Abstract
Parasitoid wasps are fundamental insects for the biological control of agricultural pests. Despite the importance of wasps as natural enemies for more sustainable and healthy agriculture, the factors that could impact their species richness, abundance, and fitness, such as viral diseases, remain almost unexplored. Parasitoid wasps have been studied with regard to the endogenization of viral elements and the transmission of endogenous viral proteins that facilitate parasitism. However, circulating viruses are poorly characterized. Here, RNA viromes of six parasitoid wasp species are studied using public libraries of next-generation sequencing through an integrative bioinformatics pipeline. Our analyses led to the identification of 18 viruses classified into 10 families (Iflaviridae, Endornaviridae, Mitoviridae, Partitiviridae, Virgaviridae, Rhabdoviridae, Chuviridae, Orthomyxoviridae, Xinmoviridae, and Narnaviridae) and into the Bunyavirales order. Of these, 16 elements were described for the first time. We also found a known virus previously identified on a wasp prey which suggests viral transmission between the insects. Altogether, our results highlight the importance of virus surveillance in wasps as its service disruption can affect ecology, agriculture and pest management, impacting the economy and threatening human food security.
Keywords:
virus; biological control; insect; Hymenoptera; Aculeata; natural enemy; RNA-seq; Braconidae; Crabronidae 1. Introduction
The study of insect viruses holds significant value for numerous reasons. Firstly, it enables the prediction of the risk of pathogenic virus spillover events that could harm economically important beneficial insects and endemic species []. Additionally, it aids in solving issues related to viral taxonomy and evolution [,] as well as understanding the evolutionary relationships between viruses and their hosts []. Furthermore, it may facilitate the identification of causal agents of disease outbreaks in animals and plants []. It should be noted that research on insect viruses has primarily focused on medical research, including hematophagous insect species that directly transmit vector-borne diseases []. Nevertheless, insects are the most diverse group of the animal kingdom, with approximately 5.5 million species on Earth []. Despite over one million insect species being scientifically identified, there are significant knowledge and research gaps concerning their viromes.
Parasitoid wasps are essential insects for the biological control of agricultural pests []. They lay eggs inside the body of their arthropod hosts, where the immature offspring will develop and ultimately kill their parasitized prey []. In addition, they can pollinate, disperse seeds, and help in the decomposition and recycling of nutrients of vertebrate dead bodies (reviewed in []). Nonetheless, global insect decline has been intensively evidenced in the last few decades [,], and little attention has been paid to this group of insects. In 2018, a declining trend for 11 out of 48 (23%) cuckoo wasp species in Finland was described, probably due to habitat loss []. However, important ecosystem service providers of Hymenoptera, like predatory and parasitoid wasps, are still understudied [].
To begin unraveling the viral diversity and dynamics in parasitoid wasps, six species widely distributed across the planet were selected for this study (Figure 1). First, Ampulex compressa (Ampulicidae), native to Ethiopian and Oriental regions [], is a specialist parasitoid of the American cockroach Periplaneta americana [], a cosmopolitan invasive household pest. The second is Cotesia vestalis (Braconidae), a specialist endoparasitoid of the diamondback moth (Plutella xylostella), which is a serious pest of brassica vegetables of global concern []. This host of C. vestalis larvae causes critical economic losses worldwide []. Third, Diachasma alloeum (Braconidae) is a parasitoid of the apple maggot fly (Rhagoletis pomonella), which is an economically important agricultural pest of apple crops in North America []. Fourth, Ectemnius lituratus (Crabronidae) is a parasitoid digger wasp from Europe that nests in burrows in a variety of dead wood. Flies of medium size are collected as prey to nourish nest cells []. Fifth, Pemphredon lugubris (Crabronidae) is a parasitoid digger wasp widely distributed throughout the northern hemisphere. It nests in dead and decaying wood []. Its prey is primarily aphids, which are very damaging pests of a wide range of crops of economic importance, including cereal crops [,]. Finally, Telenomus podisi (Platygastridae), an egg-parasitoid wasp found in Brazil and in the United States [], has been shown to be important in controlling its preferential host Euchistus heros, an abundant pest of soybean crops [,].
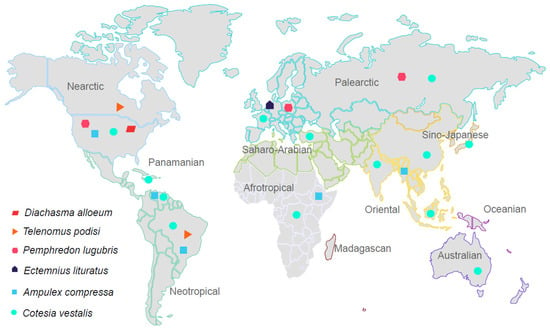
Figure 1.
Map of the geographical distribution of the six selected parasitoid species. Each symbol represents a wasp species and refers to the large biogeographical regions instead of single countries. The zoogeographic realms and regions were divided and are represented according to Holt et al., 2013.
Previous research focused on parasitoid wasps’ virome revealed the presence of members of Reoviridae, Iflaviridae [], Dicistroviridae [,], Lispiviridae, Rhabdoviridae [], and Nyamiviridae []. However, exogenous RNA viruses of these insects are still poorly investigated. On the other hand, parasitoid wasps have been substantially studied with regard to the endogenization of viral elements and the transmission of endogenous viral proteins that facilitate parasitism; thus, the integration of viral sequences into their genome is a well-explored process [,,,,,,]. Endogenous viral elements have functional roles in the parasitism success of these wasps, as well as being inheritable through wasp generations, an example of convergent evolution between DNA viruses and their hosts [].
This study aims to investigate the viral diversity of six parasitoid wasps through the analysis of fourteen publicly available RNA deep sequencing libraries. The data provided in our study highlight the importance of virus surveillance in wasps as its service disruption can affect ecology, agriculture, and pest management, impacting the economy and threatening human food security.
2. Materials and Methods
- Recovery and processing of RNA-seq libraries
Paired-end public libraries of long RNAs from six species of parasitoid wasps (Ampulex compressa, Cotesia vestalis, Diachasma alloeum, Ectemnius lituratus, Pemphredon lugubris, and Telenomus podisi) were retrieved from the NCBI Sequence Read Archive (SRA) repository (https://www.ncbi.nlm.nih.gov/sra), accessed on 4 October 2022. The accession numbers of the 14 selected libraries grouped by species are A. compressa (SRR14607675), C. vestalis (SRR6706566, SRR13704978, SRR13704979, SRR13704980, SRR13704991, SRR13704992, and SRR13704971), D. alloeum (SRR2041626 and SRR2040481), E. lituratus (ERR6054901), P. lugubris (ERR8571638), and T. podisi (SRR1274857 and SRR1274858). In addition, the geographical origin of the samples used for RNA library construction is represented in Supplementary Figure S1 and Supplementary Table S1. All libraries were processed using tools implemented on the Galaxy Australia web-based platform []. The raw reads’ quality was assessed through FastQC version 0.73 []. Then, for quality filtering, Illumina adapters and bases with poor quality scores (<20) were removed by sliding window trimming operation in Trimmomatic v. 0.36.6 []. The remaining reads were mapped against each wasp genome (except T. podisi which does not have an available reference genome) using Bowtie2 v. 2.4.5 []. The unaligned sequences were used as input to assemble the putative viral genomes with SPAdes v. 3.15.4 [] (Supplementary Table S1). Furthermore, the assembled transcripts were queried against the complete viral RefSeq (protein) database retrieved from the NCBI (https://ftp.ncbi.nlm.nih.gov/refseq/release/viral/), accessed on 6 October 2022, and uploaded to Galaxy using DIAMOND BLASTx v. 2.0.15 [], setting E-values ≤ 10−5 and standard parameters. For the metagenomics analyses, TaxonKit [] was applied. An overview of the sequence similarity results is available in Supplementary Table S2.
- Manual curation and improvement of putative viral genomes
We discarded all hits showing similarity to DNA viruses, retroviruses, and transposons since they mostly represent false positives in virome studies. The remaining sequences were double-checked regarding their viral origin using the online NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (accessed on 7 October 2022), specifically BLASTn (nucleotide collection database) and BLASTx (non-redundant protein sequences database). The sequences showing similarity to RNA viruses were further analyzed. For each putative viral sequence, genomic RNAs were manually inspected using Expasy translate (https://web.expasy.org/translate/), and ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) (accessed on 10 October 2022). The ORFs larger than 100 nt were chosen for the analyses of conserved domains using profile hidden Markov models with HMMER (https://www.ebi.ac.uk/Tools/hmmer/) (accessed on 10 October 2022) [] and NCBI Conserved Domains search tools https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 10 October 2022) []. Putative viral sequences that did not match the length or ORF structure in comparison to their closest relative in the NCBI nucleic acid databases were further investigated, and libraries from which the virus genome originated were submitted to a new round of assembly using an integrative strategy composed of multiple assemblers (SPAdes [], Trinity [], metaSPAdes [], rnaviralSPAdes, metaviralSPAdes v. 3.15.5 [], Oases v. 0.1.2 [], and MEGAHIT v. 1.2.9 []) followed by transcript consolidation with Cap3 [] as described by Espinal et al., 2023 [].
- Phylogenetic analyses
For the phylogenetic analyses, we first selected complete genomes (length ≥ 90% of related viruses’ genome size); second, fragments of viral genomes (length ≥ 500 nt, which were identified as RNA-dependent RNA polymerase (RdRp) by BLAST and presented conserved domains); third, other RdRp fragments (length ≥ 1000 nt, with the longest ORF ≥ 70% of the full-size sequence, with conserved domains). For the construction of phylogenetic trees, we obtained related virus sequences using BLASTn or BLASTx, according to the highest percentages of similarity and query coverage. The trees of putative new viruses were built with protein sequences that we retrieved from online BLASTx in association with sequences of viruses recognized by the International Committee on Taxonomy of Viruses (ICTV) when possible. The species were selected based on closely related genera or members of the same taxonomic family. Members of other families or distant genera, classified by the ICTV, were chosen as outgroups. More details on the phylogenetic trees’ construction are available in Supplementary Table S3. The selected ORFs (aa) or complete genome sequences (nt) were aligned by MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/) (accessed on 11 October 2022). Minimal manual adjustments and end trimming were carried out using AliView v. 1.28 []. The statistical selection of best-fit models of nucleotide and protein substitution was determined based on the Akaike information criterion (AIC) using ModelTest-NG on XSEDE v. 0.1.7 []. After, maximum likelihood trees were inferred by RAxML-HPC BlackBox v. 8.2.12 [], with 1000 bootstrap replicates. Both tools were implemented on CIPRES Science Gateway []. Finally, the generated phylogenetic trees were visualized and edited with FigTree v. 1.4.4 [] and iTol v.6.7.2 [].
- Quantification of viral sequences
The transcriptome assembled for each wasp species was evaluated with TransDecoder (https://github.com/TransDecoder/TransDecoder) (accessed on 15 March 2023) to identify the most likely coding sequences (CDSs). The resulting host transcripts were then added to the assembled viral transcripts to estimate the viral abundance in comparison to host mRNAs using the software Salmon v.1.9.0 []. For comparison with viral quantification, endogenous and standard genes were chosen; the hosts’ mitochondrial cytochrome b (cytb) and nuclear calmodulin were identified via sequence similarity searches (by BlastN) using Vespa velutina orthologous genes (MW401001.1 and XM_047509146.1, respectively) as references.
3. Results
3.1. Metagenomics Analyses
The initial metagenomics analyses revealed the presence of genetic material derived from several microorganisms classified into different kingdoms. As expected, we detected several transcripts matching Eukaryotic species, with higher abundance for transcripts matching insect sequences likely due to the lack of well annotated reference genomes for the species analyzed (Figure 2). We also observed transcripts derived from Bacteria in all libraries analyzed (A. compressa: 4%; C. vestalis: 7.1–10.1%; D. alloeum: 11.4%; E. lituratus: 21.1%; P. lugubris: 14.2%; and T. podisi: 2.7%). Overall, the most frequent families of bacteria were Streptomycetaceae, Pseudomonadaceae, Lactobacillaceae, Enterobacteriaceae, Burkholderiaceae, Rickettsiaceae, and Anaplasmataceae (Figure 2). In addition, we detected Fungal transcripts (A. compressa: 8.9%; C. vestalis: 0.5–2.4%; D. alloeum: 35.7%; E. lituratus: 4%; P. lugubris: 5.8%; and T. podisi: 1.8%). Some of the fungal families identified were: Saccharomycetaceae, Nectriaceae, Mucoraceae, Hypocreaceae, Glomerellaceae, Clavicipitaceae, and Aspergillaceae (Figure 2). Finally, transcripts showing similarity with viruses were distributed among Endornaviridae, Mitoviridae, Partitiviridae, Narnaviridae, Iflaviridae, Rhabdoviridae, Orthomyxoviridae, Chuviridae, Phenuiviridae, and Virgaviridae (A. compressa: 1%; C. vestalis: 3.5–9.4%; D. alloeum: 2.4%; E. lituratus: 0.8%; P. lugubris: 3.1%; and T. podisi: 2.9%). Detailed results grouped by species or for each library are shown in Supplementary Figures S2 and S3. The transcripts showing similarity to viruses were further analyzed.
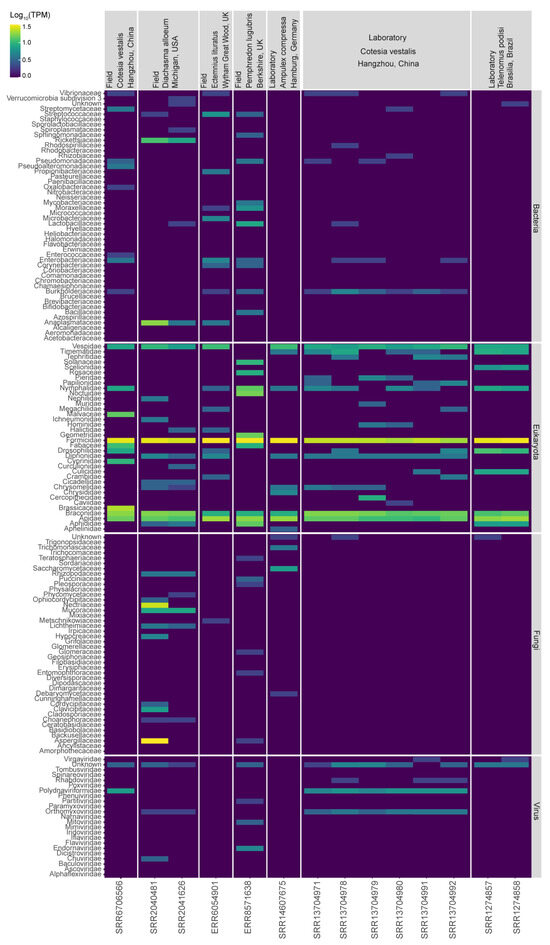
Figure 2.
Metatranscriptomics analysis for the six parasitoid wasps investigated in our study. Taxonomic classifications for bacteria, Eukaryota, fungi, and viruses are given by the library and wasp species. The geographical origins and collection sites are indicated in the top rows.
3.2. Characterization of the Wasps’ Virome
- Ampulex compressa
A library of the whole body of an adult Ampulex compressa (SRR14607675), from Germany, showed a viral sequence (Ac_Contig1) of 3732 nt, which presented sequence similarity at the nucleotide level (Table 1) to the Xiangshan narna-like virus hypothetical protein gene. Analysis at the protein level indicated that it is similar to a hypothetical protein of the same virus (UDL13948.1) that, despite being annotated as hypothetical, has an RdRp domain (cl40470), indicating that this protein represents the viral replicase (Table 1). Our assembled sequence has an ORF of 3114 nt|1037 aa, which is similar to its closest relative, the aforementioned Xiangshan narna-like virus (ORF: 3183 nt|1060 aa) (Figure 3A—left panel and Supplementary Figure S2). Based on the phylogenetic analysis, this putative virus was grouped with sequences of unclassified genera within Narnaviridae and was closely related to Xiangshan narna-like virus with 100% bootstrap (Figure 3B). Using NCBI Conserved Domain search, we were able to identify the RdRP domain cl40470 in our assembled narnavirus sequence (Figure 3A). To reflect the virus host and family, this sequence was named Ampulexvirus narnaviri, in accordance with recent ICTV guidelines [].

Table 1.
Overview of the wasp viral transcript best hits identified by sequence similarity searches.
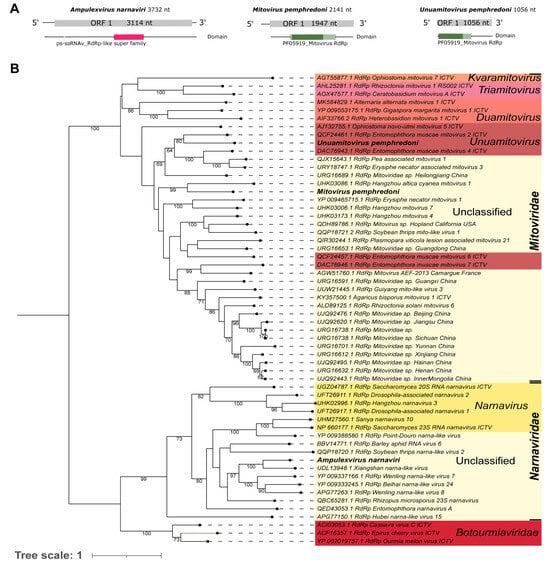
Figure 3.
Characterization of viral sequences related to elements from the Mitoviridae and Narnaviridae families. (A) ORF pattern and conserved domains of Ampulexvirus narnaviri (left panel), Mitovirus pemphredoni (middle panel), and Unuamitovirus pemphredoni (right panel). (B) Maximum likelihood tree of elements from the Mitoviridae and Narnaviridae families. The viral genomes assembled in our work are highlighted in bold. Bootstrap values under 60 are not shown.
- Cotesia vestalis
We analyzed seven libraries from China of whole-body Cotesia vestalis (field adult and laboratory-reared pupae) (Supplementary Table S2). All libraries contained viral sequences showing similarity at the amino acid level to elements of three families: Virgaviridae (one putative virus, 9075 nt, detected in four libraries), Orthomyxoviridae (five segments of one putative virus, detected in three libraries), and Rhabdoviridae (one putative virus, 12,294 nt, detected in four libraries). Furthermore, three segments of a putative unclassified virus of the Bunyavirales order were identified (RdRp with 6636 nt; glycoprotein with 1579 nt; and a nucleocapsid with 868 nt). Thus, considering the presence of polymerases that lack similarity at the nucleotide level to known viruses, we detected four possible new species infecting Cotesia vestalis.
First, Cv_Contig1, of 9075 nt, matched at the nucleotide level to the Abisko virus, complete genome, of 10,187 nt (Table 1). Its best hit at the amino acid level was the RdRp of Sanya virga-like virus 1 (Table 1). Due to low query coverage and identity to known viruses, this sequence likely represents a new virus that has similar ORF (7866 nt|2621 aa) and domains (PF01660_viral methyltransferase, PF01443_viral helicase, and PF00978_RdRp 2) (Figure 4A—top panel) to its best hit, the above-mentioned Sanya virga-like virus 1 (ORF 6267 nt) (Supplementary Figure S4). According to the phylogenetic analysis, this putative new virus clustered with Megastigmus ssRNA virus and Pemphredonvirus anglici, another Virgaviridae virus we describe in this work, with 75% bootstrap (Figure 4B). This new virus was named Cotesiavirus virgavi.
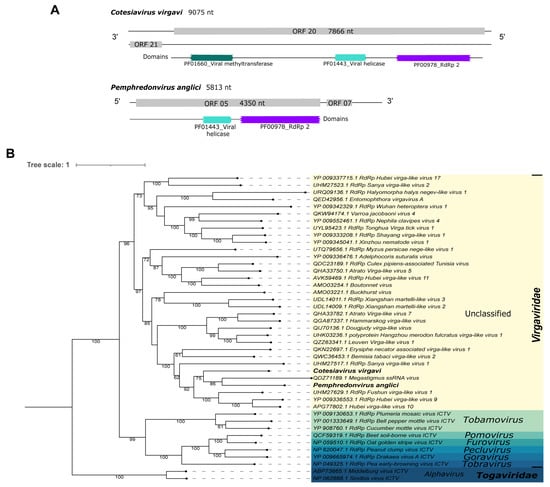
Figure 4.
Characterization of viral sequences related to elements from the Virgaviridae family. (A) ORF pattern and conserved domains of Cotesiavirus virgavi (upper panel) and Pemphredonvirus anglici (lower panel). (B) Maximum likelihood tree of elements from the Virgaviridae family. The viral genomes assembled in our work are highlighted in bold. Bootstrap values under 60 are not shown.
Second, five segments of an unclassified Orthomyxovirus were identified using sequence similarity search at the amino acid level as follows: Cv_RNA_segment_1 of 2499 nt matched to polymerase PB1 (Phasmatodean orthomyxo-related virus OKIAV172); Cv_RNA_segment_2 of 2470 nt was similar to polymerase PB2, partial (Phasmatodean orthomyxo-related virus OKIAV172); Cv_RNA_segment_3 of 2256 nt matched to polymerase PA, partial (Hymenopteran orthomyxo-related virus OKIAV171); Cv_RNA_segment_4 of 1577 nt presented similarity to hemagglutinin (Hymenopteran orthomyxo-related virus OKIAV173); and Cv_RNA_segment_5 of 1518 nt matched to the nucleocapsid protein (Blattodean orthomyxo-related virus OKIAV181) (Table 1). Four segments have the expected sizes and ORFs, according to close orthomyxoviruses (Supplementary Figure S4). Furthermore, these segments have domains that match to those identified in the closely related above-mentioned orthomyxoviruses (PF00602_Influenza RdRp subunit PB1, PF00604_Influenza RdRp subunit PB2, PF00603_Influenza RdRp subunit PA, and the PF03273_baculovirus gp64 envelope glycoprotein family), as illustrated in Figure 5A—left panel. Because PB1 is the polymerase subunit most conserved among orthomyxoviruses [], and it has been used in other studies [,], we selected this segment to perform the phylogenetic analysis of the orthomyxoviruses identified. Cv_RNA_segment_1 (PB1 RdRp) clustered with Diachasmavirus orthomyxi (Figure 5B), another orthomyxovirus we describe in this work, with 100% bootstrap (Figure 5A,B—right panel). Also, they clustered with sequences of unclassified genera in the Orthomyxoviridae family. To indicate the original host and viral family, this new virus was named Cotesiavirus orthomyxi.
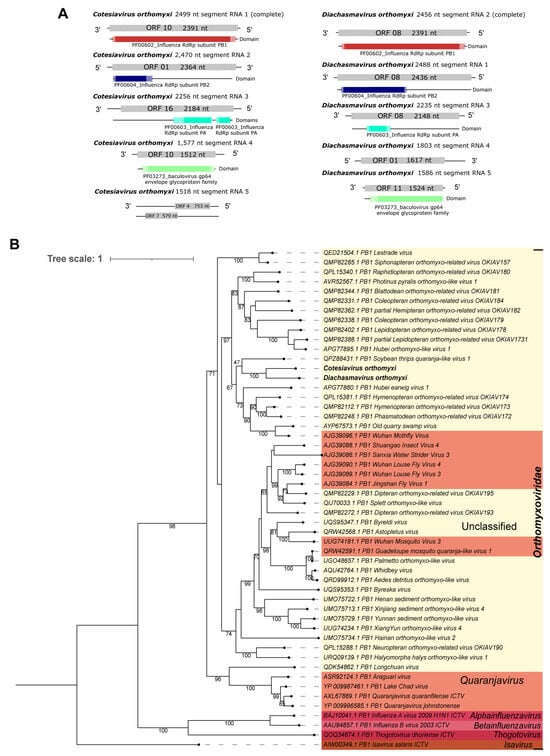
Figure 5.
Characterization of viral sequences related to elements from the Orthomyxoviridae family. (A) Segments, ORF pattern, and conserved domains of Cotesiavirus orthomyxi (left panel) and Diachasmavirus orthomyxi (right panel). (B) Maximum likelihood tree of elements from the Orthomyxoviridae family. The viral genomes assembled in our work are highlighted in bold. Bootstrap values under 60 are not shown.
Third, Cv_Contig2, of 12,294 nt, presented limited similarity at the nucleotide level (Table 1) to San Gabriel mononegavirus, of 12,620 nt. Its two best hits at the amino acid level were the RdRp (Hymenopteran rhabdo-related virus) and RdRp Wuhan Ant virus (Table 1). This new virus also has the longest ORF (6378 nt|2125 aa) and domains (PF00946_Mononegavirales RdRp, PF14318_Mononegavirales mRNA-cap V, and PF14314_virus-capping methyltransferase) (Figure 6A—top panel) according to the expected ORF of its best hit, Wuhan ant virus (ORF 6357 nt) (Supplementary Figure S4). Based on the phylogenetic analysis, this virus clustered with sequences of unclassified genera of Rhabdoviridae family and was closely related to Wuhan ant virus with 78% bootstrap (Figure 6B). This new virus was named Cotesiavirus rhabdovi, reflecting the host and viral family.
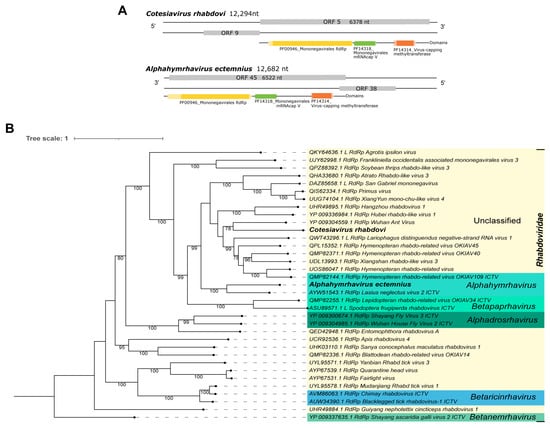
Figure 6.
Characterization of viral sequences related to elements from the Rhabdoviridae family. (A) ORF pattern and conserved domains of Cotesiavirus rhabdovi (upper panel) and Alphahymrhavirus ectemnius (lower panel). (B) Maximum likelihood tree of elements from the Rhabdoviridae family. The viral genomes assembled in our work are highlighted in bold. Bootstrap values under 60 are not shown.
Fourth, Cv_Contig3, of 6636 nt in length, is one of the three sequences that shows similarity to elements from the Bunyaviridae family. The best hit of Cv_Contig3 at the nucleotide level was the Wuhan insect virus 16 RdRp gene. Also, at the amino acid level, it matched with the polymerase of the same virus (Table 1). Its longest ORF (6438 nt|2145 aa) is as expected based on its closest virus, Wuhan insect virus 16 (ORF 6417 nt). In addition, there are two domains (PF15518_L-protein N-terminus and PF04196_Bunyavirus RdRp) that reinforce its classification as a member of Bunyavirales (Figure 7A). The sequence showing similarity to a glycoprotein, of 1579 nt, matched with Hymenopteran phenui-related virus OKIAV282 while the sequence identified as nucleoprotein, of 868 nt, was closely related to Hymenopteran phenui-related virus OKIAV275 (Table 1). They are represented in Figure 7A but did not show any conserved domain similar to the closest viral sequences (Supplementary Figure S4). Based on the phylogenetic analysis using the polymerase segment, this putative new virus is related to unclassified elements from the Bunyavirales order. Its closest virus was Wuhan insect virus 16 with 79% bootstrap (Figure 7A). This new virus was named Cotesiavirus chinense.
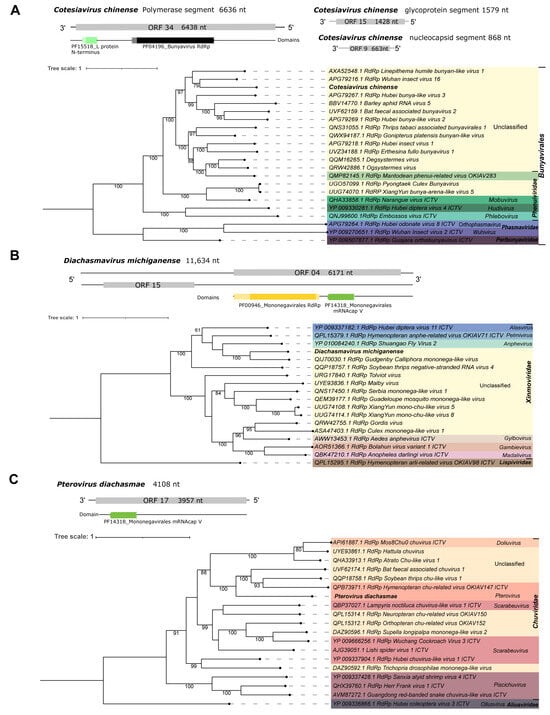
Figure 7.
Characterization of viral sequences related to elements from the Bunyavirales, Xinmoviridae, and Chuviridae families. (A) Segments, ORF pattern, conserved domains, and maximum likelihood tree of Cotesiavirus chinense. (B) ORF pattern, conserved domains, and maximum likelihood tree of Diachasmavirus michiganense. (C) ORF pattern, conserved domain, and maximum likelihood tree of Pterovirus diachasmae. The viral genomes assembled in our work are highlighted in bold. Bootstrap values under 60 are not shown.
- Diachasma alloeum
Two libraries of adult whole-body Diachasma alloeum from the United States revealed new viruses classified into at least three families: Xinmoviridae (one putative virus, 11,634 nt, repeated in two libraries), Orthomyxoviridae (five segments of one putative virus, repeated in two libraries), and Chuviridae (one putative virus, 4108 nt).
First, Da_Contig1, of 11,634 nt, presented similarity at the nucleotide level to Gudgenby Calliphora mononega-like virus, 12,763 nt (Table 1). Its best hit at the amino acid level was the RdRp of the same virus (Table 1). Due to low similarity, it is probably a new virus with the longest ORF (6171 nt|2056 aa) and domains (PF00946_Mononegavirales RdRp and PF14318_Mononegavirales mRNA-cap V) as expected through comparison with its closest aforementioned virus (Figure 7B and Supplementary Figure S4). The phylogenetic analysis pointed to this virus as an unclassified member of the Xinmoviridae family and the Mononegavirales order. Also, this virus clustered with Gudgenby Calliphora mononega-like virus with 100% bootstrap (Figure 7B). This putative new virus was named Diachasmavirus michiganense to describe its original host and geographical origin.
Second, a putative new orthomyxovirus had five segments identified through sequence similarity at the amino acid level as follows: Da_RNA_segment_2 of 2456 nt matched polymerase PB1, Hymenopteran orthomyxo-related virus OKIAV173. Da_RNA_segment_1, of 2488 nt, matched polymerase PB2, Phasmatodean orthomyxo-related virus OKIAV172; Da_RNA_segment_3, of 2235 nt, matched polymerase PA, Hymenopteran orthomyxo-related virus OKIAV173; Da_RNA_segment_4, of 1803 nt, matched putative nucleocapsid, Old quarry swamp virus; and Da_RNA_segment_5, of 1586 nt, matched hemagglutinin, Hymenopteran orthomyxo-related virus OKIAV173 (Table 1). The five segments presented expected sizes and domains (PF00602_Influenza RdRp subunit PB1, PF00604_Influenza RdRp subunit PB2, PF00603_Influenza RdRp subunit PA, and the PF03273_baculovirus gp64 envelope glycoprotein family) similar to their closest orthomyxoviruses (Figure 5A—right panel and Supplementary Figure S4). Based on the phylogenetic analysis of the PB1 segment, this virus clustered with sequences of an unclassified genus of the Orthomyxoviridae family. Also, its closest related virus was the Cotesiavirus orthomyxi already described in this work (100% bootstrap) (Figure 5B). Together, the two viruses clustered with Soybean thrips quaranja-like virus 1, although it occurred with low bootstrap (47%). This putative new virus was named Diachasmavirus orthomyxi based on its original host and viral family. It is worth noting that, we compared the two PB1 segments of Cotesiavirus orthomyxi and Diachasmavirus orthomyxi (22% query cover; e-value = 3 × 10−24; 290/446 (65%) id) using BlastN, and these sequences are unlikely from the same virus. Although, they seem to be phylogenetically close, occupying the same clade in the phylogenetic tree (Figure 5B). This may occur either due to the lower sampling of wasp viruses or because they are substantially divergent from the other viruses in the tree (Figure 5B).
Third, Da_Contig2, of 4108 nt, presented similarity at the amino acid level to the RdRp of Hymenopteran chu-related virus OKIAV147 (Table 1). Based on the phylogenetic analysis, this virus clustered with 100% bootstrap with the same virus, classified as Pterovirus, and two others unclassified at the genus level. This is a putative new Pterovirus due to the low similarity of the sequence, even though it is incomplete. The longest ORF should be 6606 nt, with Mononegavirales RdRp and mRNA capping domains, according to the best hit mentioned above (Figure 7C and Supplementary Figure S4). Here, we obtained a smaller ORF (3957 nt|1318 aa) and just the PF14318_Mononegavirales mRNA cap V domain for this viral fragment. Still, the phylogenetic tree characterized it as being from Pterovirus genus of the Chuviridae family (Figure 7C). To indicate the viral genus and original host, this new virus was named Pterovirus diachasmae.
- Ectemnius lituratus
For the single library of Ectemnius lituratus’ head and thorax (ERR6054901) included in this study, from the United Kingdom, we detected El_Contig1. This is a 12,682 nt-long sequence that showed similarity at the nucleotide level to Hymenopteran rhabdo-related virus OKIAV38, 12,376 nt. Also, it was identified at the amino acid level as the RdRp of Lasius neglectus virus 2 (Table 1). It has low similarity with the above-mentioned sequences, but the longest ORF (6522 nt|2173 aa) and domains (PF00946_Mononegavirales RdRp, PF14318_Mononegavirales mRNAcap V, and PF14314_virus-capping methyltransferase), illustrated in Figure 6A—bottom panel, which are concordant with its closest related virus, Lasius neglectus virus 2 (Supplementary Figure S4). Based on the phylogenetic analysis, this virus clustered with sequences of the Alphahymrhavirus genus of Rhabdoviridae. Its closest virus relative is Lasius neglectus virus 2, with 100% bootstrap (Figure 6B). We suggest that this is a new complete virus, named Alphahymrhavirus ectemnius due to its viral genus and original host.
- Pemphredon lugubris
The single library of the abdomen of an adult Pemphredon lugubris (ERR8571638), from the United Kingdom, revealed the largest number of new viruses for a wasp species. We detected seven putative new viruses and one known mitovirus infecting P. lugubris. The viruses were distributed among at least four families: Virgaviridae (one putative new virus sequence), Partitiviridae (one new virus sequence), Endornaviridae (three putative new virus sequences), and Mitoviridae (two new virus sequences and one known virus sequence).
Pl_Contig1, of 5813 nt, presented similarity at the amino acid level to the hypothetical protein of Megastigmus ssRNA virus (Table 1). Its longest ORF has 4350 nt|1449 aa long and two domains (PF01443_viral helicase and PF00978_RdRp 2) (Figure 4A—bottom panel). This viral sequence is incomplete since its closest virus mentioned previously has a genome of 12,061 nt with two ORFs (2985 nt and 4980 nt) and four domains (PF01660_viral methyltransferase and PF01728_FtsJ-like methyltransferase; PF01443_viral helicase and PF00978_RdRp 2, respectively) (Supplementary Figure S4). The phylogenetic analysis classified this viral sequence as a member of the Virgaviridae family and it clustered with sequences of unclassified genera. It was closely related to Megastigmus ssRNA virus, with 86% bootstrap (Figure 4B). This virus was named Pemphredonvirus anglici to indicate its original host and geographical origin.
Pl_Contig2, of 2397 nt, is a putative two-segmented new virus identified by sequence similarity at the nucleotide level as Dill cryptic virus 2 isolate IPP_hortorum segment RNA 1, complete sequence (Table 1). It has the longest ORF (2241 nt|746 aa) with a PF00680_RdRp 1 domain. The second segment of this virus (Pl_Contig2.2) is 2271 nt long and was also identified at the nucleotide level as Dill cryptic virus 2 isolate IPP_hortorum segment RNA 2, complete sequence (Table 1), with the longest ORF of 2022 nt in length, without domains (Figure 8A– bottom panel). The ORFs and RdRp domain we detected are concordant with those expected for the closest virus identified at the nucleotide level (Supplementary Figure S4). The phylogenetic analysis classified this virus as a member of the Betapartitivirus genus within the Partitiviridae family. This virus also clustered with Dill cryptic virus 2 with 100% bootstrap (Figure 8A). It was named Betapartitivirus pemphredoni.
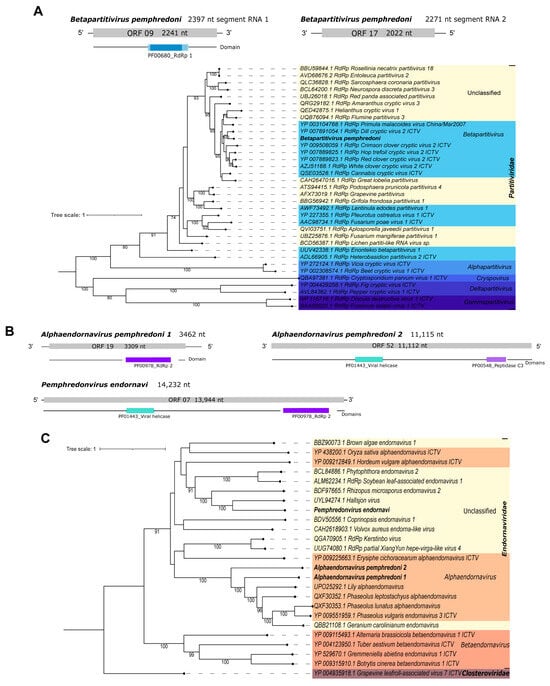
Figure 8.
Characterization of viral sequences related to elements from the Partitiviridae and Endornaviridae families. (A) Segments, ORF pattern, conserved domain, and maximum likelihood tree of Betapartitivirus pemphredoni. (B) ORF pattern and conserved domains of Alphaendornavirus pemphredoni 1 (upper left panel), Alphaendornavirus pemphredoni 2 (upper right panel), and Pemphredonvirus endornavi (lower panel). (C) Maximum likelihood tree of elements from the Endornaviridae family. The viral genomes assembled in our work are highlighted in bold. Bootstrap values under 60 are not shown.
Pl_Contig3, of 3462 nt, is a putative new virus identified by the similarity of its sequence, at the nucleotide level, to Phaseolus lunatus alphaendornavirus isolate SN35. Also, it matched at the amino acid level to Lily alphaendornavirus (Table 1). This is an incomplete viral sequence because its closest virus is 16,483 nt long. Despite this, this sequence has an ORF (3309 nt|1102 aa) that contains a PF00978_RdRp 2 domain (Figure 8B—left panel), as expected for viruses of this group (Figure 8B and Supplementary Figure S4). The phylogenetic analysis classified this virus as a member of the Alphaendornavirus genus of the Endornaviridae family. Also, this virus clustered with other sequences of the same genus with 100% bootstrap (Figure 8C). To indicate its viral genus and original host, this virus was named Alphaendornavirus pemphredoni 1.
Pl_Contig4, of 11,115 nt, did not present any related sequence at the nucleotide level. This putative new virus showed similarity at the amino acid level to Geranium carolinianum endornavirus polyprotein (Table 1). The longest ORF of this virus is 11,112 nt|3703 aa, and it contains two domains (PF01443_viral helicase and PF00548_peptidase C3) (Figure 8B—right panel). The viral sequence Pl_Contig4 is probably incomplete since the closest virus identified at the amino acid level is 14,625 nt long. However, the domains are concordant with endornaviruses (Figure 8B and Supplementary Figure S4). According to the phylogenetic analysis, this virus was classified as Alphaendornavirus within Endornaviridae (Figure 8B). This sequence clustered with other alphaendornaviruses including its closest hit, Alphaendornavirus pemphredoni 1, described above in this work, with 100% bootstrap (Figure 8B,C). The comparison of the two Alphaendornaviruses of P. lugubris at the nucleotide level did no show significant sequence similarity, suggesting that they are distinct viruses. This virus was named Alphaendornavirus pemphredoni 2.
Pl_Contig5, of 14,232 nt, had the best hit at the amino acid level with Hallsjon virus putative polyprotein (Table 1). Its longest ORF is 13,944 nt|4647 aa long, and it has two domains (PF01443_viral helicase and PF00978_RdRp 2) (Figure 8B—bottom panel). The sizes of the genome and the longest ORF are as expected for endornaviruses like Hallsjon virus (Supplementary Figure S4). However, in the phylogenetic tree, this putative new virus is an unclassified Endornaviridae member because it clustered with sequences of unclassified genera of this family (Figure 8C). In addition, this virus clustered with Hallsjon virus with 100% bootstrap. To mention its original host and viral family, this putative new virus was named Pemphredonvirus endornavi. What is noteworthy is that Pemphredonvirus endornavi has no similarity at the nucleotide level to Alphaendornavirus pemphredoni 1, and it had a very small hit with Alphaendornavirus pemphredoni 2 (0% query cover; e-value = 0.047; 23/27(85%) id). Therefore, we suggest that they are three distinct endornaviruses infecting the same wasp species.
Pl_Contig6, of 2141 nt, did not presented hits at the nucleotide level. Its best hit was at the amino acid level with the RdRp of Hangzhou altica cyanea mitovirus 1 (Table 1). Its longest ORF, translated with genetic code 4, is 1947 nt|648 aa long. Also, this ORF has a PF05919_Mitovirus RdRp domain (Figure 3A—middle panel). As expected for similar mitoviruses, like Hangzhou mitovirus 4 (its second hit in BlastX), the genome and ORF sizes agree for this group of viruses (Supplementary Figure S4). Thus, due to low similarity with the already described mitoviruses and based on the phylogenetic analysis, we suggest that this is a new mitovirus species. It clustered with sequences of unclassified genera of the Mitoviridae family and its closest virus was Hangzhou altica cyanea mitovirus 1, with 99% bootstrap (Figure 3B). To reflect the viral family and host, this putative new virus was named Mitovirus pemphredoni.
Pl_Contig7, of 1056 nt, did not present similarity at the nucleotide level with known viruses. Its best hit at the amino acid level was the RdRp of Entomophthora muscae mitovirus 2 (Table 1). Its longest ORF, translated with genetic code 4, is 1056 nt|352 aa long and it has a partial PF05919_Mitovirus RdRp domain (Figure 3A—right panel). Its closest virus, Entomophthora muscae mitovirus 2, has an ORF of 2070 nt in length and a complete domain PF05919_Mitovirus RdRp (Supplementary Figure S4). Based on the phylogenetic analysis, this putative virus clustered with sequences from the genus Unuamitovirus (Mitoviridae family) and was closely related to Entomophthora muscae mitovirus 2, with 80% bootstrap (Figure 3B). To indicate its viral genus and original host, this virus was named Unuamitovirus pemphredoni.
The comparison of Mitovirus pemphredoni 1 (2141 nt) and Unuamitovirus pemphredoni (1056 nt) showed very limited similarity at the nucleotide level with only a fraction of the sequences with similarity to each other (2% query cover; e-value = 0.010; 36/50 (72%) id). Thus, we suggest that they are two distinct species of novel mitoviruses infecting the same wasp species (P. lugubris).
Finally, Pl_Contig10, the known mitovirus of P. lugubris is a sequence of 1143 nt long and was identified at the nucleotide level as Hubei narna-like virus 25 strain SCM51430 of 2375 nt (100% query cover; e-value = 0.0; 1054/1144 (92%) id) (Table 1 and Supplementary Figure S4). The assembled sequence is likely incomplete, but it presented an ORF translated by the genetic code 4 (888 nt|295 aa), which contains a PF05919_Mitovirus RdRp domain similar to the reference virus. This virus was firstly isolated from a Dipteran host in China [].
- Telenomus podisi
The Telenomus podisi’s whole-body libraries from Brazil [] that we included in this study did not reveal new viral sequences. On the other hand, we assembled Tp_Contig1, a sequence of 8285 nt with sequence similarity to Halyomorpha halys virus isolate Beltsville, complete genome, of 9263 nt, at the nucleotide level (100% query cover; e-value = 0.0; 7351/7518 (98%) id) (Table 1 and Supplementary Figure S4). This iflavirus was isolated for the first time in 2013 from the brown marmorated stink bug, Halyomorpha halys, at Beltsville, MD, USA []. Telenomus podisi is a natural enemy of stink bug species (Hemiptera: Pentatomidae) and also occurs at Maryland []. Therefore, the identification of the same virus in prey and predator suggest this may be a case of virus transmission from ‘prey to predator’ or vice-versa.
3.3. Quantification of Viral Transcripts
In order to quantify the transcriptional activity of the virus identified in each species, we compared the raw reads from each library against viral genomes and the constitutive markers genes cytochrome b and calmodulin. The transcriptional activity of both constitutive genes was detected for all wasp libraries (Figure 9).
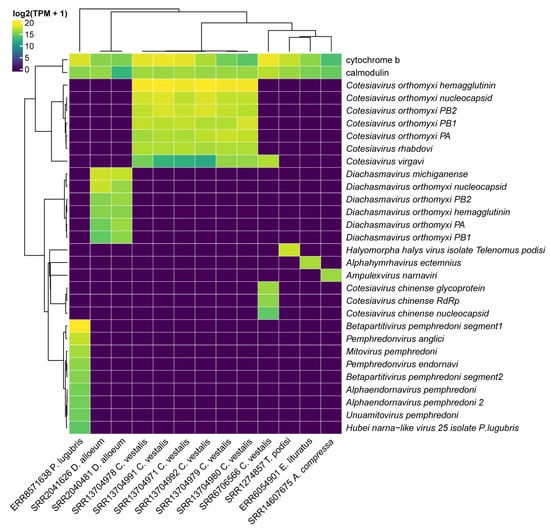
Figure 9.
Assessment of the transcriptional activity of viral sequences identified in parasitoid wasps. Abundance was estimated in transcripts per million (TPM). The viral segments are shown in rows while the RNA deep sequencing libraries and its respective wasp origin are indicated in the columns. The abundance of the constitutive genes (cytochrome b and calmodulin) are shown for each of the parasitoid wasp species.
As expected, for each parasitoid species, there is a distinct pattern of virome since different viruses were detected. The differences are likely due to the species origin. Indeed, libraries of laboratory-reared C. vestalis differ in their viruses’ composition from field-collected wasp specimens. In the first case, six libraries had Cotesiavirus rhabdovi (2536–4679 TPM), and Cotesiavirus orthomyxi segments (1712–10,800 TPM) were the most abundant viral transcripts (Figure 9). On the other hand, we observed Cotesiavirus chinense exclusively in field-collected C. vestalis samples; Cotesiavirus virgavi was detected in samples derived from both origins.
Diachasmavirus michiganense (15,235 TPM) was the most abundant virus for D. alloeum wasps, followed by Diachasmavirus orthomyxi segments, while Pterovirus diachasmae presented the lowest abundance. Alphahymrhavirus ectemnius from E. lituratus had 2902 TPM. Regarding P. lugubris, the most abundant virus was Betapartitivirus pemphredoni segment1 (38,302 TPM), followed by Pemphredonvirus anglici (7327 TPM) and Mitovirus pemphredoni (2900 TPM). Finally, Halyomorpha halys virus isolate Telenomus podisi had 5438 TPM.
Assessment of the transcriptional activity of the viral segments indicated consistency in the abundance of segments from the same viral species, such as those identified in P. lugubris (Betapartitivirus pemphredoni), C. vestalis (Cotesiavirus orthomyxi), and D. alloeum (Diachasmavirus orthomyxi), which contained viruses with segmented genomes (Figure 9).
4. Discussion
On the evolutionary history of the parasitoids, they diffused among a richness of hosts and parasitoid ecological niches, such as egg-parasitoidism, hyperparasitoidism, kleptoparasitoidism, and polyembryony []. On many occasions, this was possible due to the cooptation of the viruses to subjugate their hosts []. In addition, the probability of parasitoids expanding their geographical distribution depends on factors such as suitable host species and propitious environmental conditions (climate, host plants, host habitat, etc.) []. To illustrate, climate change may influence predator–prey relationships by modifying the behavior or distribution of the species involved []. Due to the wide geographical distribution of the selected parasitoid species (Figure 1), for instance, it is possible for wasp viruses to spread to other beneficial insects, such as pollinators [,]. Of equal importance, current RNA viruses circulating in ecologically important parasitoid wasps may synergistically affect the ecological interactions [,] of parasitoid hosts, biodiversity, ecosystem services dispensed, and the safety of using such wasps as biocontrol agents. Additionally, non-pathogenic viruses may establish mutualistic interactions with their hosts []. For those reasons, it is important to know which viruses compose the microbiome of these highly diverse ecosystem service providers. Consequently, this knowledge may help to mitigate the loss of biodiversity and unwatched viral spillover.
Here, we found 18 viruses that could be classified into 10 families (Iflaviridae, Endornaviridae, Mitoviridae, Partitiviridae, Virgaviridae, Rhabdoviridae, Chuviridae, Orthomyxoviridae, Xinmoviridae, and Narnaviridae) and in the Bunyavirales order (Figure 10). Sixteen of them likely represent novel viral species.
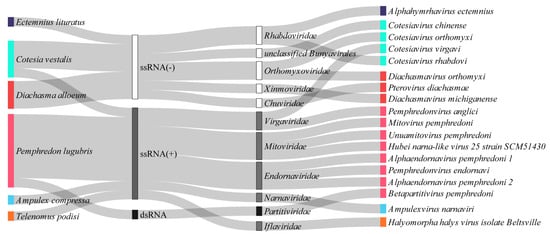
Figure 10.
Baltimore and viral family classification of the RNA viral species identified in the parasitoid wasps. Each viral sequence is represented with the same color of its source host. The viral diversity is represented by Baltimore classification: ssRNA(−) white bar, ssRNA(+) gray bar, and dsRNA black bar. Viral families are represented next to their Baltimore categories.
Viruses of Iflaviridae are non-enveloped with monopartite, positive-stranded RNA genomes of 9–11 kb, and infections can be asymptomatic or symptomatic (behavioral changes, premature mortality, and malformations) []. They have been found in Arthropoda hosts, such as honey bees, wasps, Varroa destructor [], and are transmitted mainly by the ingestion of contaminated food []. In this study, we found a previously described iflavirus Halyomorpha halys virus (TP_Contig1) in Telenomus podisi libraries from Brasília, Brazil; the original study published in 2015 contains important details to elucidate whether the virus is circulating in the parasitoid wasp. It is interesting that the study was conducted with three different species of stink bugs because in Brazil the major pests of Pentatomidae are Euchistus heros, Chinavia ubica, and Dichelops melacanthus, with them principally attacking soybean crops []. Moreover, in 2019, another study was published reporting an iflavirus of four species of stink bugs, and it was conducted with the same three pest species and libraries already published in 2015 []. Therefore, they reported the same Halyomorpha halys virus species asymptomatically circulating in the original host and the other three South American stink bugs []. Here, we detected the same virus in the parasitoid of those Pentatomidae species. It is possible that the virus passed from the prey to the parasitoid during its development in the laboratory and remained active after the adult’s eclosion. What is noteworthy is that the adult wasps obtained from laboratory colonies were maintained separated from their prey and nourished with pure honey and, for reproduction, they received E. heros eggs to parasitize []. Since 20-day-old wasps were used for RNA extraction, it is unlikely that the virus originated from E. heros’ egg remnants. Therefore, the prey’s virus passed to its parasitoid wasp previously and is probably circulating in T. podisi as well.
Three new putative viruses of Endornaviridae (Alphaendornavirus pemphredoni 1, Pemphredonvirus endornavi, and Alphaendornavirus pemphredoni 2) were identified in P. lugubris. As stated by the ICTV, Endornaviruses are positive-sense, single-stranded RNA viruses with genomes of 9.7 to 17.6 kb. They have been found in fungi, oomycetes, and plants []. However, recently, two Alphaendornaviruses, a strain of Hallsjon virus, firstly described in Culex torrentium [], and a novel virus named Tvarminne alphaendornavirus [] were detected in mosquitoes from Finland. Here, Pemphredonvirus endornavi had sequence similarity at the amino acid level to Hallsjon virus, but with low identity.
Pl_Contig10, one previously described mitovirus, and two new putative mitoviruses (Mitovirus pemphredoni and Unuamitovirus pemphredoni) were detected in P. lugubris. Mitoviridae viruses are RNA viruses with positive-sense, single-stranded, adenine–uracil (AU) rich genomes. These are capsidless virusesand their genomes range from 2151 to 4955 nt, with one ORF coding an RdRp domain that ranges from 636 to 1137 aa []. Thus far, several putative mitoviruses have been described from a diversity of fungi and Plantae species [,], and a recently published study found evidence to enlarge the host possibilities beyond them. Based on phylogenetic reconstruction, Jacquat and cols (2022) suggest the existence of a lineage of mitoviruses derived from animals because the putative mitoviruses do not cluster with species of fungal origin []. Mitovirus pemphredoni and Unuamitovirus pemphredoni clustered with other sequences from several sequences from fungus and plant viruses (Figure 3). Thus, it is not possible to ensure whether they are replicating inside the wasp’s mitochondria or are derived from an external source. through contamination. Interestingly, Unuamitovirus pemphredoni clustered with 80% bootstrap with Entomophthora muscae mitovirus 2 (Figure 3B), a virus from an entomopathogenic fungus that infects, manipulates, and kills its Dipteran hosts to favor its dispersal through spores []. Entomophthoraceae appeared in our metagenomics analysis (Figure 2), pointing out that P. lugubris may be a new host for it. What is noteworthy is that the fungi of this family have already been detected in aphid pests in Argentina [], the main prey of this parasitoid wasp. More studies are needed to elucidate if such mitoviruses contribute somehow to Entomophthora muscae’s successful parasitoid strategy. Equally, further analysis should be performed to confirm the relationship between Entomophthoraceae species and P. lugubris.
Another family related to mitoviruses is the Narnaviridae family, which contains non-encapsidated positive-stranded RNA viruses that range from 2.3 to 3.6 kb []. We found in our analysis that Ampulexvirus narnaviri clustered with Xiangshan narna-like virus, which was obtained from a mixed pool of several species of insects []. It was not possible to classify this species at the genera level since all of the closest species are also defined as unclassified elements within the Narnaviridae family (Figure 3B).
P. lugubris also had two segments of a Betapartitivirus of the Partitiviridae. As maintained by the ICTV, this genus has viruses from plants or fungi. Partitivirus are small and non-enveloped with two segments of double-stranded RNA genomes that range from 3 to 4.8 kb []. Metagenomic studies have shown that partitiviruses are also common in insects such as flies and mosquitoes [,]. In addition, a Partiti-like virus was found in honey bees of several apiaries across the USA, probably causing a mild or asymptomatic infection [].
Virgaviridae viruses have a positive-sense, single-stranded RNA genome of 6.3 to 13 kb in length. There are non-segmented and segmented members within this family. They are non-enveloped viruses present in plants []. However, studies have related their occurrence in insects [,]. Here, we found two members of unclassified genera from Virgaviridae. Interestingly, Cotesiavirus virgavi and Pemphredonvirus anglici clustered with Megastigmus ssRNA virus (Figure 4), a virus isolated from Megastigmus spermotrophus (Hymenoptera: Torymidae), a North-American seed parasitoid wasp present in Europe [].
Rhabdoviridae members are viruses with negative-sense RNA genomes of 10 to 16 kb. As stated by the ICTV, they can infect a wide range of hosts, including vertebrate animals, plants, and arthropods. Further, several rhabdoviruses are transmitted by arthropods and may be pathogenic to humans, livestock, fish, and farm crops []. In this study, two novel rhabdoviruses were identified. Cotesiavirus rhabdovi clustered with Wuhan Ant Virus, firstly described in China [], and isolated from Camponotus japonicus, an ant native to eastern Asia. The C. vestalis’ libraries we have studied are from the same country. Similarly, Alphahymrhavirus ectemnius clustered with a virus of an ant host, Lasius neglectus virus 2, isolated from Lasius neglectus sampled from Cambridge, UK, once again the same country of the library’s origin [].
Chuviridae is a latterly described family of negative-sense single-stranded RNA viruses that infect various arthropods, such as mosquitoes []. Here, we detected Pterovirus diachasmae that clustered with three other viral sequences from: Aphelinus abdominalis (Hymenopteran chu-related virus OKIAV147), soybean thrips or Neohydatothrips variabilis (Soybean thrips chu-like virus 1), and a bat (Bat faecal associated chuvirus 1). Aphelinus abdominalis is an aphid parasitoid wasp used for the biocontrol of lettuce crops []. Soybean thrips are worrying agricultural pests and also vectors of diverse plants’ viruses [].
According to the ICTV, Orthomyxoviridae and Xinmoviridae are families of negative-sense single-stranded RNA viruses that have been found in insects [,]. In particular, orthomyxoviruses are constituted by 6 to 8 segments and have been isolated from insect pollinators in China and Korea [,]. Here, we detected two putative orthomyxoviruses composed by five segments (Cotesiavirus orthomyxi and Diachasmavirus orthomyxi) that clustered together. Regarding xinmoviruses, we described Diachasmavirus michiganense, a putative Xinmoviridae virus that clustered with Gudgenby Calliphora mononega-like virus, which was isolated from ectoparasites (blowflies) of rabbits [].
Viruses of the Bunyavirales order have been described in many hosts (such as arthropods, plants, and mammals), since this is the most abundant RNA virus order with eight families []. In this study, we found a member of this order, named Cotesiavirus chinense, which clustered with Wuhan insect virus 16.
More studies on the virome of parasitoid wasps are needed to clarify the prevalence of the new viruses identified here at the population or species level. Identifying which viruses are present in the prey may indicate a possible viral origin. Further investigations looking for viruses in RNA-seq from different species, locations, tissues, and life stages of insects, whether predators or prey, will indicate whether the viruses establish infection or are only obtained mechanically or by contamination, without replication in more than one host. As there are still few studies covering this theme, it is not possible to discuss the scope of these viruses in depth. However, simply increasing the number of viral sequences described from parasitoid wasps will certainly facilitate the identification and classification of viral agents, favoring the development of more applied studies on this subject in the future.
To sum up, parasitoid wasps are the best adapted insects and are highly diverse, with more than 50% of all known Hymenopteran species classified within this group []. Furthermore, there is a pressing need for new methods of biological control to reduce pesticides’ use. In conclusion, a great diversity of associated viruses was registered among the parasitoid wasps analyzed in this study. Therefore, parasitoids and their viruses should be part of the research focus for the following reasons: maximizing the use of parasitoid wasps to control agricultural pests; applying wasps to fields without putting the survival of native insect species at risk (such as pollinators) by inadvertent viral spread; promoting species conservation; and increasing knowledge regarding insect viruses and their ecological and/or evolutionary relationships.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v15122448/s1. Figure S1: Geographical distribution of the libraries used in the study classified by wasp species; Figure S2: Metagenomics analysis grouped by species of parasitoid wasps; Figure S3: Metagenomics analysis for each RNA-seq library included in the study; Figure S4: Characterization of segments, ORFs, and domains of sequences closely related to the assembled viral transcripts identified in the work; Table S1: Overview of RNA-deep sequenced libraries used in the work. Table S2: Detailed result of sequence similarity searches of transcripts related to viral species. Table S3: Description of the parameters applied in the phylogenetic analyses.
Author Contributions
G.B.C.-G.: conceptualization, methodology, validation, formal analysis, investigation, data curation, writing—original draft preparation, writing—review and editing, and picture designer. V.C.S.: methodology, validation, formal analysis, investigation, data curation, and picture designer. P.L.C.F.: methodology, validation, formal analysis, and writing—review. J.P.P.d.A.: formal analysis, data curation, validation, writing—review, and picture designer. M.A.C.: conceptualization, validation, and writing—review and editing. E.R.G.R.A.: conceptualization, resources, supervision, data curation, validation, writing—original draft preparation, writing—review and editing, and picture designer. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-CAPES-(Brazil), financial code 001, which provides Ph.D. scholarships to students of the Programa de Pós-Graduação em Genética e Biologia Molecular (PPGGBM) at the Universidade Estadual de Santa Cruz. M.A.C. and E.R.G.R.A. are research fellows from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The nucleotide sequence data reported in this work are available from the Third Party Annotation Section of the DDBJ/ENA/GenBank databases under the accession numbers TPA: BK063681-BK063709.
Acknowledgments
We thank all members of the Virus Bioinformatics Laboratory–UESC for the gratifying discussions.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Purkiss, T.; Lach, L. Pathogen spillover from Apis mellifera to a stingless bee. Proc. R. Soc. B Boil. Sci. 2019, 286, 20191071. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Lauber, C. Bioinformatics of virus taxonomy: Foundations and tools for developing sequence-based hierarchical classification. Curr. Opin. Virol. 2022, 52, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-X.; Shi, M.; Tian, J.-H.; Lin, X.-D.; Kang, Y.-J.; Chen, L.-J.; Qin, X.-C.; Xu, J.; Holmes, E.C.; Zhang, Y.-Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 2015, 4, e05378. [Google Scholar] [CrossRef] [PubMed]
- Herniou, E.A.; Huguet, E.; Thézé, J.; Bézier, A.; Periquet, G.; Drezen, J.-M. When parasitic wasps hijacked viruses: Genomic and functional evolution of polydnaviruses. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20130051. [Google Scholar] [CrossRef] [PubMed]
- Blomström, A.-L. Viral metagenomics as an emerging and powerful tool in veterinary medicine. Vet. Q. 2011, 31, 107–114. [Google Scholar] [CrossRef]
- Käfer, S.; Paraskevopoulou, S.; Zirkel, F.; Wieseke, N.; Donath, A.; Petersen, M.; Jones, T.C.; Liu, S.; Zhou, X.; Middendorf, M.; et al. Re-assessing the diversity of negative strand RNA viruses in insects. PLoS Pathog. 2019, 15, e1008224. [Google Scholar] [CrossRef]
- Stork, N.E. How Many Species of Insects and Other Terrestrial Arthropods Are There on Earth? Annu. Rev. Entomol. 2018, 63, 31–45. [Google Scholar] [CrossRef]
- Ballal, C.R.; Verghese, A. Role of Parasitoids and Predators in the Management of Insect Pests. In New Horizons in Insect Science: Towards Sustainable Pest Management; Chakravarthy, A.K., Ed.; Springer: New Delhi, India, 2015. [Google Scholar] [CrossRef]
- Hiroyoshi, S.; Harvey, J.A.; Nakamatsu, Y.; Nemoto, H.; Mitsuhashi, J.; Mitsunaga, T.; Tanaka, T. Potential Host Range of the Larval Endoparasitoid Cotesia vestalis (=plutellae) (Hymenoptera: Braconidae). Int. J. Insect Sci. 2017, 9, 1179543317715623. [Google Scholar] [CrossRef]
- Brock, R.E.; Cini, A.; Sumner, S. Ecosystem services provided by aculeate wasps. Biol. Rev. 2021, 96, 1645–1675. [Google Scholar] [CrossRef]
- Raven, P.H.; Wagner, D.L. Agricultural intensification and climate change are rapidly decreasing insect biodiversity. Proc. Natl. Acad. Sci. USA 2021, 118, e2002548117. [Google Scholar] [CrossRef]
- Forister, M.L.; Pelton, E.M.; Black, S.H. Declines in insect abundance and diversity: We know enough to act now. Conserv. Sci. Pract. 2019, 1, e80. [Google Scholar] [CrossRef]
- Paukkunen, J.; Pöyry, J.; Kuussaari, M. Species traits explain long-term population trends of Finnish cuckoo wasps (Hymenoptera: Chrysididae). Insect Conserv. Divers. 2017, 11, 58–71. [Google Scholar] [CrossRef]
- Sánchez-Bayo, F.; Wyckhuys, K.A. Worldwide decline of the entomofauna: A review of its drivers. Biol. Conserv. 2019, 232, 8–27. [Google Scholar] [CrossRef]
- Bohart, R.M.; Menke, A.S. Sphecid Wasps of the World; University of California Press: Oakland, CA, USA, 1976; No. 1. [Google Scholar] [CrossRef]
- Arvidson, R.; Landa, V.; Frankenberg, S.; Adams, M.E. Life History of the Emerald Jewel Wasp Ampulex compressa. J. Hymenopt. Res. 2018, 63, 1–13. [Google Scholar] [CrossRef]
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback Moth Ecology and Management: Problems, Progress, and Prospects. Annu. Rev. Èntomol. 2013, 58, 517–541. [Google Scholar] [CrossRef]
- Forbes, A.A.; Powell, T.H.Q.; Lobo, N.F.; Noor, M.A.F.; Feder, J.L. Permanent genetic resources: Polymorphic microsatellite loci for Diachasma alloeum (Hymenoptera: Braconidae). Mol. Ecol. Resour. 2008, 8, 373–376. [Google Scholar] [CrossRef]
- Bees Wasps & Ants Recording Society, “Ectemnius lituratus”. 2014. Available online: https://www.bwars.com/wasp/crabronidae/crabroninae/ectemnius-lituratus (accessed on 11 February 2023).
- Bees Wasps & Ants Recording Society, “Pemphredon lugubris”. Available online: https://www.bwars.com/wasp/crabronidae/inae/pemphredon-lugubris (accessed on 11 February 2023).
- Nalam, V.; Louis, J.; Shah, J. Plant defense against aphids, the pest extraordinaire. Plant Sci. 2018, 279, 96–107. [Google Scholar] [CrossRef]
- Schmidt, M.H.; Lauer, A.; Purtauf, T.; Thies, C.; Schaefer, M.; Tscharntke, T. Relative importance of predators and parasitoids for cereal aphid control. Proc. R. Soc. B Biol. Sci. 2003, 270, 1905–1909. [Google Scholar] [CrossRef]
- Borges, M.; Colazza, S.; Ramirez-Lucas, P.; Chauhan, K.R.; Moraes, M.C.B.; Aldrich, J.R. Kairomonal effect of walking traces from Euschistus heros (Heteroptera: Pentatomidae) on two strains of Telenomus podisi (Hymenoptera: Scelionidae). Physiol. Èntomol. 2003, 28, 349–355. [Google Scholar] [CrossRef]
- Parra, J.R.P. Controle Biológico na Agricultura Brasileira. Entomol. Commun. 2019, 1, ec01002. [Google Scholar] [CrossRef]
- Weber, I.D.; Garcia, A.G.; Bueno, A.d.F.; de Oliveira, R.C.; Godoy, W.A.C. Release strategies of Telenomus podisi for control of Euschistus heros: A computational modeling approach. Pest Manag. Sci. 2022, 78, 4544–4556. [Google Scholar] [CrossRef] [PubMed]
- Lüthi, M.N.; Vorburger, C.; Dennis, A.B. A Novel RNA Virus in the Parasitoid Wasp Lysiphlebus fabarum: Genomic Structure, Prevalence, and Transmission. Viruses 2020, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Izraeli, Y.; Lepetit, D.; Atias, S.; Mozes-Daube, N.; Wodowski, G.; Lachman, O.; Luria, N.; Steinberg, S.; Varaldi, J.; Zchori-Fein, E.; et al. Genomic characterization of viruses associated with the parasitoid Anagyrus vladimiri (Hymenoptera: Encyrtidae). J. Gen. Virol. 2022, 103, 001810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, F.; Yuan, B.; Yang, L.; Yang, Y.; Fang, Q.; Kuhn, J.H.; Song, Q.; Ye, G. A novel cripavirus of an ectoparasitoid wasp increases pupal duration and fecundity of the wasp’s Drosophila melanogaster host. ISME J. 2021, 15, 3239–3257. [Google Scholar] [CrossRef]
- Wang, F.; Yuan, B.; Xiao, S.; Zhang, J.; Jia, W.; Fang, Q.; Wang, F.; Song, Q.; Ye, G. Diverse RNA Viruses Discovered in Three Parasitoid Wasps of the Rice Weevil Sitophilus oryzae. mSphere 2021, 6, e00331-21. [Google Scholar] [CrossRef]
- Wang, F.; Fang, Q.; Wang, B.; Yan, Z.; Hong, J.; Bao, Y.; Kuhn, J.H.; Werren, J.H.; Song, Q.; Ye, G. A novel negative-stranded RNA virus mediates sex ratio in its parasitoid host. PLoS Pathog. 2017, 13, e1006201. [Google Scholar] [CrossRef]
- Gauthier, J.; Drezen, J.-M.; Herniou, E.A. The recurrent domestication of viruses: Major evolutionary transitions in parasitic wasps. Parasitology 2017, 145, 713–723. [Google Scholar] [CrossRef]
- Drezen, J.-M.; Bézier, A.; Burke, G.R.; Strand, M.R. Bracoviruses, ichnoviruses, and virus-like particles from parasitoid wasps retain many features of their virus ancestors. Curr. Opin. Insect Sci. 2021, 49, 93–100. [Google Scholar] [CrossRef]
- Bézier, A.; Annaheim, M.; Herbinière, J.; Wetterwald, C.; Gyapay, G.; Bernard-Samain, S.; Wincker, P.; Roditi, I.; Heller, M.; Belghazi, M.; et al. Polydnaviruses of Braconid Wasps Derive from an Ancestral Nudivirus. Science 2009, 323, 926–930. [Google Scholar] [CrossRef]
- Burke, G.R.; Hines, H.M.; Sharanowski, B.J. The Presence of Ancient Core Genes Reveals Endogenization from Diverse Viral Ancestors in Parasitoid Wasps. Genome Biol. Evol. 2021, 13, evab105. [Google Scholar] [CrossRef]
- Legeai, F.; Santos, B.F.; Robin, S.; Bretaudeau, A.; Dikow, R.B.; Lemaitre, C.; Jouan, V.; Ravallec, M.; Drezen, J.-M.; Tagu, D.; et al. Genomic architecture of endogenous ichnoviruses reveals distinct evolutionary pathways leading to virus domestication in parasitic wasps. BMC Biol. 2020, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Santos, B.F.; Klopfstein, S.; Whitfield, J.B.; Sharanowski, B.J. Many evolutionary roads led to virus domestication in ichneumonoid parasitoid wasps. Curr. Opin. Insect Sci. 2021, 50, 100861. [Google Scholar] [CrossRef] [PubMed]
- Coffman, K.A.; Harrell, T.C.; Burke, G.R. A Mutualistic Poxvirus Exhibits Convergent Evolution with Other Heritable Viruses in Parasitoid Wasps. J. Virol. 2020, 94, e02059-19. [Google Scholar] [CrossRef]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020, 48, W395–W402. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraha.ac.uk/projects/fastqc (accessed on 1 October 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Shen, W.; Ren, H. TaxonKit: A practical and efficient NCBI taxonomy toolkit. J. Genet. Genom. 2021, 48, 844–850. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2015, 32, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.H.; Zerbino, D.R.; Vingron, M.; Birney, E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012, 28, 1086–1092. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Espinal, R.B.A.; de Santana, S.F.; Santos, V.C.; Lizardo, G.N.R.; Silva, R.J.S.; Corrêa, R.X.; Loguercio, L.L.; Góes-Neto, A.; Pirovani, C.P.; Fonseca, P.L.C.; et al. Uncovering a Complex Virome Associated with the Cacao Pathogens Ceratocystis cacaofunesta and Ceratocystis fimbriata. Pathogens 2023, 12, 287. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2019, 37, 291–294. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.4: Tree Figure Drawing Tool. GitHub. 2019. Available online: https://github.com/rambaut/figtree/releases (accessed on 1 October 2022).
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Postler, T.S.; Rubino, L.; Adriaenssens, E.M.; Dutilh, B.E.; Harrach, B.; Junglen, S.; Kropinski, A.M.; Krupovic, M.; Wada, J.; Crane, A.; et al. Guidance for creating individual and batch latinized binomial virus species names. J. Gen. Virol. 2022, 103, 001800. [Google Scholar] [CrossRef]
- Lin, D.A.; Roychoudhury, S.; Palese, P.; Clay, W.C.; Fuller, F.J. Evolutionary relatedness of the predicted gene product of RNA segment 2 of the Tick-Borne Dhori virus and the PB1 polymerase gene of influenza viruses. Virology 1991, 182, 1–7. [Google Scholar] [CrossRef]
- Batts, W.N.; LaPatra, S.E.; Katona, R.; Leis, E.; Ng, T.F.F.; Brieuc, M.S.; Breyta, R.B.; Purcell, M.K.; Conway, C.M.; Waltzek, T.B.; et al. Molecular characterization of a novel orthomyxovirus from rainbow and steelhead trout (Oncorhynchus mykiss). Virus Res. 2017, 230, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.B.; Ballard, J.R.; Tesh, R.B.; Brown, J.D.; Ruder, M.G.; Keel, M.K.; Munk, B.A.; Mickley, R.M.; Gibbs, S.E.J.; da Rosa, A.P.A.T.; et al. Cyclic Avian Mass Mortality in the Northeastern United States Is Associated with a Novel Orthomyxovirus. J. Virol. 2015, 89, 1389–1403. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Farias, L.R.; Schimmelpfeng, P.H.C.; Togawa, R.C.; Costa, M.M.C.; Grynberg, P.; Martins, N.F.; Borges, M.; Blassioli-Moraes, M.C.; Laumann, R.A.; Báo, S.N.; et al. Transcriptome-Based Identification of Highly Similar Odorant-Binding Proteins among Neotropical Stink Bugs and Their Egg Parasitoid. PLoS ONE 2015, 10, e0132286. [Google Scholar] [CrossRef]
- Sparks, M.E.; Gundersen-Rindal, D.E.; Harrison, R.L. Complete Genome Sequence of a Novel Iflavirus from the Transcriptome of Halyomorpha halys, the Brown Marmorated Stink Bug. Genome Announc. 2013, 1, e00910-13. [Google Scholar] [CrossRef]
- Polaszek, A.; Vilhemsen, L. Biodiversity of hymenopteran parasitoids. Curr. Opin. Insect Sci. 2023, 56, 101026. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, H.M.; Vanlaerhoven, S.L.; García, M.M.; Hunt, D.W. Food web associations and effect of trophic resources and environmental factors on parasitoids expanding their host range into non-native hosts. Entomol. Exp. Appl. 2018, 166, 277–288. [Google Scholar] [CrossRef]
- Laws, A.N. Climate change effects on predator–prey interactions. Curr. Opin. Insect Sci. 2017, 23, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Manley, R.; Boots, M.; Wilfert, L. REVIEW: Emerging viral disease risk to pollinating insects: Ecological, evolutionary and anthropogenic factors. J. Appl. Ecol. 2015, 52, 331–340. [Google Scholar] [CrossRef]
- Highfield, A.; Kevill, J.; Mordecai, G.; Hunt, J.; Henderson, S.; Sauvard, D.; Feltwell, J.; Martin, S.J.; Sumner, S.; Schroeder, D.C. Detection and Replication of Moku Virus in Honey Bees and Social Wasps. Viruses 2020, 12, 607. [Google Scholar] [CrossRef]
- French, R.K.; Holmes, E.C. An Ecosystems Perspective on Virus Evolution and Emergence. Trends Microbiol. 2019, 28, 165–175. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Chen, Y.-M.; Wang, W.; Qin, X.-C.; Holmes, E.C. Expanding the RNA Virosphere by Unbiased Metagenomics. Annu. Rev. Virol. 2019, 6, 119–139. [Google Scholar] [CrossRef]
- Pradeu, T. Mutualistic viruses and the heteronomy of life. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2016, 59, 80–88. [Google Scholar] [CrossRef]
- Valles, S.M.; Chen, Y.; Firth, A.E.; Guérin, D.M.A.; Hashimoto, Y.; Herrero, S.; De Miranda, J.R.; Ryabov, E.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Iflaviridae. J. Gen. Virol. 2017, 98, 527–528. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Brettell, L.E.; Pachori, P.; Villalobos, E.M.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Moku virus; a new Iflavirus found in wasps, honey bees and Varroa. Sci. Rep. 2016, 6, 34983. [Google Scholar] [CrossRef]
- dos Santos, E.R.; Trentin, L.B.; Ecker, A.; Silva, L.A.; Borges, M.; Mowery, J.D.; Ribeiro, B.M.; Harrison, R.L.; Ardisson-Araújo, D.M. An iflavirus found in stink bugs (Hemiptera: Pentatomidae) of four different species. Virology 2019, 534, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.A.; Khalifa, M.; Okada, R.; Fukuhara, T.; Sabanadzovic, S.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Endornaviridae. J. Gen. Virol. 2019, 100, 1204–1205. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, J.H.-O.; Shi, M.; Eden, J.-S.; Holmes, E.C.; Hesson, J.C. Meta-Transcriptomic Comparison of the RNA Viromes of the Mosquito Vectors Culex pipiens and Culex torrentium in Northern Europe. Viruses 2019, 11, 1033. [Google Scholar] [CrossRef]
- Nguyen, P.T.T.; Culverwell, C.L.; Suvanto, M.T.; Korhonen, E.M.; Uusitalo, R.; Vapalahti, O.; Smura, T.; Huhtamo, E. Characterisation of the RNA Virome of Nine Ochlerotatus Species in Finland. Viruses 2022, 14, 1489. [Google Scholar] [CrossRef]
- Jacquat, A.G.; Ulla, S.B.; Debat, H.J.; Muñoz-Adalia, E.J.; Theumer, M.G.; Pedrajas, M.D.G.; Dambolena, J.S. An in silico analysis revealed a novel evolutionary lineage of putative mitoviruses. Environ. Microbiol. 2022, 24, 6463–6475. [Google Scholar] [CrossRef] [PubMed]
- Bruenn, J.A.; Warner, B.E.; Yerramsetty, P. Widespread mitovirus sequences in plant genomes. PeerJ 2015, 3, e876. [Google Scholar] [CrossRef]
- Myers, J.M.; Bonds, A.E.; Clemons, R.A.; Thapa, N.A.; Simmons, D.R.; Carter-House, D.; Ortanez, J.; Liu, P.; Miralles-Durán, A.; Desirò, A.; et al. Survey of Early-Diverging Lineages of Fungi Reveals Abundant and Diverse Mycoviruses. mBio 2020, 11, e02027-20. [Google Scholar] [CrossRef]
- Nibert, M.L.; Debat, H.J.; Manny, A.R.; Grigoriev, I.V.; Licht, H.H.D.F. Mitovirus and Mitochondrial Coding Sequences from Basal Fungus Entomophthora muscae. Viruses 2019, 11, 351. [Google Scholar] [CrossRef]
- Manfrino, R.G.; Zumoffen, L.; Salto, C.E.; Lastra, C.C.L. Natural occurrence of entomophthoroid fungi of aphid pests on Medicago sativa L. in Argentina. Rev. Argent. De Microbiol. 2014, 46, 49–52. [Google Scholar] [CrossRef]
- Hillman, B.I.; Cai, G. The Family Narnaviridae. In Advances in Virus Research, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; Volume 86, pp. 149–176. [Google Scholar] [CrossRef]
- Li, N.; Huang, Y.; Li, W.; Xu, S. Virome Analysis Reveals Diverse and Divergent RNA Viruses in Wild Insect Pollinators in Beijing, China. Viruses 2022, 14, 227. [Google Scholar] [CrossRef]
- Vainio, E.J.; Chiba, S.; Ghabrial, S.A.; Maiss, E.; Roossinck, M.; Sabanadzovic, S.; Suzuki, N.; Xie, J.; Nibert, M. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Partitiviridae. J. Gen. Virol. 2018, 99, 17–18. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.T.; Maertens, B.L.; Dunham, T.J.; Rodgers, C.P.; Brehm, A.L.; Miller, M.R.; Williams, A.M.; Foy, B.D.; Stenglein, M.D. Partitiviruses Infecting Drosophila melanogaster and Aedes aegypti Exhibit Efficient Biparental Vertical Transmission. J. Virol. 2020, 94, e01070-20. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.M.; Lopez, D.L.; Martinez, J.F.I.; Galbraith, D.A.; Rose, R.; VAN Engelsdorp, D.; Rosa, C.; Evans, J.D.; Grozinger, C.M. Distribution of recently identified bee-infecting viruses in managed honey bee (Apis mellifera) populations in the USA. Apidologie 2020, 51, 736–745. [Google Scholar] [CrossRef]
- Adams, M.J.; Adkins, S.; Bragard, C.; Gilmer, D.; Li, D.; MacFarlane, S.A.; Wong, S.-M.; Melcher, U.; Ratti, C.; Ryu, K.H.; et al. ICTV Virus Taxonomy Profile: Virgaviridae. J. Gen. Virol. 2017, 98, 1999–2000. [Google Scholar] [CrossRef]
- Kondo, H.; Chiba, S.; Maruyama, K.; Andika, I.B.; Suzuki, N. A novel insect-infecting virga/nege-like virus group and its pervasive endogenization into insect genomes. Virus Res. 2019, 262, 37–47. [Google Scholar] [CrossRef]
- Mailleux, A.-C.; Roques, A.; Molenberg, J.-M.; Grégoire, J.-C. A North American invasive seed pest, Megastigmus spermotrophus (Wachtl) (Hymenoptera: Torymidae): Its populations and parasitoids in a European introduction zone. Biol. Control 2008, 44, 137–141. [Google Scholar] [CrossRef]
- Walker, P.J.; Freitas-Astúa, J.; Bejerman, N.; Blasdell, K.R.; Breyta, R.; Dietzgen, R.G.; Fooks, A.R.; Kondo, H.; Kurath, G.; Kuzmin, I.V.; et al. ICTV Virus Taxonomy Profile: Rhabdoviridae 2022. J. Gen. Virol. 2022, 103, 001689. [Google Scholar] [CrossRef]
- Kleanthous, E.; Olendraite, I.; Lukhovitskaya, N.I.; Firth, A.E. Discovery of three RNA viruses using ant transcriptomic datasets. Arch. Virol. 2018, 164, 643–647. [Google Scholar] [CrossRef]
- Shrestha, G.; Skovgård, H.; Reddy, G.V.P.; Steenberg, T.; Enkegaard, A. Role of the aphid species and their feeding locations in parasitization behavior of Aphelinus abdominalis, a parasitoid of the lettuce aphid Nasonovia ribisnigri. PLoS ONE 2017, 12, e0184080. [Google Scholar] [CrossRef][Green Version]
- Thekke-Veetil, T.; Lagos-Kutz, D.; McCoppin, N.K.; Hartman, G.L.; Ju, H.-K.; Lim, H.-S.; Domier, L.L. Soybean Thrips (Thysanoptera: Thripidae) Harbor Highly Diverse Populations of Arthropod, Fungal and Plant Viruses. Viruses 2020, 12, 1376. [Google Scholar] [CrossRef]
- Wu, H.; Pang, R.; Cheng, T.; Xue, L.; Zeng, H.; Lei, T.; Chen, M.; Wu, S.; Ding, Y.; Zhang, J.; et al. Abundant and Diverse RNA Viruses in Insects Revealed by RNA-Seq Analysis: Ecological and Evolutionary Implications. mSystems 2020, 5, e00039-20. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Jung, C.; Kil, E.-J. Metagenomic analysis of viromes in honey bee colonies (Apis mellifera; Hymenoptera: Apidae) after mass disappearance in Korea. Front. Cell. Infect. Microbiol. 2023, 13, 1124596. [Google Scholar] [CrossRef] [PubMed]
- Mahar, J.E.; Shi, M.; Hall, R.N.; Strive, T.; Holmes, E.C. Comparative Analysis of RNA Virome Composition in Rabbits and Associated Ectoparasites. J. Virol. 2020, 94, e02119-19. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Zhang, W.; Ye, C.; Smagghe, G.; Wang, J.; Niu, J. Discovery of a widespread presence bunyavirus that may have symbiont-like relationships with different species of aphids. Insect Sci. 2021, 29, 1120–1134. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).