Design and Identification of a Novel Antiviral Affinity Peptide against Fowl Adenovirus Serotype 4 (FAdV-4) by Targeting Fiber2 Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus Strain
2.2. Reagents and Antibodies
2.3. Expression and Purification of the Knob Domain of the Fiber2 Protein
2.4. Crystallization, Data Collection, and Structural Determination of Fiber2
2.5. Analytical Ultracentrifugation
2.6. The Design and Virtual Screen of the Peptides
2.7. Immunoperoxidase Monolayer Assay
2.8. Real-Time Quantitative Polymerase Chain Reaction
2.9. Surface Plasmon Resonance Assay
2.10. Cytotoxicity Assay
2.11. TCID50 and Indirect Immunofluorescence Assay
2.12. Statistical Analysis
3. Results
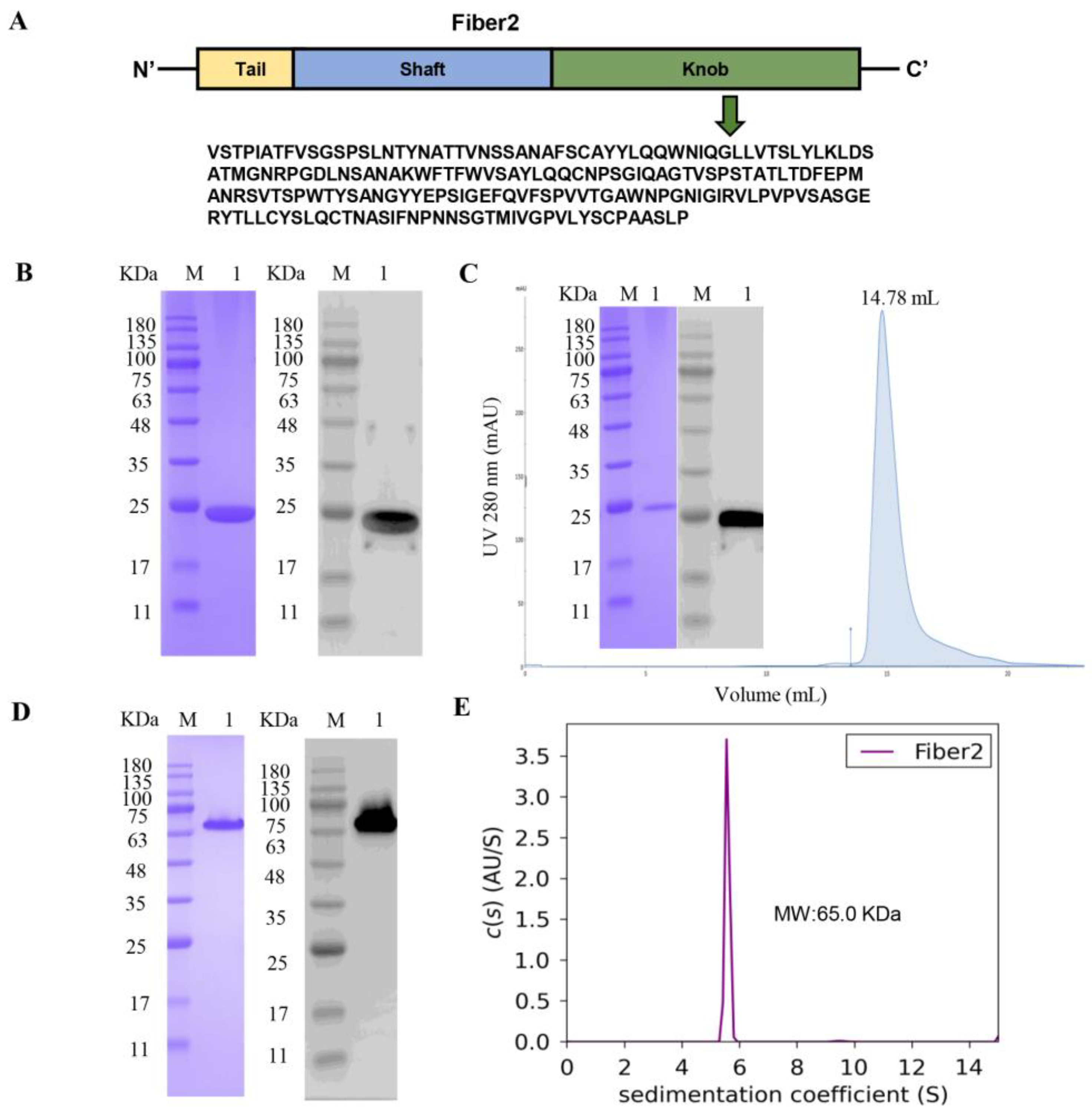
3.1. Expression and Purification of Recombinant Protein Fiber2
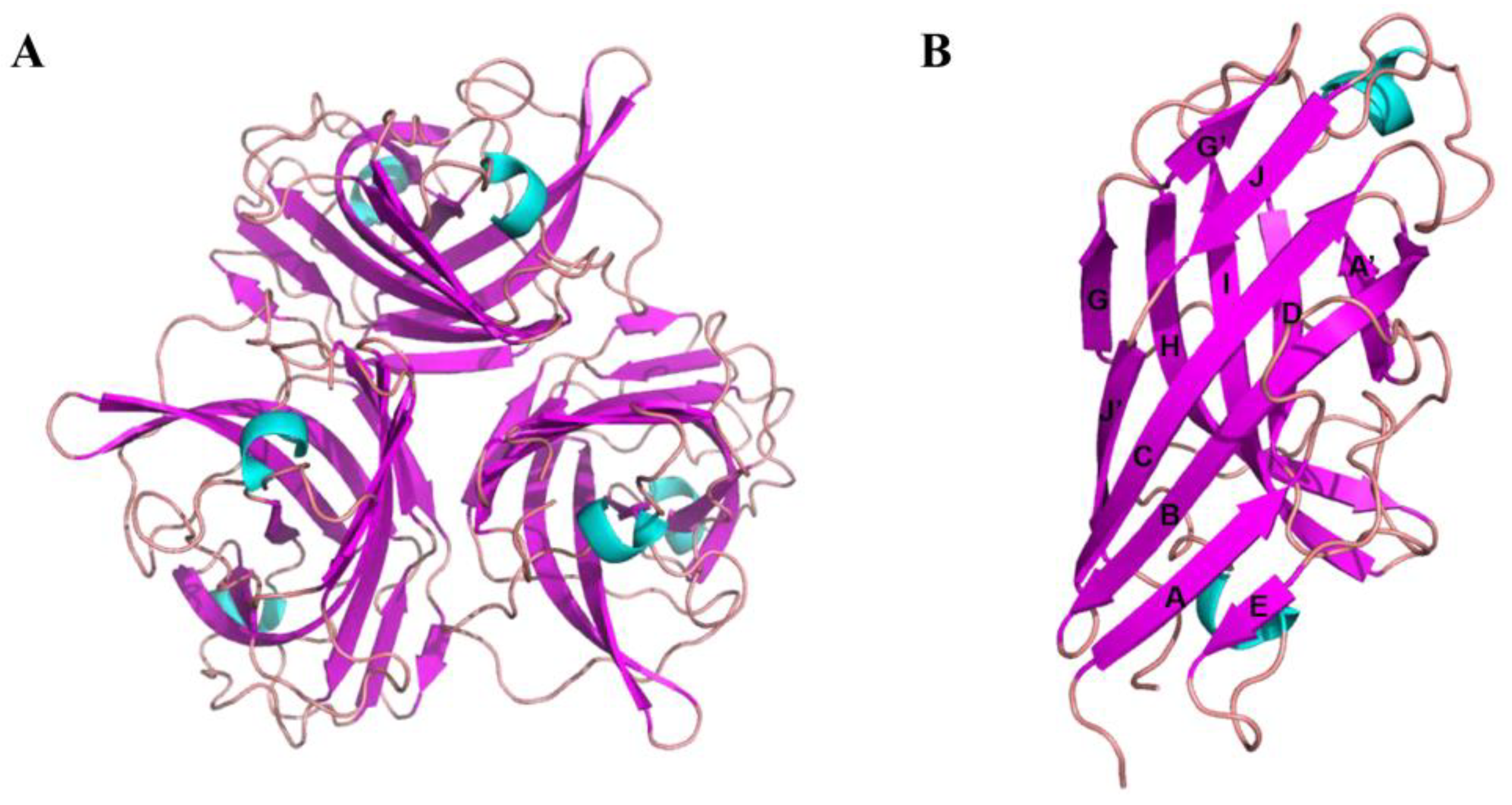
3.2. Crystal Structure of the Knob Domain of FAdV-4 Fiber2
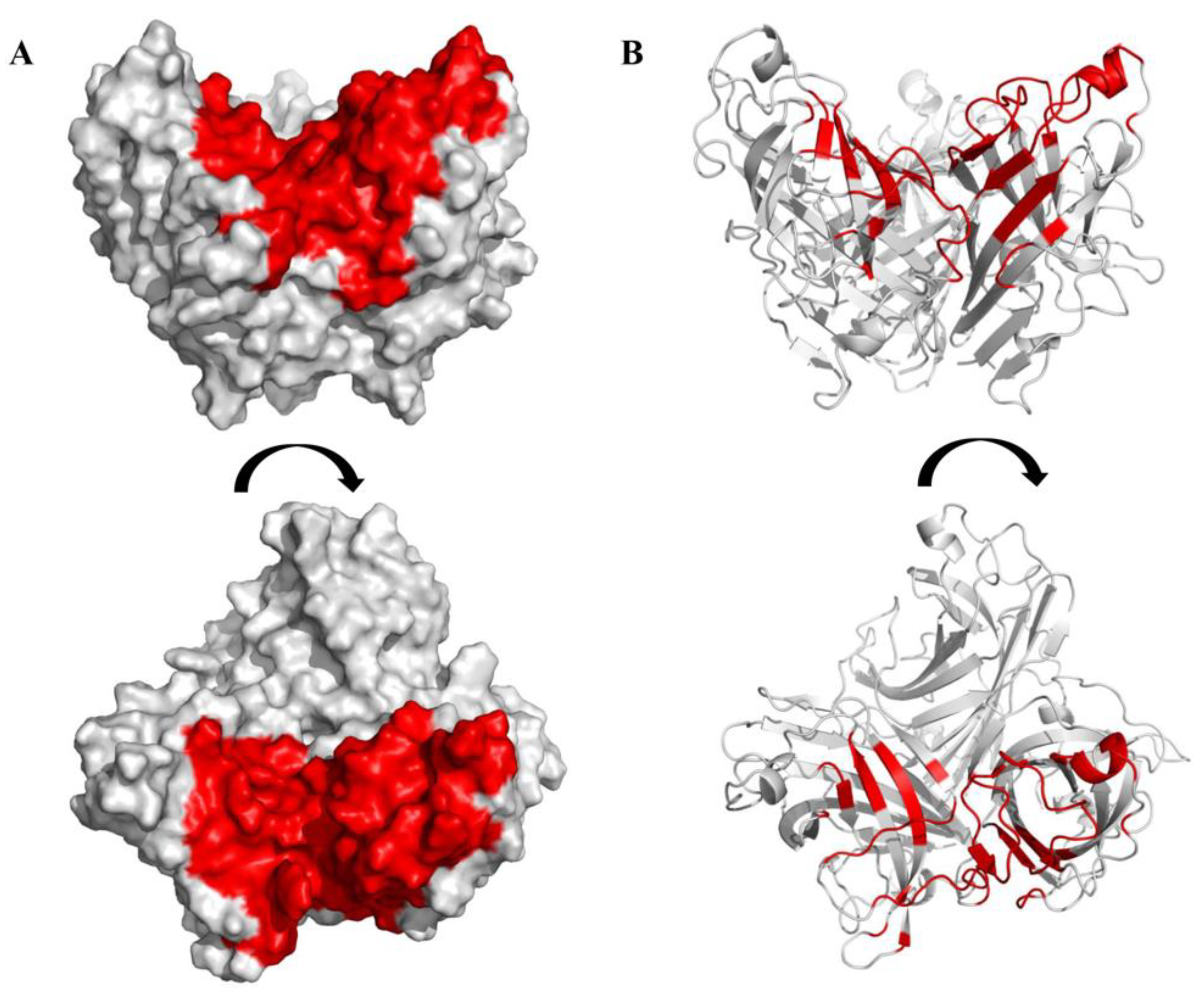
3.3. Design and Synthesis of Peptides on the Basis of the Crystal Structure of the Knob Domain of the FAdV-4 Fiber2
3.4. Screening of Peptides
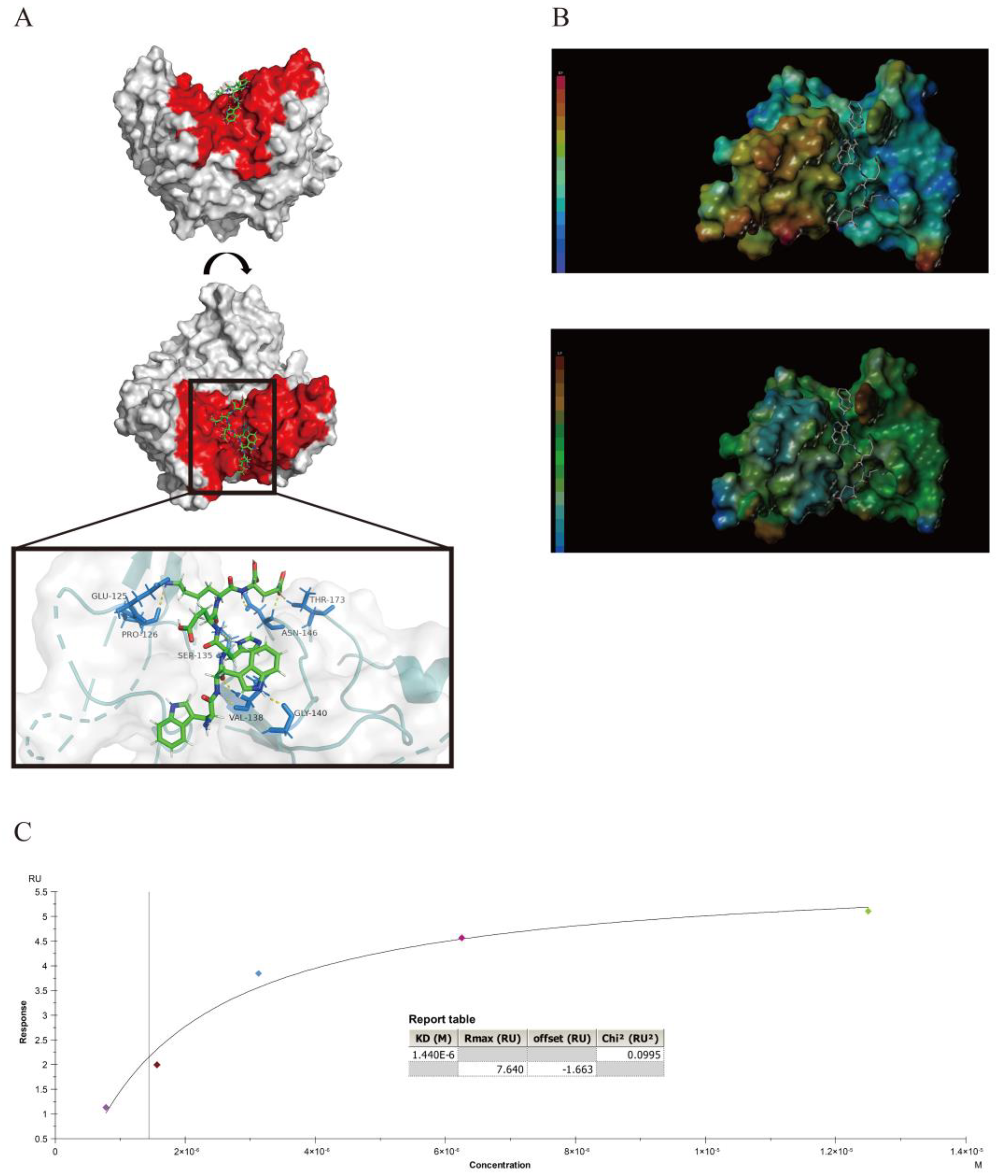
3.5. Interaction between the Peptides and the Knob Domain of the Fiber2 Protein
3.6. Cytotoxicity of P15 in LMH Cells
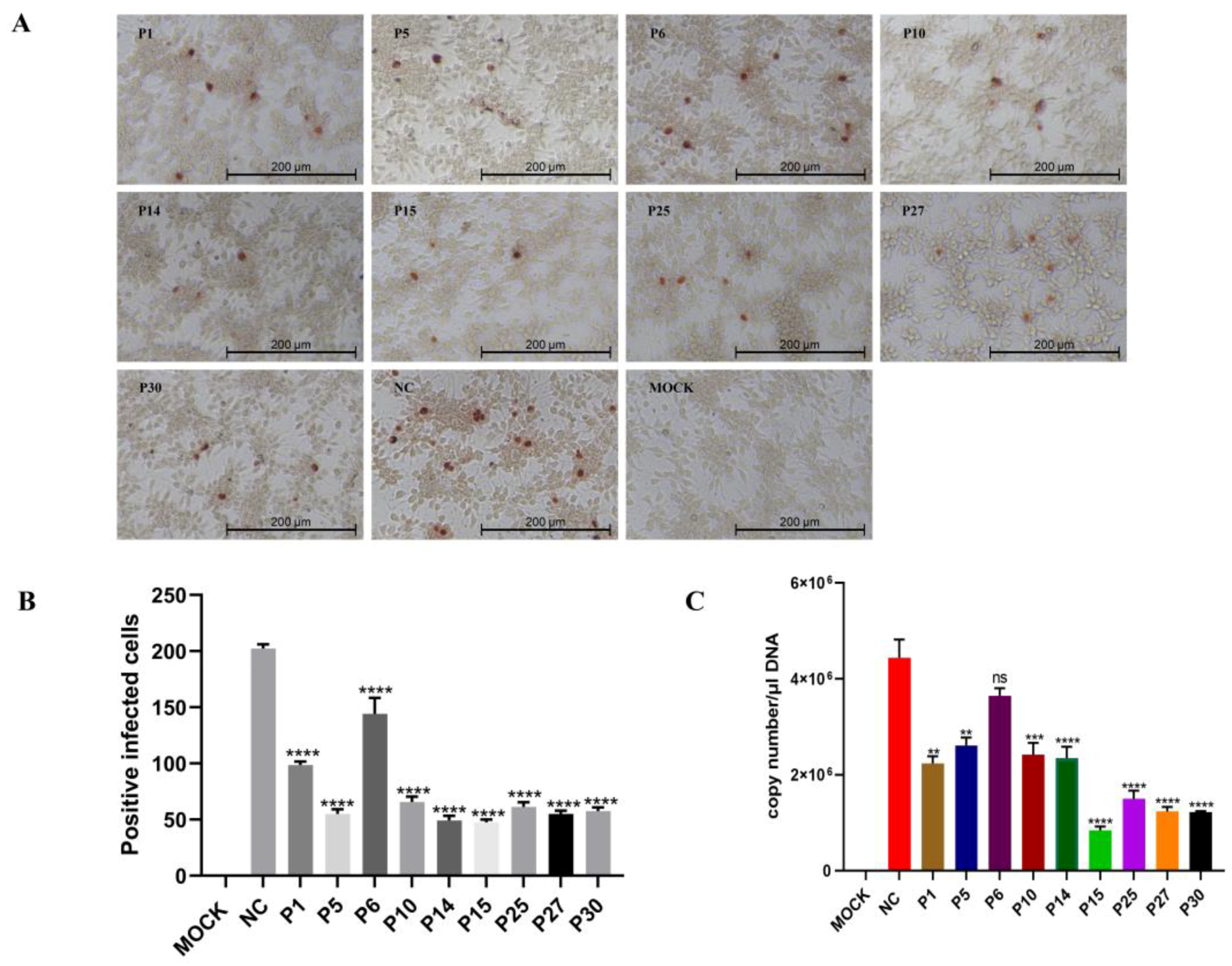
3.7. P15 Inhibits the Infectivity of FAdV-4 In Vitro
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hess, M. Detection and differentiation of avian adenoviruses: A review. Avian Pathol. 2000, 29, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Steer, P.A.; Kirkpatrick, N.C.; O’Rourke, D.; Noormohammadi, A.H. Classification of fowl adenovirus serotypes by use of high-resolution melting-curve analysis of the hexon gene region. J. Clin. Microbiol. 2009, 47, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.; Muneer, R.; Stein, G. Studies on the aetiology of hydropericardium syndrome (Angara disease) in broilers. Vet. Rec. 1991, 128, 591–593. [Google Scholar] [CrossRef] [PubMed]
- Schachner, A.; Matos, M.; Grafl, B.; Hess, M. Fowl adenovirus-induced diseases and strategies for their control—A review on the current global situation. Avian Pathol. 2018, 47, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Aziz, T.A.; al-Attar, M.A. New syndrome in Iraqi chicks. Vet. Rec. 1991, 129, 272. [Google Scholar] [CrossRef]
- Borisov, V.; Borisov, A.; Gusev, A.A. Hydropericardium syndrome in chickens in Russia. In Proceedings of the Tenth International Congress of Veterinary Poultry Association, Budapest, Hungary, 18–22 August 1997; p. 258. [Google Scholar]
- Jantosovic, J.; Konard, J.; Saly, J.; Skardova, I.; Kusev, J.; Beninghausova, K.J.V. Hydropericardium syndrome in chicks. Veterinastvi 1991, 41, 261–263. [Google Scholar]
- Mazaheri, A.; Prusas, C.; Voss, M.; Hess, M. Some strains of serotype 4 fowl adenoviruses cause inclusion body hepatitis and hydropericardium syndrome in chickens. Avian Pathol. 1998, 27, 269–276. [Google Scholar] [CrossRef]
- Toro, H.; Prusas, C.; Raue, R.; Cerda, L.; Geisse, C.; González, C.; Hess, M. Characterization of fowl adenoviruses from outbreaks of inclusion body hepatitis/hydropericardium syndrome in Chile. Avian Dis. 1999, 43, 262–270. [Google Scholar] [CrossRef]
- Abe, T.; Nakamura, K.; Tojo, H.; Mase, M.; Shibahara, T.; Yamaguchi, S.; Yuasa, N. Histology, immunohistochemistry, and ultrastructure of hydropericardium syndrome in adult broiler breeders and broiler chicks. Avian Dis. 1998, 42, 606–612. [Google Scholar] [CrossRef]
- Kim, J.N.; Byun, S.H.; Kim, M.J.; Kim, J.; Sung, H.W.; Mo, I.P. Outbreaks of hydropericardium syndrome and molecular characterization of Korean fowl adenoviral isolates. Avian Dis. 2008, 52, 526–530. [Google Scholar] [CrossRef]
- Pan, Q.; Zhang, Y.; Liu, A.; Cui, H.; Gao, Y.; Qi, X.; Liu, C.; Zhang, Y.; Li, K.; Gao, L.; et al. Development of a Novel Avian Vaccine Vector Derived from the Emerging Fowl Adenovirus 4. Front. Microbiol 2021, 12, 780978. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yin, L.; Zhou, Q.; Peng, P.; Du, Y.; Liu, L.; Zhang, Y.; Xue, C.; Cao, Y. Epidemiological investigation of fowl adenovirus infections in poultry in China during 2015–2018. BMC Vet. Res. 2019, 15, 271. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Liu, M.; Wang, C.; Zhou, X.; Li, F.; Song, J.; Pu, J.; Sun, Y.; Wang, M.; Shahid, M.; et al. Characterization of fowl adenovirus serotype 4 circulating in chickens in China. Vet. Microbiol. 2019, 238, 108427. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Xiang, B.; Li, S.; Ren, X.; Hong, Y.; Liao, J.; Yu, D.; Ren, T.; Liao, M.; Xu, C. Genetic characterization of fowl adenovirus serotype 4 isolates in Southern China reveals potential cross-species transmission. Infect. Genet. Evol. 2019, 75, 103928. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Xie, Z.; Zhang, L.; Luan, Y.; Luo, S.; Deng, X.; Xie, L.; Xie, Z.; Fan, Q. Genetic characterization of fowl aviadenovirus 4 isolates from Guangxi, China, during 2017–2019. Poult. Sci. 2020, 99, 4166–4173. [Google Scholar] [CrossRef]
- Liu, Y.; Wan, W.; Gao, D.; Li, Y.; Yang, X.; Liu, H.; Yao, H.; Chen, L.; Wang, C.; Zhao, J. Genetic characterization of novel fowl aviadenovirus 4 isolates from outbreaks of hepatitis-hydropericardium syndrome in broiler chickens in China. Emerg. Microbes Infect. 2016, 5, e117. [Google Scholar] [CrossRef]
- Pan, Q.; Liu, L.; Gao, Y.; Liu, C.; Qi, X.; Zhang, Y.; Wang, Y.; Li, K.; Gao, L.; Wang, X.; et al. Characterization of a hypervirulent fowl adenovirus 4 with the novel genotype newly prevalent in China and establishment of reproduction infection model of hydropericardium syndrome in chickens. Poult. Sci. 2017, 96, 1581–1588. [Google Scholar] [CrossRef]
- Mittal, D.; Jindal, N.; Tiwari, A.K.; Khokhar, R.S. Characterization of fowl adenoviruses associated with hydropericardium syndrome and inclusion body hepatitis in broiler chickens. Virus Dis. 2014, 25, 114–119. [Google Scholar] [CrossRef]
- Chen, H.; Dou, Y.; Zheng, X.; Tang, Y.; Zhang, M.; Zhang, Y.; Wang, Z.; Diao, Y. Hydropericardium Hepatitis Syndrome Emerged in Cherry Valley Ducks in China. Transbound. Emerg. Dis. 2017, 64, 1262–1267. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, H.; Diao, Y.; Li, X.; Zhang, S.; Gao, B.; Tang, Y.; Hu, J.; Diao, Y. Pathogenicity of fowl adenovirus (FAdV) serotype 4 strain SDJN in Taizhou geese. Avian Pathol. 2019, 48, 477–485. [Google Scholar] [CrossRef]
- Li, P.H.; Zheng, P.P.; Zhang, T.F.; Wen, G.Y.; Shao, H.B.; Luo, Q.P. Fowl adenovirus serotype 4: Epidemiology, pathogenesis, diagnostic detection, and vaccine strategies. Poult. Sci. 2017, 96, 2630–2640. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, Z.; Chen, H.; Niu, X.; Dou, Y.; Yang, J.; Tang, Y.; Diao, Y. Serological and Pathogenic Analyses of Fowl Adenovirus Serotype 4 (FAdV-4) Strain in Muscovy Ducks. Front. Microbiol. 2018, 9, 1163. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.L.; Bezerra, L.P.; Shawar, D.E.; Neto, N.A.; Mesquita, F.P.; da Silva, G.O.; Souza, P.N. Synthetic antiviral peptides: A new way to develop targeted antiviral drugs. Future Virol. 2022, 17, 577–591. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin B 2020, 10, 766–788. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Yadav, A.; Sarkar, P. Molecular docking and ADMET study of bioactive compounds of Glycyrrhiza glabra against main protease of SARS-CoV2. Mater. Today Proc. 2022, 49, 2999–3007. [Google Scholar] [CrossRef]
- Davella, R.; Gurrapu, S.; Mamidala, E. Phenolic compounds as promising drug candidates against COVID-19—An integrated molecular docking and dynamics simulation study. Mater. Today Proc. 2022, 51, 522–527. [Google Scholar] [CrossRef]
- Guasch, L.; Zakharov, A.V.; Tarasova, O.A.; Poroikov, V.V.; Liao, C.; Nicklaus, M.C. Novel HIV-1 Integrase Inhibitor Development by Virtual Screening Based on QSAR Models. Curr. Top. Med. Chem. 2016, 16, 441–448. [Google Scholar] [CrossRef]
- Tanbin, S.; Ahmad Fuad, F.A.; Abdul Hamid, A.A. Virtual Screening for Potential Inhibitors of Human Hexokinase II for the Development of Anti-Dengue Therapeutics. Biotech 2020, 10, 1. [Google Scholar] [CrossRef]
- Yuriev, E.; Ramsland, P.A. Latest developments in molecular docking: 2010-2011 in review. J. Mol. Recognit. 2013, 26, 215–239. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, R.; Tian, K.; Wang, Z.; Yang, X.; Gao, D.; Zhang, Y.; Fu, J.; Wang, H.; Zhao, J. Fiber2 and hexon genes are closely associated with the virulence of the emerging and highly pathogenic fowl adenovirus 4. Emerg. Microbes Infect. 2018, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wang, W.; Li, L.; Kan, Q.; Fu, H.; Geng, T.; Li, T.; Wan, Z.; Gao, W.; Shao, H.; et al. Domain in Fiber-2 interacted with KPNA3/4 significantly affects the replication and pathogenicity of the highly pathogenic FAdV-4. Virulence 2021, 12, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; He, L.; Zhu, E.; Fang, T.; Yue, J.; Wen, M.; Wang, K.; Cheng, Z. A fowl adenovirus serotype 4 (FAdV-4) Fiber2 subunit vaccine candidate provides complete protection against challenge with virulent FAdV-4 strain in chickens. Vet. Microbiol. 2021, 263, 109250. [Google Scholar] [CrossRef] [PubMed]
- Ruan, S.; Zhao, J.; Yin, X.; He, Z.; Zhang, G. A subunit vaccine based on fiber-2 protein provides full protection against fowl adenovirus serotype 4 and induces quicker and stronger immune responses than an inactivated oil-emulsion vaccine. Infect. Genet. Evol. 2018, 61, 145–150. [Google Scholar] [CrossRef]
- Tian, K.Y.; Guo, H.F.; Li, N.; Zhang, Y.H.; Wang, Z.; Wang, B.; Yang, X.; Li, Y.T.; Zhao, J. Protection of chickens against hepatitis-hydropericardium syndrome and Newcastle disease with a recombinant Newcastle disease virus vaccine expressing the fowl adenovirus serotype 4 fiber-2 protein. Vaccine 2020, 38, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Li, G.; Wang, X.; Cai, L.; Rong, M.; Li, H.; Xie, M.; Zhang, Z.; Rong, J. Development of a subunit vaccine based on fiber2 and hexon against fowl adenovirus serotype 4. Virus Res. 2021, 305, 198552. [Google Scholar] [CrossRef]
- Schachner, A.; Marek, A.; Jaskulska, B.; Bilic, I.; Hess, M. Recombinant FAdV-4 fiber-2 protein protects chickens against hepatitis-hydropericardium syndrome (HHS). Vaccine 2014, 32, 1086–1092. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, J.; Wang, W.; Li, T.; Liang, G.; Shao, H.; Gao, W.; Qin, A.; Ye, J. A novel monoclonal antibody efficiently blocks the infection of serotype 4 fowl adenovirus by targeting fiber-2. Vet. Res. 2018, 49, 29. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Hu, M.; Li, N.; Sun, X.; Xing, G.; Zheng, G.; Jin, Q.; Liu, Y.; Cui, C.; Zhang, G. Precise Assembly of Multiple Antigens on Nanoparticles with Specially Designed Affinity Peptides. ACS Appl. Mater. Interfaces 2022, 14, 39843–39857. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wang, F.; Li, N.; Xing, G.; Sun, X.; Zhang, Y.; Cao, S.; Cui, N.; Zhang, G. An antigen display system of GEM nanoparticles based on affinity peptide ligands. Int. J. Biol. Macromol. 2021, 193, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Henry, L.J.; Gerard, R.D.; Deisenhofer, J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 A resolution. Structure 1994, 2, 1259–1270. [Google Scholar] [CrossRef]
- Yin, D.; Yin, L.; Wang, J.; Shen, X.; Dai, Y.; Zhao, R.; Hu, X.; Hou, H.; Zhang, D.; Wang, G.; et al. Antiviral and Virucidal Activities of Camptothecin on Fowl Adenovirus Serotype 4 by Blocking Virus Replication. Front. Cell. Infect. Microbiol. 2022, 12, 823820. [Google Scholar] [CrossRef]
- Wang, F.; Hao, J.; Li, N.; Xing, G.; Hu, M.; Zhang, G. Integrated System for Purification and Assembly of PCV Cap Nano Vaccine Based on Targeting Peptide Ligand. Int. J. Nanomed. 2020, 15, 8507–8517. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid-phase peptide synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 1969, 32, 221–296. [Google Scholar] [CrossRef]
- Mao, J.; Akhtar, J.; Zhang, X.; Sun, L.; Guan, S.; Li, X.; Chen, G.; Liu, J.; Jeon, H.N.; Kim, M.S.; et al. Comprehensive strategies of machine-learning-based quantitative structure-activity relationship models. iScience 2021, 24, 103052. [Google Scholar] [CrossRef]
- Lyne, P.D. Structure-based virtual screening: An overview. Drug Discov. Today 2002, 7, 1047–1055. [Google Scholar] [CrossRef]
- Pinto, G.P.; Hendrikse, N.M.; Stourac, J.; Damborsky, J.; Bednar, D. Virtual screening of potential anticancer drugs based on microbial products. Semin. Cancer Biol. 2022, 86, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Chen, M.; Qin, H.; Liu, H.; Ye, Y. Pueraria lobata Potentially Treating Prostate Cancer on Single-Cell Level by Network Pharmacology and AutoDock: Clinical Findings and Drug Targets. Comput. Math. Methods Med. 2022, 2022, 3758219. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, R.; Jain, A.N. Surflex-Dock: Docking benchmarks and real-world application. J. Comput. Aided Mol. Des. 2012, 26, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Heo, L.; Lee, M.S.; Seok, C. GalaxyPepDock: A protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 2015, 43, W431–W435. [Google Scholar] [CrossRef]
- Jain, A.N. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003, 46, 499–511. [Google Scholar] [CrossRef]
- Seiradake, E.; Lortat-Jacob, H.; Billet, O.; Kremer, E.J.; Cusack, S. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fiber heads in complex with the D1 domain of coxsackie and adenovirus receptor. J. Biol. Chem. 2006, 281, 33704–33716. [Google Scholar] [CrossRef]
- Baker, A.T.; Mundy, R.M.; Davies, J.A.; Rizkallah, P.J.; Parker, A.L. Human adenovirus type 26 uses sialic acid-bearing glycans as a primary cell entry receptor. Sci. Adv. 2019, 5, eaax3567. [Google Scholar] [CrossRef]
- Lopez-Gordo, E.; Doszpoly, A.; Duffy, M.R.; Coughlan, L.; Bradshaw, A.C.; White, K.M.; Denby, L.; Nicklin, S.A.; Baker, A.H. Defining a Novel Role for the Coxsackievirus and Adenovirus Receptor in Human Adenovirus Serotype 5 Transduction In Vitro in the Presence of Mouse Serum. J. Virol. 2017, 91, 577–591. [Google Scholar] [CrossRef]
- Soudais, C.; Boutin, S.; Hong, S.S.; Chillon, M.; Danos, O.; Bergelson, J.M.; Boulanger, P.; Kremer, E.J. Canine adenovirus type 2 attachment and internalization: Coxsackievirus-adenovirus receptor, alternative receptors, and an RGD-independent pathway. J. Virol. 2000, 74, 10639–10649. [Google Scholar] [CrossRef] [PubMed]
- Vassal-Stermann, E.; Effantin, G.; Zubieta, C.; Burmeister, W.; Iseni, F.; Wang, H.; Lieber, A.; Schoehn, G.; Fender, P. CryoEM structure of adenovirus type 3 fibre with desmoglein 2 shows an unusual mode of receptor engagement. Nat. Commun. 2019, 10, 1181. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, W.P.; Guilligay, D.; Cusack, S.; Wadell, G.; Arnberg, N. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J. Virol. 2004, 78, 7727–7736. [Google Scholar] [CrossRef] [PubMed]
- Roelvink, P.W.; Lizonova, A.; Lee, J.G.; Li, Y.; Bergelson, J.M.; Finberg, R.W.; Brough, D.E.; Kovesdi, I.; Wickham, T.J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 1998, 72, 7909–7915. [Google Scholar] [CrossRef]
- Arbi, M.; Larbi, I.; Nsiri, J.; Behi, I.E.; Rejeb, A.; Miled, K.; Ghram, A.; Houimel, M. Inhibition of avian influenza virus H9N2 infection by antiviral hexapeptides that target viral attachment to epithelial cells. Virus Res. 2022, 313, 198745. [Google Scholar] [CrossRef]
- Lv, X.; Zhang, M.; Yu, S.; Zhang, C.; Fang, T.; Liu, D.; Jia, B.; Zhu, M.; Wang, B.; Wang, Q.; et al. Antiviral and virucidal activities of lycorine on duck tembusu virus in vitro by blocking viral internalization and entry. Poult. Sci. 2021, 100, 101404. [Google Scholar] [CrossRef]
- Lee, J.L.; Loe, M.W.C.; Lee, R.C.H.; Chu, J.J.H. Antiviral activity of pinocembrin against Zika virus replication. Antivir. Res. 2019, 167, 13–24. [Google Scholar] [CrossRef]
- Long, F.; Zhang, M.; Yang, X.; Liang, X.; Su, L.; An, T.; Zhang, G.; Zeng, Z.; Liu, Y.; Chen, W.; et al. The Antimalaria Drug Artesunate Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication by Activating AMPK and Nrf2/HO-1 Signaling Pathways. J. Virol. 2022, 96, e0148721. [Google Scholar] [CrossRef]
- Baggen, J.; Thibaut, H.J.; Strating, J.; van Kuppeveld, F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018, 16, 368–381. [Google Scholar] [CrossRef]


| Crystal | Knob Domain of FAdV-4 Fiber2 |
|---|---|
| Data Collection | |
| Space group | P63 |
| Cell dimensions (length Å; angle °) | a = 60.34, b = 60.34, c = 192.60; α = β = 90, γ = 120 |
| Resolution (Å) | 50.48–1.29 (1.32–1.29) b |
| Rmerge (%) a | 9.1 (77.8) b |
| Unique reflections | 98,150 (6455) b |
| Completeness (%) | 98.8 (88.3) b |
| <I/σ(I)> | 16.8 (2.0) b |
| Redundancy | 17.3 (6.4) b |
| Refinement | |
| Resolution (Å) | 50.48–1.29 (1.32–1.29) b |
| Rwork/Rfree (%) | 18.5/22.1 |
| RMSD bond length (Å)/angle (°) | 0.008/0.996 |
| No. of molecules per asymmetric unit | 2 |
| No. of atoms | |
| Protein | 3004 |
| Water | 782 |
| Average B factors (Å2) | 16.56 |
| Ramachandran plot, residues in (%) | |
| Most favored regions | 96.7 |
| Additional allowed regions | 2.5 |
| Disallowed regions | 0.8 |
| PDB ID | 7W83 |
| Primer Name | Sequence (5’-3’) | Product Size/bp |
|---|---|---|
| FAdV-4 Hexon F | CGAGGACTACGACGATTA | 95 |
| FAdV-4 Hexon R | CGTGATACAGCAGGTTAATG |
| Peptide No. | Sequence | Total Score |
|---|---|---|
| P1 | QWKKHI | 16.3328 |
| P2 | MQKKWR | 15.2680 |
| P3 | QWKKWW | 14.8531 |
| P4 | QWKWMW | 14.7913 |
| P5 | QWKIEK | 14.7873 |
| P6 | QWKIHR | 14.7709 |
| P7 | MQKEWQ | 14.5475 |
| P8 | WWHQWD | 14.5184 |
| P9 | WWHWEF | 14.4116 |
| P10 | QWKNCK | 14.3470 |
| P11 | WWHLPV | 14.3400 |
| P12 | WWHEWQ | 14.3068 |
| P13 | WWHEWG | 14.2490 |
| P14 | QWKARV | 14.1350 |
| P15 | WWHEKE | 14.0258 |
| P16 | WWHSYT | 14.0035 |
| P17 | QWKFEY | 14.0002 |
| P18 | WWHHHC | 13.9934 |
| P19 | WWHVEK | 13.9661 |
| P20 | MQKRWK | 13.9108 |
| P21 | QWKQKW | 13.9061 |
| P22 | QWKAFG | 13.8920 |
| P23 | WWHGHM | 13.8756 |
| P24 | MQKADM | 13.3874 |
| P25 | MQKHEW | 13.2421 |
| P26 | MQKYTY | 13.1730 |
| P27 | MQKHCG | 13.1302 |
| P28 | MQKLWW | 13.0898 |
| P29 | MQKQWI | 12.9858 |
| P30 | MQKKFI | 12.9694 |
| Peptide No. | KD (M) |
|---|---|
| P1 | 6.161 × 10−6 |
| P5 | 1.199 × 10−6 |
| P6 | 2.779 × 10−6 |
| P10 | 1.021 × 10−6 |
| P14 | 2.397 × 10−5 |
| P25 | 1.367 × 10−6 |
| P27 | 5.725 × 10−7 |
| P30 | 9.070 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Wei, Q.; Si, F.; Wang, F.; Lu, Q.; Guo, Z.; Chai, Y.; Zhu, R.; Xing, G.; Jin, Q.; et al. Design and Identification of a Novel Antiviral Affinity Peptide against Fowl Adenovirus Serotype 4 (FAdV-4) by Targeting Fiber2 Protein. Viruses 2023, 15, 821. https://doi.org/10.3390/v15040821
Chen X, Wei Q, Si F, Wang F, Lu Q, Guo Z, Chai Y, Zhu R, Xing G, Jin Q, et al. Design and Identification of a Novel Antiviral Affinity Peptide against Fowl Adenovirus Serotype 4 (FAdV-4) by Targeting Fiber2 Protein. Viruses. 2023; 15(4):821. https://doi.org/10.3390/v15040821
Chicago/Turabian StyleChen, Xiao, Qiang Wei, Fusheng Si, Fangyu Wang, Qingxia Lu, Zhenhua Guo, Yongxiao Chai, Rongfang Zhu, Guangxu Xing, Qianyue Jin, and et al. 2023. "Design and Identification of a Novel Antiviral Affinity Peptide against Fowl Adenovirus Serotype 4 (FAdV-4) by Targeting Fiber2 Protein" Viruses 15, no. 4: 821. https://doi.org/10.3390/v15040821
APA StyleChen, X., Wei, Q., Si, F., Wang, F., Lu, Q., Guo, Z., Chai, Y., Zhu, R., Xing, G., Jin, Q., & Zhang, G. (2023). Design and Identification of a Novel Antiviral Affinity Peptide against Fowl Adenovirus Serotype 4 (FAdV-4) by Targeting Fiber2 Protein. Viruses, 15(4), 821. https://doi.org/10.3390/v15040821







