and classical human astroviruses in stool and cerebrospinal fluid: Comprehensive screening in a tertiary care hospital, Switzerland. Emerg. Microbes Infect. 2017, 6, e84. [Google Scholar] [CrossRef]
- Piret, J.; Boivin, G. Antiviral Drugs Against Herpesviruses. In Antiviral Drug Discovery and Development; Springer: Singapore, 2021; Volume 1322, pp. 1–30. [Google Scholar] [CrossRef]
- Hatakeyama, N.; Suzuki, N.; Kudoh, T.; Hori, T.; Mizue, N.; Tsutsumi, H. Successful cidofovir treatment of adenovirus-associated hemorrhagic cystitis and renal dysfunction after allogenic bone marrow transplant. Pediatr. Infect. Dis. J. 2003, 22, 928–929. [Google Scholar] [CrossRef]
- Kadambi, P.V.; Josephson, M.A.; Williams, J.; Corey, L.; Jerome, K.R.; Meehan, S.M.; Limaye, A.P. Treatment of refractory BK virus-associated nephropathy with cidofovir. Am. J. Transplant. 2003, 3, 186–191. [Google Scholar] [CrossRef]
- Vats, A.; Shapiro, R.; Singh Randhawa, P.; Scantlebury, V.; Tuzuner, A.; Saxena, M.; Moritz, M.L.; Beattie, T.J.; Gonwa, T.; Green, M.D.; et al. Quantitative viral load monitoring and cidofovir therapy for the management of BK virus-associated nephropathy in children and adults. Transplantation 2003, 75, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Canavan, T.N.; Baddley, J.W.; Pavlidakey, P.; Tallaj, J.A.; Elewski, B.E. Human polyomavirus-7-associated eruption successfully treated with acitretin. Am. J. Transplant. 2018, 18, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
- Aubert, M.; Boyle, N.M.; Stone, D.; Stensland, L.; Huang, M.-L.; Magaret, A.S.; Galetto, R.; Rawlings, D.J.; Scharenberg, A.M.; Jerome, K.R. In vitro Inactivation of Latent HSV by Targeted Mutagenesis Using an HSV-specific Homing Endonuclease. Mol. Ther. Nucleic Acids 2014, 3, e146. [Google Scholar] [CrossRef] [PubMed]
- Corey, L.; Huang, M.L.; Selke, S.; Wald, A. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time taqman PCR assay. J. Med. Virol. 2005, 76, 350–355. [Google Scholar] [CrossRef]
- Weidmann, M.; Meyer-Konig, U.; Hufert, F.T. Rapid detection of herpes simplex virus and varicella-zoster virus infections by real-time PCR. J. Clin. Microbiol. 2003, 41, 1565–1568. [Google Scholar] [CrossRef]
- Rockett, R.J.; Sloots, T.P.; Bowes, S.; O’Neill, N.; Ye, S.; Robson, J.; Whiley, D.M.; Lambert, S.; Wang, D.; Nissen, M.; et al. Detection of novel polyomaviruses, TSPyV, HPyV6, HPyV7, HPyV9 and MWPyV in feces, urine, blood, respiratory swabs and cerebrospinal fluid. PLoS ONE 2013, 8, e62764. [Google Scholar] [CrossRef]
- Verheyen, J.; Timmen-Wego, M.; Laudien, R.; Boussaad, I.; Sen, S.; Koc, A.; Uesbeck, A.; Mazou, F.; Pfister, H. Detection of adenoviruses and rotaviruses in drinking water sources used in rural areas of Benin, West Africa. Appl. Environ. Microbiol. 2009, 75, 2798–2801. [Google Scholar] [CrossRef]
- Masouridi-Levrat, S.; Pradier, A.; Simonetta, F.; Kaiser, L.; Chalandon, Y.; Roosnek, E. Torque teno virus in patients undergoing allogeneic hematopoietic stem cell transplantation for hematological malignancies. Bone Marrow Transplant. 2016, 51, 440–442. [Google Scholar] [CrossRef]
- Chivero, E.T.; Bhattarai, N.; Rydze, R.T.; Winters, M.A.; Holodniy, M.; Stapleton, J.T. Human pegivirus RNA is found in multiple blood mononuclear cells in vivo and serum-derived viral RNA-containing particles are infectious in vitro. J. Gen. Virol. 2014, 95, 1307–1319. [Google Scholar] [CrossRef]
- Frankel, M.; Forberg, K.; Coller, K.E.; Berg, M.G.; Hackett, J.; Cloherty, G.; Dawson, G.J. Development of a high-throughput multiplexed real time RT-PCR assay for detection of human pegivirus 1 and 2. J. Virol. Methods. 2017, 241, 34–40. [Google Scholar] [CrossRef]
- Cordey, S.; Junier, T.; Gerlach, D.; Gobbini, F.; Farinelli, L.; Zdobnov, E.M.; Winther, B.; Tapparel, C.; Kaiser, L. Rhinovirus genome evolution during experimental human infection. PLoS ONE 2010, 5, e10588. [Google Scholar] [CrossRef]
- Kantola, K.; Sadeghi, M.; Antikainen, J.; Kirveskari, J.; Delwart, E.; Hedman, K.; Söderlund-Venermo, M. Real-time quantitative PCR detection of four human bocaviruses. J. Clin. Microbiol. 2010, 48, 4044–4050. [Google Scholar] [CrossRef]
- Arvia, R.; Sollai, M.; Pierucci, F.; Urso, C.; Massi, D.; Zakrzewska, K. Droplet digital PCR (ddPCR) vs quantitative real-time PCR (qPCR) approach for detection and quantification of Merkel cell polyomavirus (MCPyV) DNA in formalin fixed paraffin embedded (FFPE) cutaneous biopsies. J. Virol. Methods. 2017, 246, 15–20. [Google Scholar] [CrossRef]
- Tapparel, C.; Cordey, S.; Van Belle, S.; Turin, L.; Lee, W.-M.; Regamey, N.; Meylan, P.; Mühlemann, K.; Gobbini, F.; Kaiser, L. New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J. Clin. Microbiol. 2009, 47, 1742–1749. [Google Scholar] [CrossRef]
- Antonsson, A.; Bialasiewicz, S.; Rockett, R.J.; Jacob, K.; Bennett, I.C.; Sloots, T.P. Exploring the prevalence of ten polyomaviruses and two herpes viruses in breast cancer. PLoS ONE 2012, 7, e39842. [Google Scholar] [CrossRef]
- Urbano, P.R.; Nali, L.H.; Bicalho, C.S.; Pierrotti, L.C.; David-Neto, E.; Pannuti, C.S.; Romano, C.M. New findings about trichodysplasia spinulosa-associated polyomavirus (TSPyV)--novel qPCR detects TSPyV-DNA in blood samples. Diagn. Microbiol. Infect. Dis. 2016, 84, 123–124. [Google Scholar] [CrossRef]


| Total N = 109 | |
|---|---|
| Demographics Sex (male), n (%) | 72 (66) |
| Age, median (IQR) | 56 (18) |
| Transplant source, n (%) | |
| Peripheral blood cells | 96 (88) |
| Bone marrow | 13 (12) |
| Underlying disease, n (%) | |
| Acute myeloid leukemia | 59 (54) |
| MDS/MDPS | 27 (25) |
| Acute lymphoid leukemia | 10 (9) |
| Myeloproliferative syndrome | 4 (4) |
| Lymphoma | 4 (4) |
| Chronic lymphocytic leukemia | 2 (2) |
| Myeloma | 2 (2) |
| Chronic myelogenous leukemia | 1 (1) |
| Disease risk index *, n (%) | |
| Low | 5 (5) |
| Intermediate | 71 (65) |
| High | 29 (27) |
| Very high | 4 (4) |
| EBMT risk score, n (%) | |
| 1 | 5 (5) |
| 2 | 10 (9) |
| 3 | 46 (42) |
| 4 | 19 (17) |
| 5 | 19 (17) |
| 6 | 10 (9) |
| EBV donor/recipient constellation, n (%) | |
| +/+ | 96 (88) |
| −/+ | 8 (7) |
| +/− | 3 (3) |
| −/− | 2 (2) |
| CMV donor/recipient constellation, n (%) | |
| +/+ | 46 (42) |
| +/− | 10 (9) |
| −/− | 40 (37) |
| −/+ | 13 (12) |
| Conditioning regimen, n (%) | |
| Reduced intensity conditioning | 74 (68) |
| Myeloablative conditioning | 43 (39) |
| Ex vivo T-cell depletion, n (%) | 19 (17) |
| Donor type, n (%) | |
| HLA identical sibling donor | 28 (26) |
| HLA matched unrelated donor | 47 (43) |
| Haploidentical donor | 24 (22) |
| HLA mismatched unrelated donor | 10 (9) |
| Patients, N | |||||
|---|---|---|---|---|---|
| Day 0 | Day 30 | Month 3 | Month 6 | Year 1 | |
| Group 1 (13 virus) | 103 | 102 | 101 | 89 | 64 |
| Group 2 (17 virus) | 74 | 72 | 73 | 66 | 50 |
| Group 3 (20 virus) | 65 | 65 | 64 | 58 | 44 |
| Missing data * | 6 | 7 | 8 | 20 | 45 |
| Viral Specie | Viral Species Detected N (%) | Viral Species Undetectedor Not Tested N (%) |
|---|---|---|
| TTV | 104 (95) | 5 (5) |
| BKPyV | 57 (52) | 52 (48) |
| HPgV-1 | 51 (47) | 58 (53) |
| JCPyV | 41 (38) | 68 (62) |
| CMV | 36 (33) | 73 (67) |
| HHV-6 | 26 (24) | 83 (76) |
| HPyV6 | 26 (24) | 83 (76) |
| MCPyV | 23 (21) | 86 (79) |
| HPyV7 | 15 (14) | 94 (86) |
| EBV | 12 (11) | 97 (89) |
| HSV | 5 (5) | 104 (95) |
| HAdV | 5 (5) | 104 (95) |
| HHV-7 | 4 (4) | 105 (96) |
| B19V | 3 (3) | 106 (97) |
| VZV | 2 (2) | 107 (98) |
| HPyV9 | - | 109 (100) |
| HPgV-2 | - | 109 (100) |
| HBoV | - | 109 (100) |
| EV | - | 109 (100) |
| TSPyV | - | 109 (100) |
| Viral Species | r(RT-)PCR Result | Time-Point | ||||
|---|---|---|---|---|---|---|
| Day 0 | Day 30 | Month 3 | Month 6 | Year 1 | ||
| TTV | detected | 49 (48%) | 49 (49%) | 97 (96%) | 86 (97%) | 55 (90%) |
| not detected | 54 (52%) | 52 (51%) | 4 (4%) | 3 (3%) | 6 (10%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| HPgV-1 | detected | 29 (28%) | 26 (26%) | 34 (34%) | 32 (36%) | 18 (30%) |
| not detected | 74 (72%) | 75 (74%) | 67 (66%) | 57 (64%) | 43 (70%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| BKPyV | detected | 10 (10%) | 41 (41%) | 22 (22%) | 11 (12%) | 5 (8%) |
| not detected | 93 (90%) | 60 (59%) | 79 (78%) | 78 (88%) | 56 (92%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| JCPyV | detected | 9 (9%) | 13 (13%) | 12 (12%) | 12 (13%) | 14 (23%) |
| not detected | 94 (91%) | 88 (87%) | 89 (88%) | 77 (87%) | 47 (77%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| MCPyV | detected | 5 (7%) | 6 (8%) | 5 (7%) | 8 (12%) | 4 (9%) |
| not detected | 69 (93%) | 65 (92%) | 68 (93%) | 58 (88%) | 39 (91%) | |
| NA | 35 | 38 | 36 | 43 | 66 | |
| HPyV6 | detected | 6 (9%) | 8 (12%) | 17 (27%) | 6 (10%) | 4 (11%) |
| not detected | 59 (91%) | 56 (88%) | 47 (73%) | 52 (90%) | 34 (89%) | |
| NA | 44 | 45 | 45 | 51 | 71 | |
| HPyV7 | detected | 2 (3%) | 2 (3%) | 8 (12%) | 4 (7%) | 0 (0%) |
| not detected | 63 (97%) | 62 (97%) | 56 (88%) | 54 (93%) | 38 (100%) | |
| NA | 44 | 45 | 45 | 51 | 71 | |
| CMV | detected | 9 (9%) | 27 (27%) | 11 (11%) | 9 (10%) | 2 (3%) |
| not detected | 92 (91%) | 74 (73%) | 90 (89%) | 80 (90%) | 59 (97%) | |
| NA | 8 | 8 | 8 | 20 | 48 | |
| HHV-6 | detected | 2 (2%) | 19 (19%) | 4 (4%) | 1 (1%) | 1 (2%) |
| not detected | 101 (98%) | 82 (81%) | 97 (96%) | 88 (99%) | 60 (98%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| HSV-1/2 | detected | 2 (2%) | 2 (2%) | 0 (0%) | 0 (0%) | 1 (2%) |
| not detected | 101 (98%) | 99 (98%) | 101 (100%) | 89 (100%) | 60 (98%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| VZV | detected | 0 (0%) | 0 (0%) | 0 (0%) | 2 (2%) | 0 (0%) |
| not detected | 103 (100%) | 101 (100%) | 101 (100%) | 87 (98%) | 61 (100%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| EBV | detected | 2 (2%) | 5 (5%) | 1 (1%) | 4 (4%) | 3 (5%) |
| not detected | 99 (98%) | 96 (95%) | 100 (99%) | 85 (96%) | 58 (95%) | |
| NA | 8 | 8 | 8 | 20 | 48 | |
| HHV-7 | detected | 0 (0%) | 2 (2%) | 1 (1%) | 0 (0%) | 1 (2%) |
| not detected | 103 (100%) | 99 (98%) | 100 (99%) | 89 (100%) | 60 (98%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| HAdV | detected | 1 (1%) | 3 (3%) | 1 (1%) | 1 (1%) | 1 (2%) |
| not detected | 102 (99%) | 98 (97%) | 100 (99%) | 88 (99%) | 60 (98%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| B19V | detected | 1 (1%) | 2 (3%) | 1 (1%) | 1 (2%) | 2 (5%) |
| not detected | 73 (99%) | 69 (97%) | 72 (99%) | 65 (98%) | 41 (95%) | |
| NA | 35 | 38 | 36 | 43 | 66 | |
| TSPyV | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 65 (100%) | 64 (100%) | 64 (100%) | 58 (100%) | 38 (100%) | |
| NA | 44 | 45 | 45 | 51 | 71 | |
| HPyV9 | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 103 (100%) | 101 (100%) | 101 (100%) | 89 (100%) | 61 (100%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| HBoV | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 74 (100%) | 71 (100%) | 73 (100%) | 66 (100%) | 43 (100%) | |
| NA | 35 | 38 | 36 | 43 | 66 | |
| HPgV-2 | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 103 (100%) | 101 (100%) | 101 (100%) | 89 (100%) | 61 (100%) | |
| NA | 6 | 8 | 8 | 20 | 48 | |
| EV | detected | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| not detected | 74 (100%) | 71 (100%) | 73 (100%) | 66 (100%) | 43 (100%) | |
| NA | 35 | 38 | 36 | 43 | 66 | |
| Plasma Viral Load * | ||||
|---|---|---|---|---|
| Time-Point | Viral Species | Patients N | Median (IQ) | Range |
| Day 0 | TTV | 42 | 6.35 × 103 (1.15 × 103–3.92 × 104) | 2.76 × 102–1.73 × 107 |
| HPgV-1 | 23 | 5.96 × 105 (1.16 × 105–4.39 × 106) | 2.79 × 103–2.83 × 107 | |
| Day 30 | TTV | 37 | 2.70 × 104 (9.40 × 102–2.19 × 105) | 3.39 × 102–1.31 × 107 |
| HPgV-1 | 21 | 2.71 × 105 (8.39 × 104–1.76 × 106) | 1.73 × 104–1.89 × 107 | |
| CMV | 16 | 2.02 × 102 (1.18 × 102–3.30 × 102) | 5.8 × 101–5.70 × 103 | |
| HHV-6 | 13 | 1.16 × 103 (8.42 × 102–4.03 × 103) | 4.71 × 102–9.88 × 103 | |
| BKPyV | 5 | 1.17 × 103 (1.11 × 103–1.21 × 103) | 1.04 × 103–1.41 × 103 | |
| Month 3 | TTV | 93 | 3.29 × 105 (4.66 × 104–6.65 × 106) | 3.37 × 102–4.06 × 109 |
| HPgV-1 | 27 | 1.18 × 106 (1.15 × 105–3.60 × 106) | 2.61 × 103–4.49 × 107 | |
| HPyV6 | 7 | 1.3 × 103 (4.53 × 102–2.59 × 103) | 2.64 × 102–7.68 × 103 | |
| HPyV7 | 5 | 1.63 × 103 (1.10 × 103–5.39 × 103) | 2.73 × 102–1.71 × 104 | |
| Month 6 | TTV | 79 | 9.58 × 104 (7.74 × 103–1.57 × 106) | 3.33 × 102–1.18 × 109 |
| HPgV-1 | 28 | 4.90 × 105 (1.10 × 105–2.41 × 106) | 7.45 × 103–3.74 × 107 | |
| JCPyV | 8 | 1.17 × 103 (4.21 × 102–1.62 × 103) | 1.34 × 102–2.76 × 103 | |
| Year 1 | TTV | 48 | 1.58 × 104 (2.92 × 103–4.97 × 104) | 2.62 × 102–1.23 × 108 |
| HPgV-1 | 16 | 4.64 × 105 (8.05 × 104–3.61 × 106) | 6.01 × 103–5.98 × 107 | |
| Number of Viral Species Detected | Patients, N (%) | ||||
|---|---|---|---|---|---|
| Day 0 (N = 65) | Day 30 (N = 65) | Month 3 (N = 64) | Month 6 (N = 58) | Year 1 (N = 44) | |
| 0 | 15 (23.1%) | 6 (9.2%) | 1 (1.6%) | 1 (17.2%) | 1 (2.3%) |
| 1 | 23 (35.4%) | 20 (30.8%) | 17 (26.6%) | 20 (34.5%) | 15 (34.1%) |
| 2 | 18 (27.7%) | 12 (18.5%) | 23 (35.4%) | 21 (36.2%) | 13 (29.5%) |
| 3 | 5 (7.7%) | 18 (27.7%) | 14 (21.9%) | 8 (13.8%) | 4 (9.1%) |
| 4 | 1 (1.5%) | 7 (10.8%) | 5 (7.8%) | 6 (10.3%) | 5 (11.4%) |
| 5 | 0 | 0 | 2 (3.1%) | 2 (3.5%) | 0 |
| 6 | 0 | 1 (1.5%) | 2 (3.1%) | 0 | 0 |
| Missing data * | 3 (4.6%) | 1 (1.5%) | 0 | 0 | 6 (13.6%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanella, M.-C.; Vu, D.-L.; Hosszu-Fellous, K.; Neofytos, D.; Van Delden, C.; Turin, L.; Poncet, A.; Simonetta, F.; Masouridi-Levrat, S.; Chalandon, Y.; et al. Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma. Viruses 2023, 15, 928. https://doi.org/10.3390/v15040928
Zanella M-C, Vu D-L, Hosszu-Fellous K, Neofytos D, Van Delden C, Turin L, Poncet A, Simonetta F, Masouridi-Levrat S, Chalandon Y, et al. Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma. Viruses. 2023; 15(4):928. https://doi.org/10.3390/v15040928
Chicago/Turabian StyleZanella, Marie-Céline, Diem-Lan Vu, Krisztina Hosszu-Fellous, Dionysios Neofytos, Chistian Van Delden, Lara Turin, Antoine Poncet, Federico Simonetta, Stavroula Masouridi-Levrat, Yves Chalandon, and et al. 2023. "Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma" Viruses 15, no. 4: 928. https://doi.org/10.3390/v15040928
APA StyleZanella, M.-C., Vu, D.-L., Hosszu-Fellous, K., Neofytos, D., Van Delden, C., Turin, L., Poncet, A., Simonetta, F., Masouridi-Levrat, S., Chalandon, Y., Cordey, S., & Kaiser, L. (2023). Longitudinal Detection of Twenty DNA and RNA Viruses in Allogeneic Hematopoietic Stem Cell Transplant Recipients Plasma. Viruses, 15(4), 928. https://doi.org/10.3390/v15040928







