Experimental Pathogenicity of H9N2 Avian Influenza Viruses Harboring a Tri-Basic Hemagglutinin Cleavage Site in Sonali and Broiler Chickens
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Virus Detection by Real Time RT-PCR (RT-qPCR)
2.2. Virus Isolation
2.3. Sequencing and Phylogenetic Analysis
2.4. Experimental Pathogenesis Study of H9N2 AIVs
2.4.1. Virus Titration and EID50 Calculation
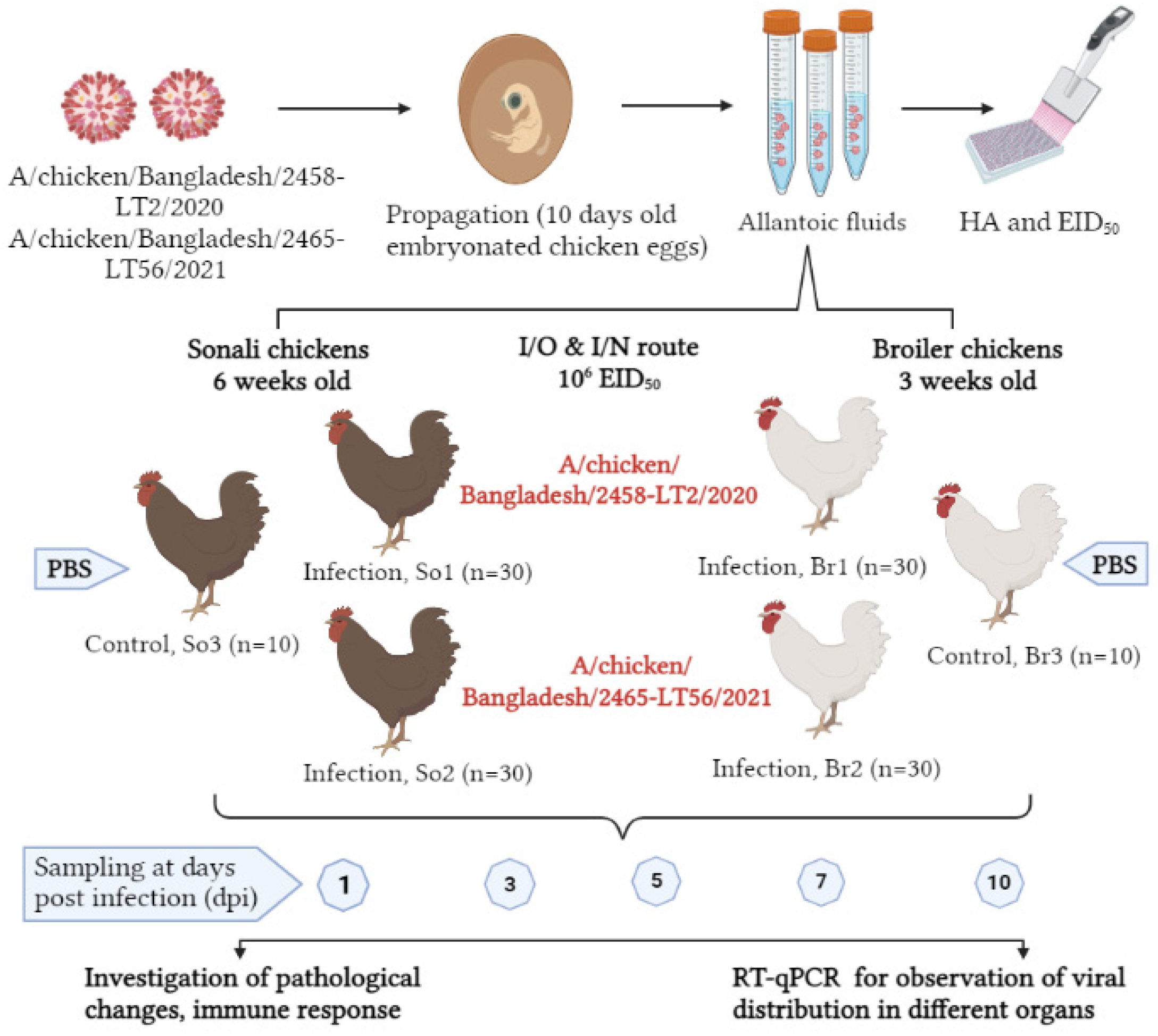
2.4.2. Experimental Design
2.4.3. Clinical and Pathological Investigation
2.4.4. Viral Distribution and Shedding
2.4.5. Serology
2.4.6. Statistical Analyses
3. Results
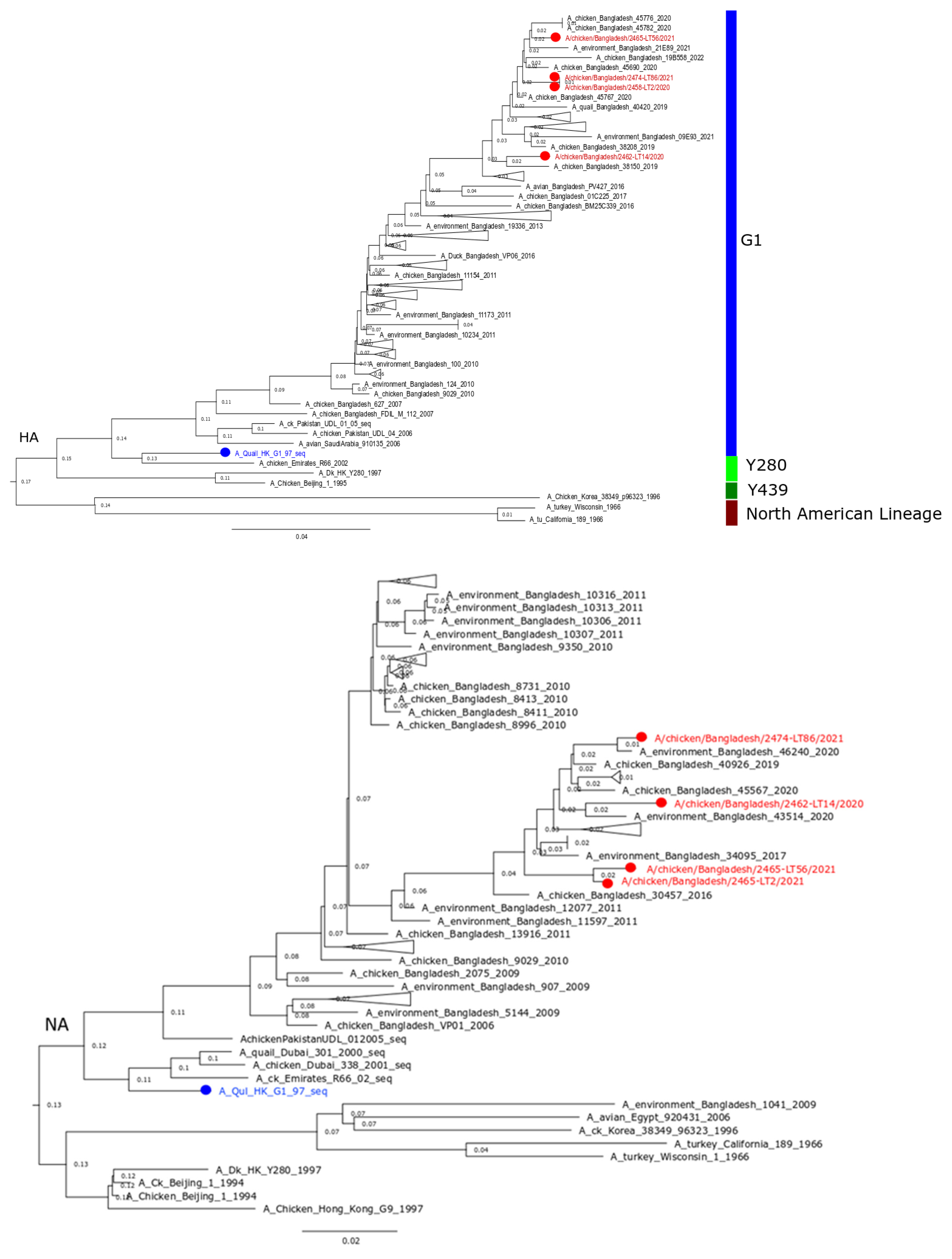
3.1. Genetic Background of Recently Circulating H9N2 Viruses in Bangladesh
3.2. Experimental Pathogenicity Study
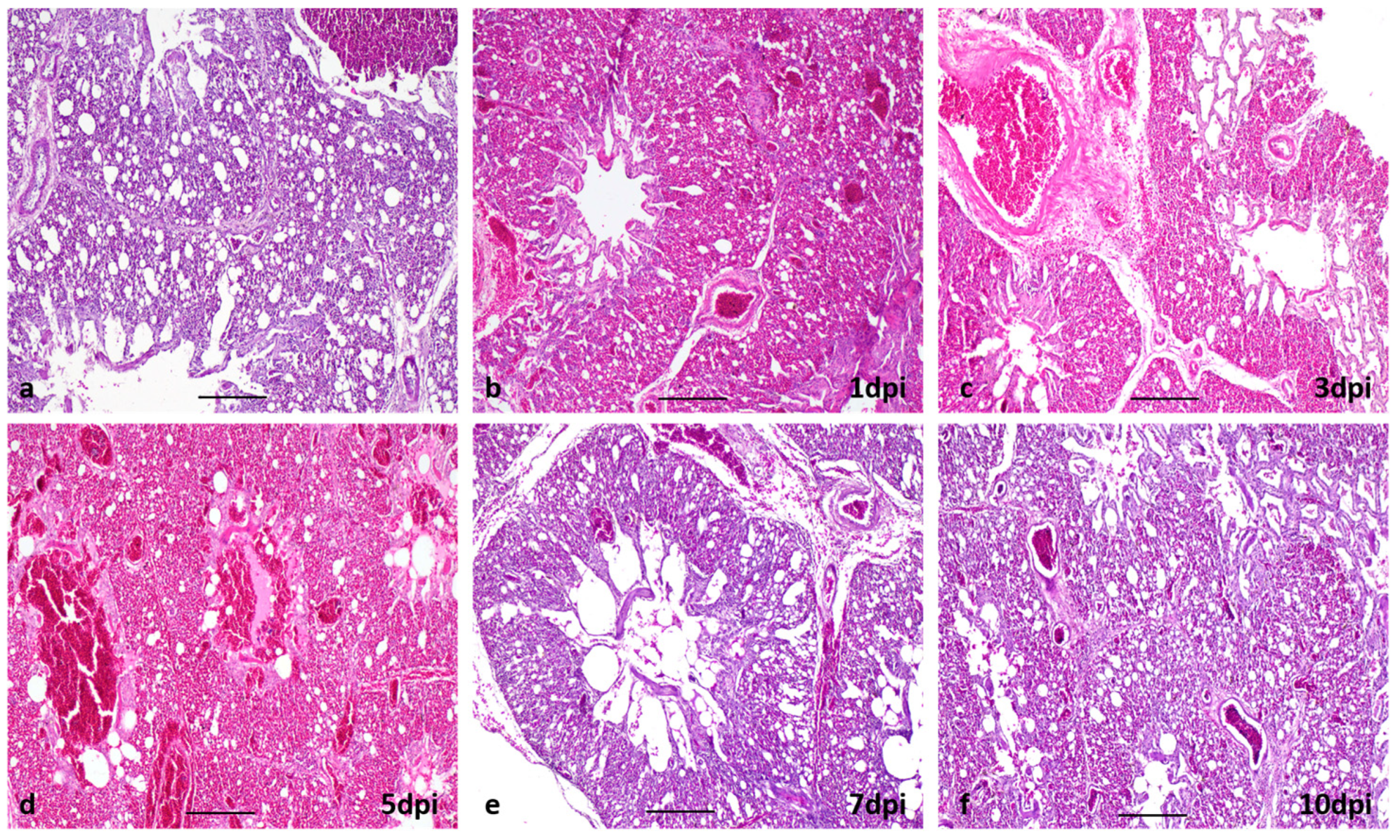
3.2.1. Clinical Signs, Gross and Histopathological Lesions
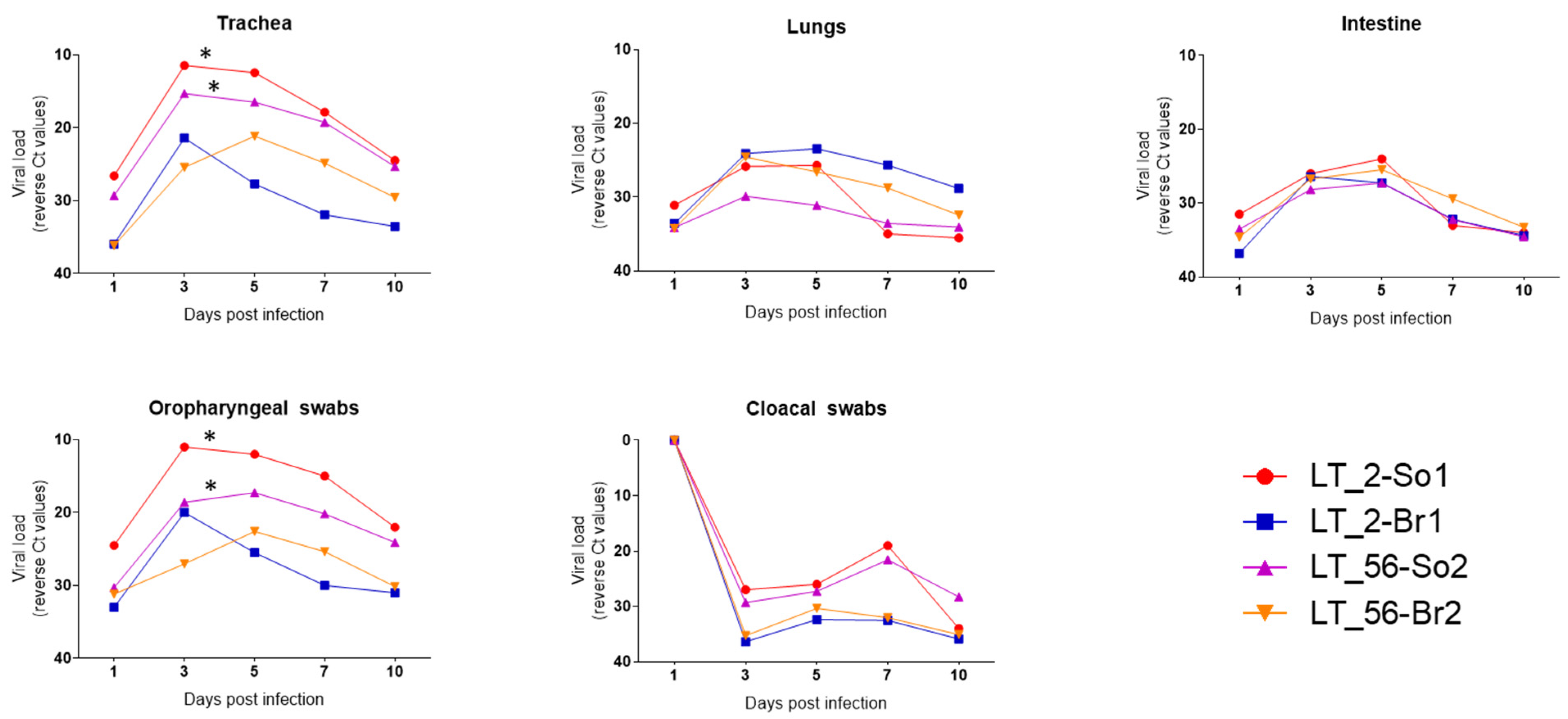
3.2.2. Respiratory and Intestinal Distribution and Shedding of the H9N2 Viruses
3.2.3. Seroconversion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jannat, N.; Chowdhury, E.; Parvin, R.; Begum, J.; Giasuddin, M.; Mahna, K. Investigation of an outbreak of low pathogenic avian influenza in poultry in Bangladesh. Intl. J. Livest. Res. 2013, 3, 21–32. Available online: https://www.bibliomed.org/?mno=41919 (accessed on 16 September 2022).
- Kariithi, H.M.; Welch, C.N.; Ferreira, H.L.; Pusch, E.A.; Ateya, L.O.; Binepal, Y.S.; Apopo, A.A.; Dulu, T.D.; Afonso, C.L.; Suarez, D.L. Genetic characterization and pathogenesis of the first H9N2 low pathogenic avian influenza viruses isolated from chickens in Kenyan live bird markets. Infect. Genet. Evol. 2020, 78, 104074. [Google Scholar] [CrossRef]
- Kye, S.J.; Park, M.J.; Kim, N.Y.; Lee, Y.N.; Heo, G.B.; Baek, Y.K.; Shin, J.I.; Lee, M.H.; Lee, Y.J. Pathogenicity of H9N2 low pathogenic avian influenza viruses of different lineages isolated from live bird markets tested in three animal models: SPF chickens, Korean native chickens, and ducks. Poult. Sci. 2021, 100, 101318. [Google Scholar] [CrossRef]
- Nili, H.; Asasi, K. Natural cases and an experimental study of H9N2 avian influenza in commercial broiler chickens of Iran. Avian Pathol. 2002, 31, 247–252. [Google Scholar] [CrossRef]
- Pusch, E.A.; Suarez, D.L. The Multifaceted Zoonotic Risk of H9N2 Avian Influenza. Vet. Sci. 2018, 5, 82. [Google Scholar] [CrossRef]
- Parvin, R.; Begum, J.A.; Chowdhury, E.H.; Islam, M.R.; Beer, M.; Harder, T. Co-subsistence of avian influenza virus subtypes of low and high pathogenicity in Bangladesh: Challenges for diagnosis, risk assessment and control. Sci. Rep. 2019, 9, 8306. [Google Scholar] [CrossRef]
- Rimi, N.A.; Hassan, M.Z.; Chowdhury, S.; Rahman, M.; Sultana, R.; Biswas, P.K.; Debnath, N.C.; Islam, S.S.; Ross, A.G. A Decade of Avian Influenza in Bangladesh: Where Are We Now? Trop. Med. Infect. Dis. 2019, 4, 119. [Google Scholar] [CrossRef]
- Shanmuganatham, K.; Feeroz, M.M.; Jones-Engel, L.; Smith, G.J.; Fourment, M.; Walker, D.; McClenaghan, L.; Alam, S.M.; Hasan, M.K.; Seiler, P.; et al. Antigenic and molecular characterization of avian influenza A(H9N2) viruses, Bangladesh. Emerg. Infect. Dis. 2013, 19, 1393–1402. [Google Scholar] [CrossRef]
- Gerloff, N.A.; Khan, S.U.; Zanders, N.; Balish, A.; Haider, N.; Islam, A.; Chowdhury, S.; Rahman, M.Z.; Haque, A.; Hosseini, P.; et al. Genetically Diverse Low Pathogenicity Avian Influenza A Virus Subtypes Co-Circulate among Poultry in Bangladesh. PLoS ONE 2016, 11, e0152131. [Google Scholar] [CrossRef]
- Khan, S.U.; Gurley, E.S.; Gerloff, N.; Rahman, M.Z.; Simpson, N.; Rahman, M.; Haider, N.; Chowdhury, S.; Balish, A.; Zaman, R.U.; et al. Avian influenza surveillance in domestic waterfowl and environment of live bird markets in Bangladesh, 2007–2012. Sci. Rep. 2018, 8, 9396. [Google Scholar] [CrossRef]
- Parvin, R.; Heenemann, K.; Halami, M.Y.; Chowdhury, E.H.; Islam, M.; Vahlenkamp, T.W. Full-genome analysis of avian influenza virus H9N2 from Bangladesh reveals internal gene reassortments with two distinct highly pathogenic avian influenza viruses. Arch. Virol. 2014, 159, 1651–1661. [Google Scholar] [CrossRef]
- Parvin, R.; Begum, J.A.; Nooruzzaman, M.; Chowdhury, E.H.; Islam, M.R.; Vahlenkamp, T.W. Review analysis and impact of co-circulating H5N1 and H9N2 avian influenza viruses in Bangladesh. Epidemiol. Infect. 2018, 146, 1259–1266. [Google Scholar] [CrossRef]
- Parvin, R.; Nooruzzaman, M.; Kabiraj, C.K.; Begum, J.A.; Chowdhury, E.H.; Islam, M.R.; Harder, T. Controlling avian influenza virus in Bangladesh: Challenges and recommendations. Viruses 2020, 12, 751. [Google Scholar] [CrossRef]
- Parvin, R.; Schinkoethe, J.; Grund, C.; Ulrich, R.; Bönte, F.; Behr, K.P.; Voss, M.; Samad, M.A.; Hassan, K.E.; Luttermann, C.; et al. Comparison of pathogenicity of subtype H9 avian influenza wild-type viruses from a wide geographic origin expressing mono-, di-, or tri-basic hemagglutinin cleavage sites. Vet. Res. 2020, 51, 48. [Google Scholar] [CrossRef]
- Bhuiyan, A.; Bhuiyan, M.; Deb, G. Indigenous chicken genetic resources in Bangladesh: Current status and future outlook. Anim. Genet. Resour. 2005, 36, 73–84. [Google Scholar] [CrossRef]
- Hamid, M.; Rahman, M.; Ahmed, S.; Hossain, K. Status of poultry industry in Bangladesh and the role of private sector for its development. Asian J. Poult. Sci. 2017, 11, 1–13. Available online: https://scialert.net/abstract/?doi=ajpsaj.2017.1.13 (accessed on 25 October 2022). [CrossRef]
- Rahman, M.M.; Nooruzzaman, M.; Kabiraj, C.K.; Mumu, T.T.; Das, P.M.; Chowdhury, E.H.; Islam, M.R. Surveillance on respiratory diseases reveals enzootic circulation of both H5 and H9 avian influenza viruses in small-scale commercial layer farms of Bangladesh. Zoonoses Public Health 2021, 68, 896–907. [Google Scholar] [CrossRef]
- Spackman, E.; Senne, D.A.; Myers, T.; Bulaga, L.L.; Garber, L.P.; Perdue, M.L.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002, 40, 3256–3260. [Google Scholar] [CrossRef]
- Hoffmann, B.; Hoffmann, D.; Henritzi, D.; Beer, M.; Harder, T.C. Riems influenza a typing array (RITA): An RT-qPCR-based low density array for subtyping avian and mammalian influenza a viruses. Sci. Rep. 2016, 6, 27211. [Google Scholar] [CrossRef]
- OIE, Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2018, Chapter 3.3.4. Avian Influenza (Infection with Avian Influenza Viruses). 2021. Available online: https://www.woah.org/fileadmin/Home/fr/Health_standards/tahm/3.03.04_AI.pdf (accessed on 12 August 2022).
- King, J.; Harder, T.; Beer, M.; Pohlmann, A. Rapid multiplex MinION nanopore sequencing workflow for Influenza A viruses. BMC Infect. Dis. 2020, 20, 648. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- The Intravenous Pathogenicity Index Test for Avian Influenza. Available online: https://science.vla.gov.uk/flu-lab-net/docs/protocol_IntravenousPathogenicity.pdf (accessed on 24 July 2022).
- Manual of Histologic Staining Methods. Available online: http://webs.fcm.unc.edu.ar/wp-content/blogs.dir/75/files/2018/07/MANUAL-OF-HISTOLOGIC-STAINING-METHODS-THIRD-EDITION-Mc-GRAW-HILL.pdf (accessed on 20 July 2022).
- Iqbal, M.; Yaqub, T.; Mukhtar, N.; Shabbir, M.Z.; McCauley, J.W. Infectivity and transmissibility of H9N2 avian influenza virus in chickens and wild terrestrial birds. Vet. Res. 2013, 44, 100. [Google Scholar] [CrossRef]
- Arnold, M.E.; Slomka, M.J.; Coward, V.J.; Mahmood, S.; Raleigh, P.J.; Brown, I.H. Evaluation of the pooling of swabs for real-time PCR detection of low titre shedding of low pathogenicity avian influenza in turkeys. Epidemiol. Infect. 2013, 141, 1286–1297. [Google Scholar] [CrossRef]
- Suttie, A.; Deng, Y.M.; Greenhill, A.R.; Dussart, P.; Horwood, P.F.; Karlsson, E.A. Inventory of molecular markers affecting biological characteristics of avian influenza A viruses. Virus Genes 2019, 55, 739–768. [Google Scholar] [CrossRef]
- Zhu, R.; Xu, S.; Sun, W.; Li, Q.; Wang, S.; Shi, H.; Liu, X. HA gene amino acid mutations contribute to antigenic variation and immune escape of H9N2 influenza virus. Vet. Res. 2022, 53, 43. [Google Scholar] [CrossRef]
- Subtain, S.M.; Chaudhry, Z.I.; Anjum, A.A.; Maqbool, A.; Sadique, U. Study on pathogenesis of low pathogenic avian influenza virus H9 in broiler chickens. Pak. J. Zool. 2011, 43, 999–1008. Available online: https://www.zsp.com.pk/pdf43/999-1008%20(25)%20PJZ-549-10.pdf (accessed on 20 May 2022).
- Kim, J.A.; Cho, S.H.; Kim, H.S.; Seo, S.H. H9N2 influenza viruses isolated from poultry in Korean live bird markets continuously evolve and cause the severe clinical signs in layers. Vet. Microbiol. 2006, 118, 169–176. [Google Scholar] [CrossRef]
- Monne, I.; Hussein, H.A.; Fusaro, A.; Valastro, V.; Hamoud, M.M.; Khalefa, R.A.; Dardir, S.N.; Radwan, M.I.; Capua, I.; Cattoli, G. H9N2 influenza A virus circulates in H5N1 endemically infected poultry population in Egypt. Influenza Other Respir Viruses 2013, 7, 240–243. [Google Scholar] [CrossRef]
- Hassan, K.E.; Ali, A.; Shany, S.A.; El-Kady, M.F. Experimental co-infection of infectious bronchitis and low pathogenic avian influenza H9N2 viruses in commercial broiler chickens. Res. Vet. Sci. 2017, 115, 356–362. [Google Scholar] [CrossRef]
- Hablolvarid, M.H.; Sohraby Haghdost, I.; Pourbakhsh, S.A.; Gholami, M.R. Histopathological study of intranasally inoculate A/Chicken/Iran/259/1998 (H9N2) influenza virus in chicken. Arch. Razi Inst. 2004, 58, 51–62. Available online: https://archrazi.areeo.ac.ir/article_103825_3d56ffa45fac1e202f843547dd3db366.pdf (accessed on 15 May 2022).
- Slemons, R.D.; Condobery, P.K.; Swayne, D.E. Assessing pathogenicity potential of waterfowl-origin type A influenza viruses in chickens. Avian Dis. 1991, 35, 210–215. [Google Scholar] [CrossRef]
- Slemons, R.D.; Locke, L.N.; Sheerar, M.G.; Duncan, R.M.; Hinshaw, V.S.; Easterday, B.C. Kidney lesions associated with mortality in chickens inoculated with waterfowl influenza viruses. Avian Dis. 1990, 34, 120–128. [Google Scholar] [CrossRef]
- Swayne, D.E. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis. 2007, 51 (Suppl. S1), 242–249. [Google Scholar] [CrossRef]
- Böttcher-Friebertshäuser, E.; Klenk, H.D.; Garten, W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 2013, 69, 87–100. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, Y.; Chen, L.; Zhang, B.; Zhang, M.; Wang, J.; Jiang, Y.; Yang, C.; Jiang, T. Genetic Characteristics and Pathogenicity Analysis in Chickens and Mice of Three H9N2 Avian Influenza Viruses. Viruses 2019, 11, 1127. [Google Scholar] [CrossRef]


| Sequence Accession & ID | Potential Antigenic Sites | RBS | HACS | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H9 Numbering | 66 | 90 | 147 | 149 | 153 | 167 | 168 | 196 | 198 | 216 | 224 | 282 | 283 | 166 | 234 | 399 | 335–338 |
| EPI2187437_LT_86/2021 | H | G | T | K | N | S | L | D | A | D | L | N | S | N | L | K | KSKR |
| EPI2187429_LT_56/2021 | R | . | . | . | D | . | . | . | . | . | . | . | . | . | . | . | KSKR |
| EPI2187421_LT_14/2020 | . | . | . | . | D | . | . | . | T | . | . | . | . | . | . | . | KSKR |
| EPI2187413_LT_2/2020 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | KSKR |
| EPI1778204_40818/2019 | P | . | . | . | D | G | Q | . | . | N | . | . | . | . | . | . | KSKR |
| EPI1581761_35417/2018 | R | . | . | . | D | G | . | . | . | N | . | K | . | . | . | . | KSKR |
| EPI1777263_NRL3238/2017 | R | . | . | . | D | G | . | . | . | N | . | K | . | . | . | . | KSKR |
| EPI1508528_AR11758/2016 | Q | . | . | . | D | G | Q | . | . | N | . | . | . | . | . | . | KSKR |
| EPI965466_24249/2015 | R | . | . | . | D | G | . | . | . | N | . | . | . | . | . | . | KSKR |
| EPI963330_23727/2014 | . | E | . | . | D | S | Q | . | . | N | . | R | . | . | . | . | KSKR |
| EPI528468_19870/2013 | . | . | . | . | D | G | . | . | . | N | . | . | . | . | . | . | KSKR |
| EPI528702_18224/2012 | . | . | . | . | D | G | . | . | . | N | . | . | I | . | . | . | KSKR |
| EPI462772_11309/2011 | . | . | . | . | D | G | . | . | . | N | . | . | . | . | . | . | KSKR |
| EPI462691_8415/2010 | . | . | . | . | D | G | . | . | . | N | . | . | . | . | . | . | KSSR |
| EPI462631_5209/2009 | . | . | . | . | D | G | . | . | . | N | . | . | . | . | . | . | KSSR |
| EPI383796_FDIL112/2007 | . | . | . | . | D | G | . | . | . | N | . | . | . | . | . | . | RSSR |
| EPI453588_VP01/2006 | . | . | . | . | D | G | . | . | . | N | . | . | . | . | . | . | KSSR |
| EPI985025_G1/1997 | . | E | I | R | G | G | F | Y | E | . | V | K | . | S | . | . | RSSR |
| Infected Groups | Sonali Chickens | Broiler Chickens | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lesions/Dpi | 1 | 3 | 5 | 7 | 10 | 15 | 1 | 3 | 5 | 7 | 10 |
| Trachea | |||||||||||
| Proliferation of goblet cells | + | + | + | + | + | + | +/− | + | − | ++ | ++ |
| Loss of epithelial layer | + | + | +++ | +++ | +++ | +++ | − | + | +++ | − | − |
| Loss of cilia | + | + | +++ | +++ | +++ | +++ | − | + | +++ | − | − |
| Inflammation | +/− | + | ++ | +++ | +++ | +++ | − | + | +++ | − | − |
| Congestion and | − | − | − | − | − | − | + | − | − | − | − |
| Hemorrhages | − | − | − | − | − | − | − | − | − | − | − |
| Lung | |||||||||||
| Congestion | + | ++ | ++ | +++ | + | +/− | ++ | ++ | +++ | + | + |
| Hemorrhage | + | + | ++ | +++ | + | +/− | ++ | + | +++ | + | +/− |
| Presence of inflammatory cells | +/− | +/− | + | + | + | +/− | +/− | +/− | ++ | + | + |
| Collapsing of alveoli | + | + | + | ++ | + | +/− | + | ++ | ++ | − | − |
| Rupture of alveoli | − | − | + | ++ | + | +/− | + | ++ | ++ | − | − |
| Loss of Pneumocyte−I | +++ | +++ | +++ | +++ | + | + | + | + | + | − | − |
| Increased proliferation of pneumocyte−I | − | − | − | − | + | + | − | − | − | − | − |
| Proliferation of pneumocyte−II | + | ++ | +++ | + | + | + | + | + | + | − | − |
| Intestine | |||||||||||
| Shortening of villi | − | ++ | − | − | − | − | − | + | − | − | − |
| Desquamation of villus epithelium | − | − | +++ | − | − | − | − | − | ++ | − | − |
| Infiltration of inflammatory cells | − | − | ++ | − | − | − | − | − | + | − | − |
| Glandular proliferation of crypt epithelium | − | − | ++ | − | − | − | − | − | + | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Begum, J.A.; Hossain, I.; Nooruzzaman, M.; King, J.; Chowdhury, E.H.; Harder, T.C.; Parvin, R. Experimental Pathogenicity of H9N2 Avian Influenza Viruses Harboring a Tri-Basic Hemagglutinin Cleavage Site in Sonali and Broiler Chickens. Viruses 2023, 15, 461. https://doi.org/10.3390/v15020461
Begum JA, Hossain I, Nooruzzaman M, King J, Chowdhury EH, Harder TC, Parvin R. Experimental Pathogenicity of H9N2 Avian Influenza Viruses Harboring a Tri-Basic Hemagglutinin Cleavage Site in Sonali and Broiler Chickens. Viruses. 2023; 15(2):461. https://doi.org/10.3390/v15020461
Chicago/Turabian StyleBegum, Jahan Ara, Ismail Hossain, Mohammed Nooruzzaman, Jacqueline King, Emdadul Haque Chowdhury, Timm C. Harder, and Rokshana Parvin. 2023. "Experimental Pathogenicity of H9N2 Avian Influenza Viruses Harboring a Tri-Basic Hemagglutinin Cleavage Site in Sonali and Broiler Chickens" Viruses 15, no. 2: 461. https://doi.org/10.3390/v15020461
APA StyleBegum, J. A., Hossain, I., Nooruzzaman, M., King, J., Chowdhury, E. H., Harder, T. C., & Parvin, R. (2023). Experimental Pathogenicity of H9N2 Avian Influenza Viruses Harboring a Tri-Basic Hemagglutinin Cleavage Site in Sonali and Broiler Chickens. Viruses, 15(2), 461. https://doi.org/10.3390/v15020461









