Abstract
The Picornaviridae family comprises a large group of non-enveloped viruses with enormous impact on human and animal health. The picornaviral genome contains one open reading frame encoding a single polyprotein that can be processed by viral proteases. The picornaviral 3C proteases share similar three-dimensional structures and play a significant role in the viral life cycle and virus–host interactions. Picornaviral 3C proteins also have conserved RNA-binding activities that contribute to the assembly of the viral RNA replication complex. The 3C protease is important for regulating the host cell response through the cleavage of critical host cell proteins, acting to selectively ‘hijack’ host factors involved in gene expression, promoting picornavirus replication, and inactivating key factors in innate immunity signaling pathways. The protease and RNA-binding activities of 3C are involved in viral polyprotein processing and the initiation of viral RNA synthesis. Most importantly, 3C modifies critical molecules in host organelles and maintains virus infection by subtly subverting host cell death through the blocking of transcription, translation, and nucleocytoplasmic trafficking to modulate cell physiology for viral replication. Here, we discuss the molecular mechanisms through which 3C mediates physiological processes involved in promoting virus infection, replication, and release.
1. Introduction
The positive-strand RNA viruses from Picornaviridae have tremendous medical, veterinary, and agricultural importance [1,2,3,4]. These viruses affect the digestive and respiratory systems, skin, liver, and heart in humans and animals. This family of monocistronic positive-strand RNA viruses consists of 63 genera and 285 different virus species, including enterovirus, rhinovirus, hepatovirus, and cardiovirus [5].
The picornaviral virion and genome structures are highly conserved in nature. The genome consists of one open reading frame coding for a single polyprotein that can be targeted by viral proteases [3,6,7,8]. The P1 region encodes for the capsid proteins of the virus, while the P2 and P3 portions encode the non-structural proteins (Figure 1). The genome incorporates an open reading frame (ORF), a 5′ untranslated region (5′ UTR), and a 3′ untranslated region (3′ UTR) [9,10,11,12]. There is a polyadenylated (poly(A)) tail in the 3′ UTR. These viruses are non-enveloped and confine single-stranded RNA (length: 7–10 kb) with an icosahedral symmetric structure, having a diameter of about 30 nm (Figure 1) [13,14]. A covalently attached viral nucleotidylylated protein that caps a genome (VPg or, 3B) is present at the 5′ end of 5′ UTR [9]. The RNA genome carries an internal ribosome entry site (IRES) segment within the 5′ UTR to direct viral protein synthesis while employing ribosomes and other host factors [15,16].
The monocistronic single-strand RNA encodes only one polyprotein that is proteolytically cleaved by virally encoded proteases into several mature proteins, varying upon the genus [17,18]. The N-terminal region of the polyprotein is joined to a leader protein (L) in some viruses from various genera, including cardiovirus and aphthovirus [19,20,21,22]. The co- and post-translation processing of the polyprotein results in the expression of structural proteins 1A to 1D (VP4, VP2, VP3 and VP1), non-structural proteins (2A, 2B, 2C, 3A, 3B, 3C, 3D), or their intermediates (e.g., 3ABCD, 3ABC, 3BCD and 3CD) [1,23,24]. The 3C or its 3CD precursor are predominantly responsible for the proteolytic transformation of the polyprotein [25,26].
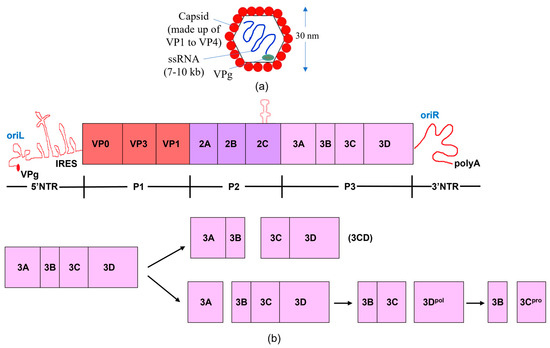
Figure 1.
The genome and proteome of poliovirus. (a) Structure of poliovirus. (b) The processing of polyproteins and the poliovirus genome are shown schematically. The poly (A) tail, broad open reading frame, and 3′ and 5′ non-translated regions (NTR) comprise the poliovirus genome. The type II internal ribosome entry site (IRES) and a cloverleaf structure comprise the 5′NTR. The virus-encoded 3B (VPg) protein is attached to the 5′ end of the RNA. The viral genome encodes a single polyprotein; the P1 region encodes the structural proteins of the virus, while the P2 and P3 portions encode the non-structural proteins. The P3 region is cleaved by viral proteases via two main pathways, resulting in the production of the 3C and 3D proteins, either separately or collectively as 3CD. The RNA-dependent RNA polymerase 3D replicates the viral RNA, while 3C, a protease, cleaves at specific, conserved motifs located within flexible linkers that separate discrete proteins within the viral polyprotein. Because 3CD lacks polymerase activity and has a unique protease specificity that enables it to cleave the P1 capsid region differently than 3C, the functional activity changes when the 3C and 3D proteins are combined to produce 3CD [27,28,29,30].
Almost fifty years ago, proteolytic processing was demonstrated to be crucial for picornavirus capsid protein synthesis and the construction of virions [27,28,29,30]. The 3C protein is recruited to perform this activity [31]. The 3C protein participates in the processing of polypeptides, binding of RNA, initialization of protein-priming of RNA synthesis, and viral translation to replication steps [1,3,19]. Antiviral therapeutic targets for 3C and similar viral proteases have been already identified [32]. In this Review, we emphasize various functional roles of 3C, including in the regulation of RNA replication, transcription, and translation, as well as its ability to inhibit nucleocytoplasmic transport and trigger apoptosis. Considering all these vast functions of 3C, this protease acts as an important virulence factor of picornaviruses [3].
2. Structure and Functions of 3C
2.1. Catalytic and RNA-Binding Sites
The 3C protease has a catalytic triad resembling those of serine proteases belonging to the trypsin-like family, although the catalytic nucleophile is a cysteine (Cys) residue rather than a serine (Ser) [33,34]. The X-ray crystal structure of human rhinovirus (3C) was determined nearly three decades ago (PDB ID:1CQQ) [35], followed by structure determination of 3C from other viruses including poliovirus (PV; PDB ID:1L1N) [36], hepatitis A virus (HAV; PDB ID:1QA7) [37], foot-and-mouth disease virus (FMDV; PDB ID:2BHG) [38,39], coxsackievirus B (CVB; PDB ID:2ZU1) [40,41,42], and enterovirus 71 (EV71; PDB ID:3OSY) [43]. All these 3C proteins are highly structurally similar.
Protein sequence similarities and phylogenetic relationships were previously established using the maximum likelihood method [44]. The catalytic triad, consisting of a Cys, His, and either Glu or Asp, was found to be highly conserved among the 3C proteases. The N-terminal alpha-helix, an important structural element involved in lipid membrane interactions, and the RNA binding region are not so conserved among picornaviral 3C proteases [45,46]. Six primary classes of 3C proteases from picornaviruses were identified by the phylogenetic study, in which the 3C proteases exhibited 38–45% amino acid similarity, a higher degree of sequence similarity than that observed in other picornaviruses.
Our own research is primarily focused on PV 3C [47,48,49]. The PV 3C protein structure has a traditional trypsin-like fold (Figure 2) [36,50], despite having less than 10% sequence identity with the trypsin protease family. It includes six β-sheets that fold into two β-barrel subdomains arranged at right angles [36,51]. The surface groove connecting these subdomains serves as the substrate-binding site, housing the central catalytic triad in the active site [36]. The 3C proteins from HRV [35], PV [36], CVB [41], and EV71 [43] have a Glu as part of the catalytic triad, whereas in HAV and FMDV it is an Asp [37,38]. The first member of the catalytic triad, the absolutely conserved Cys residue, serves as the catalytic nucleophile, which is then aided by the His general base and the carboxylate group of either the Asp or Glu residue. Despite the structural similarity to Ser proteases, mutagenesis of the active site Cys for a Ser gives an enzyme with a drastically reduced activity. Caspases also employ a Cys/His diad, but appear to be much more active than 3C [37]. As the third component of the triad (Glu or Asp) is not rigorously conserved across all 3C proteases and the side chains tend to point away from the active site His, its necessity to catalyze is not as clear.
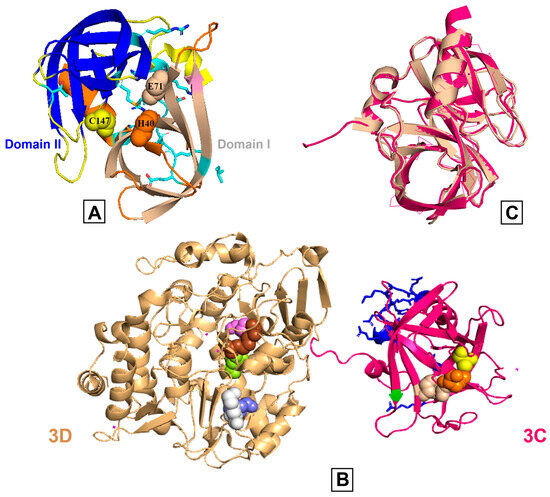
Figure 2.
Comparison of poliovirus 3C and 3CD proteins. (A) The poliovirus 3C protease X-ray crystal structure (PDB: 1L1N) is shown, and the β-barrels of domains I and II are represented in wheat and blue colors, respectively. The catalytic triad (His40 (orange), Glu71 (light wheat), Cys147 (yellow)) is shown in spheres and the substrate pockets are in different colors. Side chains are colored by heteroatoms and the RNA binding region (E81-H89) is highlighted. (B) The X-ray crystal structure of poliovirus 3CD (PDB: 2IJD); the wheat and pink colors represent 3D and 3C, respectively. The active site residues are shown on both subdomains using spheres (3C: His40 (orange), Glu71 (light wheat), Cys147 (yellow); 3D, starting at the first residue of the 3D domain: Arg174 (brown), Asp233 (slate), Ser288 (violet), Asn297 (lemon), Lys359 (white)). The RNA binding region of 3C is colored blue. (C) The 3C region of 3C (pink) and 3CD (wheat) are structurally comparable as evidenced by an overlay with an RMSD value of 0.46 angstroms.
The substrate-binding capsule is formed by a surface groove that sits between two β-barrel domains. The β-ribbons and substrates form a strong association once they are bound, enhancing catalytic effectiveness [34,36,43]. The 3C groove ensures accurate substrate binding, with the overlying β-strands directing the configuration and depth of the active-site region [36]. The X-ray crystal structure shows that the β-barrel adopts a flexible conformation to increase the likelihood of substrate identification when a substrate is absent [7,36,37,43,52]. The flexible loop that comes before the catalytic Cys most likely changes conformation in response to substrate binding [36]. The groove of 3C determines its substrate specificity, as the β-ribbon above it influences the shape and depth of the active site. In EV71 3C, Gly123 and His133, two significant residues (not part of the catalytic triad) near the β-ribbon base, function as hinges to control the ribbon inherent flexibility. Studies using structure-guided mutagenesis have demonstrated the significance of the hinge residues for the proteolytic function of EV71 3C [43,53].
2.2. Viral Polyprotein Processing and Substrate Recognition
Viral proteins are produced by the picornavirus family via the polyprotein processing. The majority of cleavage sites, including at the VP2–VP3, VP3–VP1, 2B–2C, 2C–3A, 3A–3B, 3B–3C, and 3C–3D junctions [18,28], (see Figure 1), are processed by 3C, with four exceptions: L–VP4, VP4–VP2, P1–2A, and 2A–2B [54,55]. For example, the leader protein (L) in aphthoviruses and erboviruses releases itself from VP0 [1]. In some picornaviruses (i.e., aphthoviruses, cardioviruses), ribosome skipping at the Asn–Pro–Gly–Pro site causes cleavage of 2A–2B. Almost all secondary cleavages in picornaviruses are caused by 3C [1]. Only 3C and the 3CD precursor are needed to accomplish all cleavages in the case of the Aichi virus, as L and 2Apro lack proteolytic activity. Studies on enteroviruses and apthoviruses have demonstrated that 3CDpro can cleave at VP2–VP3 and VP3–P1 more effectively than 3C [56,57]. SVV 3C protease cleaves the P1 polypeptide to provide VP0, VP1, and VP3 [58]. Most 3C proteases cleave at single sites such as (Q|G), (Q|S), (Q|R), (E|Q), and (E|G) [7] (see Table 1 for example).

Table 1.
The 3C cleavage of proteins involved in cellular translation. Cleavage site identified by underline.
Crystal structure analysis has provided some insights into substrate recognition. For poliovirus 3C, the specificity pockets are clearly delineated, and modelling investigations help explain the established substrate specificity [36]. For FMDV 3C, the distinct substrate preferences for specific residue types, as well as the relative promiscuity, are well explained by the structure of the enzyme–peptide complex [70]. For instance, a similar peptide binding mechanism is revealed by crystallographic analysis of the complex containing a modified VP1–2A peptide (APAKE|LLNFD) with a Gln-to-Glu substitution [70]. This also explains why 3C can cleave sequences containing either P1–Gln or P1–Glu.
In contrast, 3C EV71 takes on an unusually open conformation in which the active site is highly exposed to solvent because of its open β-ribbon shape. There are insufficient electron densities to identify the conformations of the active site, and none to define the side chain conformation of catalytic Glu71 [53]. This inherent flexibility may be important in recognizing diverse cleavage sites observed in virus and host protein targets.
2.3. 3C(D) Also Interacts with Virus Replication Membranes
How viral proteins specifically target genome replication sites has been a persistent question in the field of virology [71,72]. Virus infection changes the lipid and protein composition relative to membranes found in uninfected cells [73,74,75]. The field has long known that these RNA viruses manipulate host cell membranes to create specific replication sites [22], but only in the past decade or so has it been understood how/why virus proteins are relocalized to these replication membranes.
RNA viruses have been shown to induce the production of a lipid called phosphatidylinositol 4-phosphate (PI4P) in host cells. In 2010, Altan-Bonnet and colleagues found that PI4P was produced during picornavirus infection and associated with regions of genome replication [76]. The study revealed that lipids are vital for RNA replication, with viruses 3D RNA polymerase binding to specific lipids such as PI4P [76,77]. It is well known that phosphatidylinositol phosphate phospholipids (PIPs) serve as a type of “zip code” for protein sorting to the appropriate subcellular compartment [49,78,79]. For instance, the pleckstrin homology (PH) domain from phospholipase C is the most extensively studied PIP-binding determinant, which interacts with phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) [80,81,82]. These domains support latent enzymatic activity until activated by the proper PIP, limiting enzyme activation to the appropriate subcellular compartment [83,84,85,86]. These characteristics are ideal for a virus to direct its enzymes and proteins to the proper locations and regulate their functions in those areas.
PIP lipids found on the cell membrane can be used by picornaviruses to control viral RNA replication. PV 3C binds PI4P through its dynamic N-terminal α-helix, which competes with RNA binding due to the overlap between the PIP- and RNA-binding sites [49]. PIPs may therefore be used by picornaviruses to control viral RNA replication [76]. NMR and MD simulations have helped to delineate the PIP-binding site (Figure 3) [49].
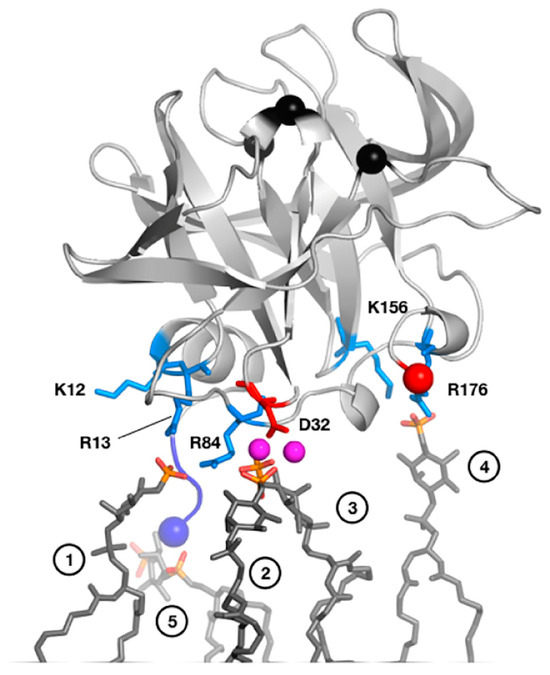
Figure 3.
MD-derived model of PV 3C interactions with a PI4P-containing lipid membrane. Residues involved in interactions include Arg13 in the N-terminal alpha helix and Arg84 in the RNA-binding region. NMR studies with short-chain, soluble PI4P lipids largely confirm this model. In this model, PV 3C interacts with five clustered PI4P lipid molecules as shown in dark grey sticks with phosphates colored orange (phosphorus) and red (oxygen). The following specific interactions are listed, with numbers in parenthesis denoting the panel’s head group: (1) R13 and R84; (2) D32, mediated by sodium ions; (3) D32 mediated by sodium; (4) K156 and R176; and (5) α-amino group of G1. Reprinted from Structure, 25(12), D. Shengjuler, Y.M. Chan, S. Sun, I. Moustafa, Z.-L. Li, D.W. Gohara, M. Buck, P.S. Cremer, D.D. Boehr, C.E. Cameron. The RNA-binding site of poliovirus 3C protein doubles as a phosphoinositide-binding domain, 1875–1886, Copyright (2017) with permission from Elsevier.
The roles of 3C in the viral replication cycle may be better understood with further research into the structural characteristics and functional significance of PV 3C and its interactions with host cell membranes. The remaining part of the review emphasizes the ability of picornaviral 3Cs, through their abilities to bind RNA and act as a protease, to hijack host cell metabolism and defenses.
3. The 3C Protease Function Intervenes in Host Cell Processes
The 3C protease cleaves the viral polyprotein and host cell proteins and also binds RNA which is essential for virus replication and inhibiting host cell responses [1,3]. Viral infection induces interferon (IFN) production inside the host cells owing to the antiviral immune response [87]. The co-evolution of viruses and host cells has led to the development of IFN-evasion techniques, allowing viruses to persist inside host cells, in which 3C plays an important role [69,87]. For instance, 3C plays a key role in blocking host signaling pathways, thus allowing the virus to circumvent host cell defenses and readily multiply inside host cells [88,89]. Picornaviruses inhibit antiviral protein synthesis and transport and block immune responses through transcription and cap-dependent translation [90,91]. Here, we discuss the multiple roles that 3C plays in the host cells, including halting transcription, inhibiting protein synthesis, blocking nucleocytoplasmic transport, and inducing cell death (see Figure 4).
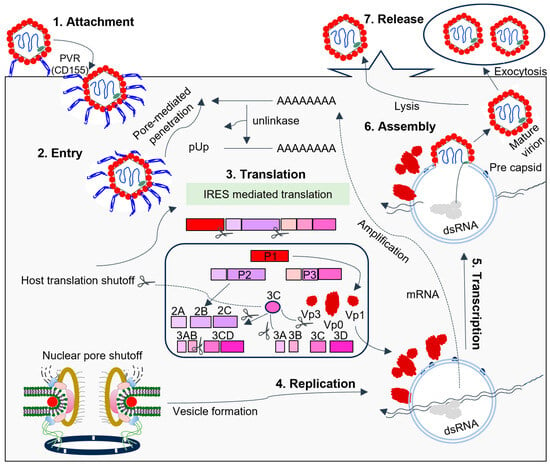
Figure 4.
The picornavirus life cycle involves 3C interfering with host components to facilitate RNA replication. Endocytosis, promoted by the attachment of the virus to host receptors, allows the virus to enter the host cell. In PV, the receptor is CD155. Viral proteases cleave translation initiation factors, halting cellular cap-dependent translation. Replication occurs in endoplasmic reticulum-derived membrane vesicles in viral factories, using genomic single-stranded positive-strand RNA (ssRNA(+)) to create a double-stranded RNA intermediate. Transcription and replication produce viral mRNAs and new ssRNA(+) genomes. The packaging of genomic RNA into prepared procapsids leads to cell death and virus release. During SVV infection, the virion first attaches itself to the host cell ANXTR1 receptor before internalizing itself through an endocytic pathway [92,93]. Unlike other picornaviruses, SVV has a distinct uncoating mechanism and binding mode with its receptor.
3.1. Proteolysis by 3C Proteases Suppresses the Host Transcription and Translation Mechanisms
3C proteins play crucial roles in suppressing the host transcription and translation mechanism, such that energy and resources are redirected towards virus replication. The picornavirus enters the host cell, followed by the release of the picornaviral genome into the cytoplasmic matrix, where it can interact with ribosomes [89,94,95,96]. The 3C protein acts to suppress transcription and cap-dependent translation of host genes [94,97]. For instance, the 3C-mediated cleavage of histone H3 at its N-terminus generates a novel polypeptide Pi in BHK cells, which persists in association with chromatin [98]. This process provides binding sites for transcription [99,100]. Therefore, the host cell transcription may be halted by the cleavage of H3 into Pi [98,101]. 3C also proteolyzes transcription factors, including IIIC, cAMP response element-binding protein-1 (CREB1), octamer binding protein-1 (OCT1 or POU2F1, p53 (TP53), TATA-binding protein-associated factor 110 (TAF110 or TAF1C), and TATA box-binding protein (TBP), resulting in the blockage of transcription initiation by host RNA polymerases [102,103,104,105,106,107]. Recent evidence also shows that EV71 3C can cleave the cellular CstF-64 protein, which subsequently halts host RNA processing and polyadenylation [108]. SVV 3C cleaves MAV, TRIF, and TANK which blocks transcription [92,109,110].
Picornaviruses inhibit host translation (see Table 1) through various pathways, including IRES-dependent translation for viral protein synthesis [18,90,111] (Figure 5). Cap-dependent protein and DNA repair processes are shut off in infected host cells due to the fragmentation of cellular components [112].
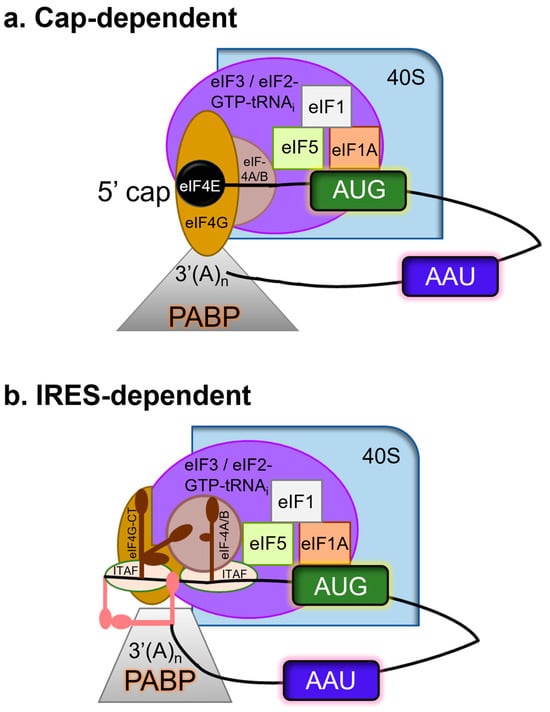
Figure 5.
Translation initiation can be (a) cap-dependent and/or (b) IRES-dependent. Using the color code, only the primary factors mentioned in the text have been depicted for simplicity. Cellular mRNAs functionally pseudo-circularize as a result of the interaction between eIF4G and PABP. Regarding the FMDV genome, the 5′-3′ long-range communication among the 3′-UTR with the S fragment (a large stem-loop formed by the folding of the terminal structure at the 5′ end), in addition to 3′ UTR-IRES interaction, permits a comparable circumstance that is probably stabilized by binding factors for RNA. IRES-driven translational initiation requires a few eIFs and IRES-transacting factors (ITAFs), i.e., PCBP2, PTB, and Germin 5 which modulate IRES activity. 3C is crucial because it guarantees the internal initiation of translation by cleaving certain eIFs and ITAFs.
Seneca Valley Virus 3C (SVV 3C) cleaves eukaryotic initiation factors (eIFs) eIF4AI and eIF4G1, while PV, CVB, and HRV 3C proteases cleave eIF5B [18,58,63,93,113]. eIF4AI inhibition alters eIF4AII protein expression [114]. eIF5B cleaves at one site (VVEQ↓G), cutting the C-terminal and N-terminal regions from the conserved GTPase domain during infection processes [59,115]. These cleavage events also affect the termination of viral translation, leading to increased RNA packaging into new viral particles. The 3C protease cleaves the viral RNA-binding protein PCBP2, causing it to lose its K-homologous domain (KH3) [62]. This 3C-processed cleavage is also observed in HAV-infected cells [116]. The polypyrimidine tract-binding protein (PTB), including PTBP1 and PTBP2, is also cleaved by HAV 3C to inhibit viral translation and improve viral genome replication (and is also observed for PV 3C) [115,117]. The FMDV and EV71 3C proteases can cleave the Sam68 nuclear RNA-binding protein, allowing it to play nonnuclear functions in the cytoplasm [59,64,118,119]. Sam68 can engage with IRES in FMDV during infection and before viral RNA translation [64].
An indirect way of promoting virus protein translation is through the 3C-directed cleavage of nuclear pore proteins. Picornaviruses produce RNA in the cytoplasm, while cellular RNA is produced in the nucleus [120]. Picornaviral proteins target five nucleoporins (Nups; Nup62, Nup98, Nup 153, Nup 214, and Nup 358) to disrupt macromolecule trafficking via the nuclear pore complex (NPC) [121]. These proteins are specifically proteolyzed by 3C (e.g., as observed with HRV 3C) [3,88,122,123], demonstrating that picornaviruses change nucleocytoplasmic shuttling to meet nuclear-resident protein requirements but obstruct cellular mRNA export [124].
3.2. 3C-Promoted Apoptosis for Virus Release
Infected cells can exhibit 3C-induced apoptosis, as shown for SVV, HAV, and CVB [17,92,125,126]. When a picornavirus infects a cell, 3C causes cell death through both caspase-dependent and caspase-independent mechanisms, interferes with the cleavage of the Golgi apparatus and MAP-4, and controls the secretory pathway and intracellular membrane trafficking [49,127,128,129]. The 3C protein regulates apoptosis through various pathways, triggering Bax and cytochrome c secretion in mitochondria [130].
The cleavage of transcription and translation initiation factors induces the production of apoptotic bodies and DNA degradation, while also degrading the transcriptional activator p53 in vivo and in vitro [1,5,89,129,131]. PV–3C is essential for p53 degradation in cells infected with PV, promoting apoptosis through transcription-independent mechanisms [5]. CVB-3C activates caspase-9-based apoptosis through caspase 3 activation [127]. 3C is essential for SVV-mediated apoptosis following caspase 3, while HAV 3C causes cell death without the aid of caspase [93,127]. VAD–fmk caspase 1 and DEVD–fmk inhibitors interfere with 3C-induced apoptosis, suggesting 3C initiates caspase cascades during late PV infection [132]. The 3C-induced apoptosis in EV 71 is related to its proteolytic activity as reported in several studies [133].
3.3. 3C Interferes with Host Cell Defenses
Host cells utilize a variety of antiviral mechanisms to combat picornavirus infection, including stress granule formation, innate inflammatory responses, autophagy, and a few others [2,125,134]. Picornaviruses must attack the cellular proteins engaged in these antiviral defenses to survive, and 3C is essential to these processes [87,88,89].
When an organism is infected with a picornavirus, 3C triggers the host innate immune system to employ host pattern recognition receptors (PRRs) to identify the presence of pathogen-associated molecular patterns [87]. These include retinoic-acid-induced gene-I (RIG-I), cytosolic RIG-like RNA helicases such as melanoma differentiation-associated gene (MDA-5), and transmembrane PRRs such as Toll-like receptors (TLRs) [135,136]. PRRs enlist various particular adaptor proteins to initiate a signaling cascade downstream and stimulate three primary pathways for the production of IFNs: the IFN regulatory factor (IRF) pathway, the mitogen-activated protein kinase (MAPK) pathway, and the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway [137]. IFNs have the ability to trigger hundreds of interferon-stimulated gene factors (ISGs) that strengthen host defenses through autocrine or paracrine signaling [138]. Table 2 summarizes the key signaling mechanisms that inhibit the generation of IFNs from picornavirus 3Cs.

Table 2.
Key signaling mechanisms in picornaviruses that inhibit the generation of IFNs from 3C.
Through its caspase-dependent protease activity, 3C degrades STAT1, STAT2, IRF9, and karyopherin1, thus inhibiting IFN-α signaling in SVV [92]. It was also discovered that 3C cleaves STAT2 at Gln758 in the transactivation domain. The cleaved products of STAT2 were found to reduce its capacity to trigger the production of IFN-stimulated genes and activate IFN-stimulated response element activity [92]. Furthermore, 3C was found to hamper the IFN-stimulated gene factor 3 complex nuclear import and formation [87]. Lastly, 3C was found to cause the degradation of karyopherin 1 to prevent STAT1/STAT2 nuclear localization. When combined, SVV 3C can disrupt the type I IFN response by focusing on STAT1–STAT2–IRF9 and karyopherin α1 signals [92,109]. This reveals a new way that SVV uses to avoid host defenses. Other viruses may operate through similar mechanisms, although this would require further research.
The recognition of viral infections in the host cell initiates the antiviral response. Double-stranded RNA, or dsRNA, is a vital ligand for 3C of picornaviruses, as it triggers the cellular immune response [143]. Cytosolic dsRNA can be sensed by RIG-I-like receptors (RLRs), which include melanoma differentiation-associated protein 5 (MDA5) [144,145,146]. The interferon regulatory factor (IRF) family of transcription factors is nuclear translocated as a result of a signaling cascade that is triggered when MDA5 binds to dsRNA [147]. Nuclear IRFs can cause the expression of antiviral interferon-stimulated genes (ISGs) in neighboring uninfected cells [148]. They can also induce the transcription of several genes with antiviral functions, such as IFIT1 and RSAD2, as well as proinflammatory cytokines, such as IFNs, for which the protein products are secreted [143].
4. RNA Binding and the Viral RNA Template Transformation
The 3C protein is a key factor for the formation of protein-RNA complexes that act toward viral replication and release from host cells. The virus requires a way to transition between RNA replication and translation [149]. Viral RNA exists in numerous copies that are manipulable on their own [150]. The two events may take place concurrently in the host cell utilizing different copies of identical RNA template. The 5′-NTR of the picornaviral genome contains cis-acting replication elements (CRE), composed of oriL (left), oriI (internal), and oriR (right) [18,151]. These three CREs are all thought to interact with 3C (or 3CD), which may also impact protease activity and/or its interactions with replication membranes [47]. It has been shown that the conformational energy landscape of 3C is altered by RNA and peptide binding, potentially leading to different protein-RNA complex sites [49]. Earlier research from our lab shows that the structural dynamics of 3C are altered by RNA/peptide binding, which has an impact on both the binding site and predicted binding site of the other ligand [47]. The conformational dynamics on the pico-to-nanosecond and micro-to-millisecond timescales are also altered by RNA binding. It has been suggested that RNA binding selects different conformations, influencing peptide interaction with 3C and that the higher energy conformation may be crucial for interacting with protein substrates. These actions may also be important for 3C contribution towards switching between translation and RNA replication processes, which must use the same RNA template [149].
Picornaviral RNA serves as a template for translation and RNA replication, enabling IRES-driven viral protein generation [66]. A complementary negative-sense RNA molecule is produced using the same template, leading to the replicative form [88,152]. This intermediate produces a multiple-stranded RNA complex, generating positive-sense RNA molecules [69,119,153]. These RNA molecules are either recycled or assembled into offspring virions, using nuclear proteins found in uninfected cells. The 3C from PV, FMDV, EMCV, CVB3, and EV71 can induce the cleavage of Ras GTPase-activating protein-binding protein 1 (G3BP), which is also crucial for innate immune responses to DNA viruses [65,154,155]. As such, cleaving G3BP1 may increase the risk of secondary DNA viral infections, promoting viral replication. During HAV, CVB3, SVV, and EV-A71 infection, 3C-induced cleavage of Toll/IL-1 receptor-domain-containing adapter-inducing interferon (TRIF) is significant [110,156,157]. Picornaviruses and host cells both possess translational regulators, and 3C protease can initiate viral RNA replication by cleaving translation-associated proteins linked to viral RNA [18,93].
6. 3C Protease Inhibitors
Drug–protein interactions can be better understood at the molecular level through computational and bioinformatics tools, which can be used to expedite the design and development of enhanced antivirals targeting important proteins [176]. A specific field of structure-based/ligand-based drug design called computer-aided drug discovery (CADD) boasts several successes, including in the field of antivirals. The development of FDA-approved medications such as Saquinavir (anti-HIV) and Oseltamivir (anti-influenza virus) has confirmed the effectiveness of these methods [176] (see Figure 6 for chemical structures). One intriguing family of flexible lead compounds among the various peptidomimetic and small-molecule inhibitors that have been tried against viral proteins is isatin and its derivatives. Isatins have demonstrated antiviral efficacy against a variety of picornaviruses through interactions with 3C proteases.
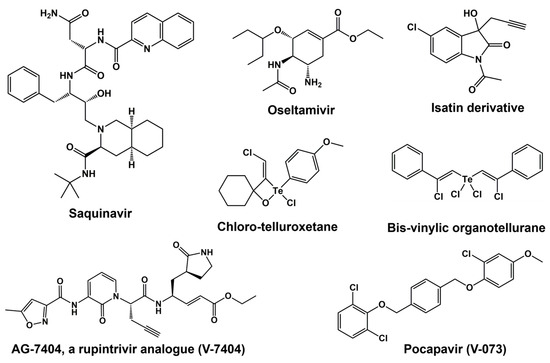
Figure 6.
Chemical structure of various 3C protease inhibitors.
Initial reports described the usage of tellurium compounds as PV 3C protease inhibitors. PV 3C was found to be rapidly inactivated by a stoichiometric and covalent reaction with both chloro-telluroxetane and bis-vinylic organotellurane [177]. Additionally, these substances have second-order rate constants for inhibition of human cathepsins B, L, S, and K that are greater than those found for PV 3C. Under low micromolar concentrations and below the lethal threshold for host cells, chloro-telluroxetane prevents the replication of PV in human embryonic rhabdomyosarcoma cells [177]. Although bis-vinylic organotellurane is a more potent antiviral drug, at 10 μM it inhibits cell viability by 20%, a dose that almost stops virus growth. This is the first account of this family of drugs antiviral properties through suppression of viral 3C proteinase.
Most clinical compounds have been rejected because of their weak selectivity between antiviral activity and cytotoxicity or their lack of potency [178]. Two compounds have been found so far in the results that satisfy the requirements for being considered poliovirus antiviral development candidates. One such substance is pocapavir (V-073), a capsid inhibitor licensed by ViroDefense Inc. and initially discovered by Schering-Plough (SCH 48973). The second substance is the 3C protease inhibitor AG7404, a rupintrivir analogue that was found by Agouron (Pfizer Inc., McPherson, KS, USA) and is currently being developed as V-7404 by ViroDefense Inc [138]. Initially created by Pfizer as an anti-rhinovirus drug, the 3C protease inhibitor V-7404 also exhibits strong in vitro anti-poliovirus action [179,180]. Although V-7404 is being positioned for combination treatment with pocapavir if drug resistance arises when treating PID patients with a single compound, pocapavir is currently being developed as a single-agent treatment. The Task Force for Childhood Development and Survival has entered into a contract with ViroDefense Inc. to assess the PV antiviral candidate V-073. In addition, ViroDefense Inc. is taking part in an NIAID program to provide specific preclinical services.
V-7404 is an orally accessible, single-dose organic compound which inhibits the human rhinovirus (HRV) 3C protease irreversibly [180]. The concentration-dependent suppression of HRV 3C-mediated polyprotein processing in infected cells by this compound directly confirms that the inhibition of 3C protease is the cause of the cell-based antiviral action. Nonclinical safety investigations conducted in vitro and in vivo have demonstrated that V-7404 is safe at the highest doses that could be administered [181]. In a phase-I-ascending single-dose research study, oral dosages of Compound 1 up to 2000 mg have been shown to be safe and well tolerated in healthy individuals.
7. Conclusions
Positive-strand RNA viruses act to maximize their genomic information content in part by encoding multifunctional proteins, as exemplified by 3C. Picornaviral 3C performs important roles in viral replication and inhibition of host cell responses through its ability to interact with RNA and act as the main protease. It is noted that many of these functions may be enacted through its polyprotein precursors (e.g., 3CD). The RNA-binding ability of 3C is crucial for initiating viral RNA synthesis. The mode and interaction specificity of 3C for binding various RNA is still under investigation.
The 3C protease intervenes in host cell transcription, translation, and replication events. It also plays critical roles in apoptosis and generation of phosphatidylserine (PS) lipid-rich vesicles that assist in virus release and spread. The binding of 3C to RNA alters the structural dynamics of 3C and controls how 3C functions with other cellular substrates. The cellular substrates of 3C have varying activities in host cells, potentially advantageous to viruses at different lifecycles [1,17]. Several newly identified substrates for PV and CVB3 3C have been found employing terminal amine isotopic labelling of substrates [182]. High-throughput technologies may help discover new cellular substrates and better understand how 3C modifies host processes and pathophysiology [183].
Picornaviruses inhibit antiviral protein synthesis and transport, suppressing immune responses through transcription and cap-dependent translation, and using mechanisms such as PARP9–DTX3L complex to regulate ISG expression and the immune response [6,24,77,83,128]. Furthermore, 3C precursors are also crucial in viral replication owing to their ability to hijack cellular resources and establish an optimal intracellular condition for the virus life cycle. The 3C-mediated cleavage of PABP disrupts mRNA circularization, and 3C also alters specific cellular components and fragments, such as eIF4G, PTB, and PCBP2 [66,184,185,186,187]. These findings highlight the critical functions that 3C performs in viral replication by ensuring translation, replication, and the transition from translation to replication.
Given the central role of 3C (or its precursors) in the many aspects of the picornaviral life cycle, there has been much effort in the development of 3C inhibitors [188,189]. To date, there are only a few treatments available for picornavirus infection [190,191]. The development of new antiviral treatments may become more accessible with a broader understanding of the roles played by picornavirus 3C. Efforts are currently ongoing to identify irreversible inhibitors for 3C, with further investigations into its molecular functions, structure, and dynamics potentially aiding in this goal [176,192].
Author Contributions
Conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation: S.M. and D.D.B.; writing—original draft preparation: S.M., G.S. and D.D.B.; writing—review and editing, visualization: S.M. and D.D.B.; supervision, project administration, funding acquisition: D.D.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NIH, grant number AI104878.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yi, J.; Peng, J.; Yang, W.; Zhu, G.; Ren, J.; Li, D.; Zheng, H. Picornavirus 3C—A Protease Ensuring Virus Replication and Subverting Host Responses. J. Cell Sci. 2021, 134, jcs253237. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.S.; Stobart, C.C.; Luo, H. Innate Immune Evasion Mediated by Picornaviral 3C Protease: Possible Lessons for Coronaviral 3C-like Protease? Rev. Med. Virol. 2021, 31, 1–22. [Google Scholar] [CrossRef]
- Sun, D.; Chen, S.; Cheng, A.; Wang, M. Roles of the Picornaviral 3C Proteinase in the Viral Life Cycle and Host Cells. Viruses 2016, 8, 82. [Google Scholar] [CrossRef] [PubMed]
- Minor, P.D. Poliovirus Biology. Structure 1996, 4, 775–778. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weidman, M.K.; Yalamanchili, P.; Ng, B.; Tsai, W.; Dasgupta, A. Poliovirus 3C Protease-Mediated Degradation of Transcriptional Activator P53 Requires a Cellular Activity. Virology 2001, 291, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Gong, P. Structural Basis of Viral RNA-Dependent RNA Polymerase Nucleotide Addition Cycle in Picornaviruses. Enzymes 2021, 49, 215–233. [Google Scholar] [CrossRef]
- Seipelt, J.; Guarné, A.; Bergmann, E.; James, M.; Sommergruber, W.; Fita, I.; Skern, T. The Structures of Picornaviral Proteinases. Virus Res. 1999, 62, 159–168. [Google Scholar] [CrossRef]
- Campagnola, G.; Peersen, O. Co-Folding and RNA Activation of Poliovirus 3Cpro Polyprotein Precursors. J. Biol. Chem. 2023, 299, 105258. [Google Scholar] [CrossRef]
- Takegami, T.; Kuhn, R.J.; Anderson, C.W.; Wimmer, E. Membrane-Dependent Uridylylation of the Genome-Linked Protein VPg of Poliovirus. Proc. Natl. Acad. Sci. USA 1983, 80, 7447–7451. [Google Scholar] [CrossRef]
- Larsen, G.R.; Dorner, A.J.; Harris, T.J.; Wimmer, E. The Structure of Poliovirus Replicative Form. Nucleic Acids Res. 1980, 8, 1217–1229. [Google Scholar] [CrossRef]
- Kitamura, N.; Adler, C.; Wimmer, E. Structure and Expression of the Picornavirus Genome. Ann. N. Y Acad. Sci. 1980, 354, 183–201. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.F.; Nomoto, A.; Detjen, B.M.; Wimmer, E. A Protein Covalently Linked to Poliovirus Genome RNA. Proc. Natl. Acad. Sci. USA 1977, 74, 59–63. [Google Scholar] [CrossRef]
- Erickson, J.W.; Frankenberger, E.A.; Rossmann, M.G.; Fout, G.S.; Medappa, K.C.; Rueckert, R.R. Crystallization of a Common Cold Virus, Human Rhinovirus 14: “Isomorphism” with Poliovirus Crystals. Proc. Natl. Acad. Sci. USA 1983, 80, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Hogle, J.M. Preliminary Studies of Crystals of Poliovirus Type I. J. Mol. Biol. 1982, 160, 663–668. [Google Scholar] [CrossRef]
- Malnou, C.E.; Pöyry, T.A.A.; Jackson, R.J.; Kean, K.M. Poliovirus Internal Ribosome Entry Segment Structure Alterations That Specifically Affect Function in Neuronal Cells: Molecular Genetic Analysis. J. Virol. 2002, 76, 10617–10626. [Google Scholar] [CrossRef]
- Jaafar, Z.A.; Kieft, J.S. Viral RNA Structure-Based Strategies to Manipulate Translation. Nat. Rev. Microbiol. 2019, 17, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Barco, A.; Feduchi, E.; Carrasco, L. Poliovirus Protease 3C(pro) Kills Cells by Apoptosis. Virology 2000, 266, 352–360. [Google Scholar] [CrossRef]
- Francisco-Velilla, R.; Embarc-Buh, A.; Abellan, S.; Martinez-Salas, E. Picornavirus Translation Strategies. FEBS Open Bio 2022, 12, 1125–1141. [Google Scholar] [CrossRef]
- Cohen, L.; Kean, K.M.; Girard, M.; Van der Werf, S. Effects of P2 Cleavage Site Mutations on Poliovirus Polyprotein Processing. Virology 1996, 224, 34–42. [Google Scholar] [CrossRef]
- Hogle, J.M.; Filman, D.J. The Antigenic Structure of Poliovirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1989, 323, 467–478. [Google Scholar] [CrossRef]
- Jiang, P.; Liu, Y.; Ma, H.-C.; Paul, A.V.; Wimmer, E. Picornavirus Morphogenesis. Microbiol. Mol. Biol. Rev. 2014, 78, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Hogle, J.M. Poliovirus Cell Entry: Common Structural Themes in Viral Cell Entry Pathways. Annu. Rev. Microbiol. 2002, 56, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Takahara, Y.; Ando, N.; Kohara, M.; Hagino-Yamagishi, K.; Nomoto, A.; Itoh, H.; Numao, N.; Kondo, K. Purification of Enzymatically Active Poliovirus Proteinase 3C Produced in Escherichia Coli. Gene 1989, 79, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Ivanoff, L.A.; Towatari, T.; Ray, J.; Korant, B.D.; Petteway, S.R.J. Expression and Site-Specific Mutagenesis of the Poliovirus 3C Protease in Escherichia Coli. Proc. Natl. Acad. Sci. USA 1986, 83, 5392–5396. [Google Scholar] [CrossRef] [PubMed]
- Winston, D.S.; Boehr, D.D. Dynamics Compared to 3C pro and 3D Pol in Functionally. Viruses 2021, 13, 442. [Google Scholar] [CrossRef]
- Kean, K.M.; Teterina, N.; Girard, M. Cleavage Specificity of the Poliovirus 3C Protease Is Not Restricted to Gln-Gly at the 3C/3D Junction. J. Gen. Virol. 1990, 71 Pt 11, 2553–2563. [Google Scholar] [CrossRef]
- Baltimore, D.; Jacobson, M.F.; Asso, J.; Huang, A.S. The Formation of Poliovirus Proteins. Cold Spring Harb. Symp. Quant. Biol. 1969, 34, 741–746. [Google Scholar] [CrossRef]
- Jacobson, M.F.; Baltimore, D. Polypeptide Cleavages in the Formation of Poliovirus Proteins. Proc. Natl. Acad. Sci. USA 1968, 61, 77–84. [Google Scholar] [CrossRef]
- Jacobson, M.F.; Baltimore, D. Morphogenesis of Poliovirus. I. Association of the Viral RNA with Coat Protein. J. Mol. Biol. 1968, 33, 369–378. [Google Scholar] [CrossRef]
- Holland, J.J.; Kiehn, E.D. Specific Cleavage of Viral Proteins as Steps in the Synthesis and Maturation of Enteroviruses. Proc. Natl. Acad. Sci. USA 1968, 60, 1015–1022. [Google Scholar] [CrossRef]
- Pallansch, M.A.; Kew, O.M.; Palmenberg, A.C.; Golini, F.; Wimmer, E.; Rueckert, R.R. Picornaviral VPg Sequences Are Contained in the Replicase Precursor. J. Virol. 1980, 35, 414–419. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Hu, L.; Huang, X.; Wang, C.; Zhang, Z.; Wang, Y.; Zhang, D.; Ye, W. Potential of Coronavirus 3C-like Protease Inhibitors for the Development of New Anti-SARS-CoV-2 Drugs: Insights from Structures of Protease and Inhibitors. Int. J. Antimicrob. Agents 2020, 56, 106055. [Google Scholar] [CrossRef] [PubMed]
- Lawson, M.A.; Semler, B.L. Poliovirus Thiol Proteinase 3C Can Utilize a Serine Nucleophile within the Putative Catalytic Triad. Proc. Natl. Acad. Sci. USA 1991, 88, 9919–9923. [Google Scholar] [CrossRef] [PubMed]
- Sárkány, Z.; Polgár, L. The Unusual Catalytic Triad of Poliovirus Protease 3C. Biochemistry 2003, 42, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.A.; Smith, W.W.; Ferre, R.A.; Condon, B.; Budahazi, G.; Slsson, W.; Villafranca, J.E.; Janson, C.A.; McElroy, H.E.; Gribskov, C.L.; et al. Structure of Human Rhinovirus 3C Protease Reveals a Trypsin-like Polypeptide Fold, RNA-Binding Site, and Means for Cleaving Precursor Polyprotein. Cell 1994, 77, 761–771. [Google Scholar] [CrossRef]
- Mosimann, S.C.; Cherney, M.M.; Sia, S.; Plotch, S.; James, M.N.G. Refined X-Ray Crystallographic Structure of the Poliovirus 3C Gene Product. J. Mol. Biol. 1997, 273, 1032–1047. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, E.M.; Mosimann, S.C.; Chernaia, M.M.; Malcolm, B.A.; James, M.N. The Refined Crystal Structure of the 3C Gene Product from Hepatitis A Virus: Specific Proteinase Activity and RNA Recognition. J. Virol. 1997, 71, 2436–2448. [Google Scholar] [CrossRef]
- Birtley, J.R.; Knox, S.R.; Jaulent, A.M.; Brick, P.; Leatherbarrow, R.J.; Curry, S. Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease: New Insights into Catalytic Mechanism and Cleavage Specificity. J. Biol. Chem. 2005, 280, 11520–11527. [Google Scholar] [CrossRef]
- Pierce, D.M.; Hayward, C.; Rowlands, D.J.; Stonehouse, N.J.; Herod, M.R. Insights into Polyprotein Processing and RNA-Protein Interactions in Foot-and-Mouth Disease Virus Genome Replication. J. Virol. 2023, 97, e0017123. [Google Scholar] [CrossRef]
- Van Raaij, M.J.; Chouin, E.; Van der Zandt, H.; Bergelson, J.M.; Cusack, S. Erratum: Dimeric Structure of the Coxsackievirus and Adenovirus Receptor D1 Domain at 1.7 Å Resolution (Structure (November 15, 2000) 8 (1147–1155)). Structure 2001, 9, 1147–1155. [Google Scholar] [CrossRef]
- Ohlenschläger, O.; Wöhnert, J.; Bucci, E.; Seitz, S.; Häfner, S.; Ramachandran, R.; Zell, R.; Görlach, M. The Structure of the Stemloop D Subdomain of Coxsackievirus B3 Cloverleaf RNA and Its Interaction with the Proteinase 3C. Structure 2004, 12, 237–248. [Google Scholar] [CrossRef]
- Lee, C.C.; Kuo, C.J.; Ko, T.P.; Hsu, M.F.; Tsui, Y.C.; Chang, S.C.; Yang, S.; Chen, S.J.; Chen, H.C.; Hsu, M.C.; et al. Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-Coordinating and Peptidomimetic Compounds. J. Biol. Chem. 2009, 284, 7646–7655. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Wang, J.; Fan, T.; Qin, B.; Guo, L.; Lei, X.; Wang, J.; Wang, M.; Jin, Q. Crystal Structure of Human Enterovirus 71 3C Protease. J. Mol. Biol. 2011, 408, 449–461. [Google Scholar] [CrossRef]
- Sun, D.; Wang, M.; Wen, X.; Mao, S.; Cheng, A.; Jia, R.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; et al. Biochemical Characterization of Recombinant Avihepatovirus 3C Protease and Its Localization. Virol. J. 2019, 16, 54. [Google Scholar] [CrossRef]
- Andino, R.; Kirkegaard, K.; Macadam, A.; Racaniello, V.R.; Rosenfield, A.B. The Picornaviridae Family: Knowledge Gaps, Animal Models, Countermeasures, and Prototype Pathogens. J. Infect. Dis. 2023, 228, S427–S445. [Google Scholar] [CrossRef] [PubMed]
- Nugent, C.I.; Kirkegaard, K. RNA Binding Properties of Poliovirus Subviral Particles. J. Virol. 1995, 69, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.M.; Moustafa, I.M.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Long-Range Communication between Different Functional Sites in the Picornaviral 3C Protein. Structure 2016, 24, 509–517. [Google Scholar] [CrossRef][Green Version]
- Tsu, B.V.; Fay, E.J.; Nguyen, K.T.; Corley, M.R.; Hosuru, B.; Dominguez, V.A.; Daugherty, M.D. Running with Scissors: Evolutionary Conflicts between Viral Proteases and the Host Immune System. Front. Immunol. 2021, 12, 769543. [Google Scholar] [CrossRef]
- Shengjuler, D.; Chan, Y.M.; Sun, S.; Moustafa, I.M.; Li, Z.L.; Gohara, D.W.; Buck, M.; Cremer, P.S.; Boehr, D.D.; Cameron, C.E. The RNA-Binding Site of Poliovirus 3C Protein Doubles as a Phosphoinositide-Binding Domain. Structure 2017, 25, 1875–1886.e7. [Google Scholar] [CrossRef]
- Bazan, J.F.; Fletterick, R.J. Viral Cysteine Proteases Are Homologous to the Trypsin-like Family of Serine Proteases: Structural and Functional Implications. Proc. Natl. Acad. Sci. USA 1988, 85, 7872–7876. [Google Scholar] [CrossRef]
- Uma, M.; Hegde, S.R.; Rao, P.P.; Nagalekshmi, K.; Gauthami, S.; Kumar, D.; Hegde, N.R. A Novel Point Mutation (L70P) Inactivates Poliovirus 3C Protease. Acta Virol. 2018, 62, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.R.; Roqué-Rosell, N.; Birtley, J.R.; Leatherbarrow, R.J.; Curry, S. Structural and Mutagenic Analysis of Foot-and-Mouth Disease Virus 3C Protease Reveals the Role of the Beta-Ribbon in Proteolysis. J. Virol. 2007, 81, 115–124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jing, W.; Tingting, F.; Xue, Y.; Zhiqiang, W.; Li, G.; Xiaobo, L.; Jianwei, W.; Meitian, W.; Qi, J.; Sheng, C. Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues. J. Virol. 2011, 85, 10021–10030. [Google Scholar] [CrossRef]
- Yalamanchili, P.; Datta, U.; Dasgupta, A. Inhibition of Host Cell Transcription by Poliovirus: Cleavage of Transcription Factor CREB by Poliovirus-Encoded Protease 3C Pro. J. Virol. 1997, 71, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wang, M.; Wen, X.; Cheng, A.; Jia, R.; Sun, K.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; et al. Cleavage of Poly(A)-Binding Protein by Duck Hepatitis A Virus 3C Protease. Sci. Rep. 2017, 7, 16261. [Google Scholar] [CrossRef] [PubMed]
- Roehl, H.H.; Parsley, T.B.; Ho, T.V.; Semler, B.L. Processing of a Cellular Polypeptide by 3CD Proteinase Is Required for Poliovirus Ribonucleoprotein Complex Formation. J. Virol. 1997, 71, 578–585. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Andino, R. Interactions of Viral Protein 3CD and Poly(RC) Binding Protein with the 5′ Untranslated Region of the Poliovirus Genome. J. Virol. 2000, 74, 2219–2226. [Google Scholar] [CrossRef]
- Wang, Q.; Meng, H.; Ge, D.; Shan, H.; Geri, L.; Liu, F. Structural and Nonstructural Proteins of Senecavirus A: Recent Research Advances, and Lessons Learned from Those of Other Picornaviruses. Virology 2023, 585, 155–163. [Google Scholar] [CrossRef]
- de Breyne, S.; Bonderoff, J.M.; Chumakov, K.M.; Lloyd, R.E.; Hellen, C.U.T. Cleavage of Eukaryotic Initiation Factor EIF5B by Enterovirus 3C Proteases. Virology 2008, 378, 118–122. [Google Scholar] [CrossRef]
- Perera, R.; Daijogo, S.; Walter, B.L.; Nguyen, J.H.C.; Semler, B.L. Cellular Protein Modification by Poliovirus: The Two Faces of Poly(RC)-Binding Protein. J. Virol. 2007, 81, 8919–8932. [Google Scholar] [CrossRef]
- Kuyumcu-Martinez, N.M.; Van Eden, M.E.; Younan, P.; Lloyd, R.E. Cleavage of Poly(A)-Binding Protein by Poliovirus 3C Protease Inhibits Host Cell Translation: A Novel Mechanism for Host Translation Shutoff. Mol. Cell Biol. 2004, 24, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ross-Smith, N.; Proud, C.G.; Belsham, G.J. Cleavage of Translation Initiation Factor 4AI (EIF4AI) but Not EIF4AII by Foot-and-Mouth Disease Virus 3C Protease: Identification of the EIF4AI Cleavage Site. FEBS Lett. 2001, 507, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Belsham, G.J.; McInerney, G.M.; Ross-Smith, N. Foot-and-Mouth Disease Virus 3C Protease Induces Cleavage of Translation Initiation Factors EIF4A and EIF4G within Infected Cells. J. Virol. 2000, 74, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, P.; Schafer, E.A.; Rieder, E. The Nuclear Protein Sam68 Is Cleaved by the FMDV 3C Protease Redistributing Sam68 to the Cytoplasm during FMDV Infection of Host Cells. Virology 2012, 425, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Fung, G.; Ng, C.S.; Zhang, J.; Shi, J.; Wong, J.; Piesik, P.; Han, L.; Chu, F.; Jagdeo, J.; Jan, E.; et al. Production of a Dominant-Negative Fragment Due to G3BP1 Cleavage Contributes to the Disruption of Mitochondria-Associated Protective Stress Granules during CVB3 Infection. PLoS ONE 2013, 8, e79546. [Google Scholar] [CrossRef]
- Zhang, B.; Seitz, S.; Kusov, Y.; Zell, R.; Gauss-Müller, V. RNA Interaction and Cleavage of Poly(C)-Binding Protein 2 by Hepatitis A Virus Protease. Biochem. Biophys. Res. Commun. 2007, 364, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Morace, G.; Gauss-Müller, V.; Kusov, Y. Poly(A) Binding Protein, C-Terminally Truncated by the Hepatitis A Virus Proteinase 3C, Inhibits Viral Translation. Nucleic Acids Res. 2007, 35, 5975–5984. [Google Scholar] [CrossRef][Green Version]
- Kobayashi, M.; Arias, C.; Garabedian, A.; Palmenberg, A.C.; Mohr, I. Site-Specific Cleavage of the Host Poly(A) Binding Protein by the Encephalomyocarditis Virus 3C Proteinase Stimulates Viral Replication. J. Virol. 2012, 86, 10686–10694. [Google Scholar] [CrossRef]
- Qian, S.; Fan, W.; Liu, T.; Wu, M.; Zhang, H.; Cui, X.; Zhou, Y.; Hu, J.; Wei, S.; Chen, H.; et al. Seneca Valley Virus Suppresses Host Type I Interferon Production by Targeting Adaptor Proteins MAVS, TRIF, and TANK for Cleavage. J. Virol. 2017, 91, e00823-17. [Google Scholar] [CrossRef]
- Zunszain, P.A.; Knox, S.R.; Sweeney, T.R.; Yang, J.; Roqué-Rosell, N.; Belsham, G.J.; Leatherbarrow, R.J.; Curry, S. Insights into Cleavage Specificity from the Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease Complexed with a Peptide Substrate. J. Mol. Biol. 2010, 395, 375–389. [Google Scholar] [CrossRef]
- Cameron, C.E.; Oh, H.S.; Moustafa, I.M. Expanding Knowledge of P3 Proteins in the Poliovirus Lifecycle. Future Microbiol. 2010, 5, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, B.; Lee, L.Y.; Lakadamyali, M.; Rust, M.J.; Zhuang, X.; Hogle, J.M. Imaging Poliovirus Entry in Live Cells. PLoS Biol. 2007, 5, e183. [Google Scholar] [CrossRef] [PubMed]
- Mazzon, M.; Mercer, J. Lipid Interactions during Virus Entry and Infection. Cell Microbiol. 2014, 16, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Lenard, J. Viral Membranes. Encycl. Virol. 2008, 308–314. [Google Scholar] [CrossRef]
- Villanueva, R.A.; Rouillé, Y.; Dubuisson, J. Interactions between Virus Proteins and Host Cell Membranes during the Viral Life Cycle. Int. Rev. Cytol. 2005, 245, 171–244. [Google Scholar] [CrossRef]
- Hsu, N.-Y.; Ilnytska, O.; Belov, G.; Santiana, M.; Chen, Y.-H.; Takvorian, P.M.; Pau, C.; van der Schaar, H.; Kaushik-Basu, N.; Balla, T.; et al. Viral Reorganization of the Secretory Pathway Generates Distinct Organelles for RNA Replication. Cell 2010, 141, 799–811. [Google Scholar] [CrossRef]
- Zhang, L.; Hong, Z.; Lin, W.; Shao, R.-X.; Goto, K.; Hsu, V.W.; Chung, R.T. ARF1 and GBF1 Generate a PI4P-Enriched Environment Supportive of Hepatitis C Virus Replication. PLoS ONE 2012, 7, e32135. [Google Scholar] [CrossRef]
- Wen, Y.; Vogt, V.M.; Feigenson, G.W. PI(4,5)P2 Clustering and Its Impact on Biological Functions. Annu. Rev. Biochem. 2021, 90, 681–707. [Google Scholar] [CrossRef]
- Arita, M. Mechanism of Poliovirus Resistance to Host Phosphatidylinositol-4 Kinase III β Inhibitor. ACS Infect. Dis. 2016, 2, 140–148. [Google Scholar] [CrossRef]
- Lemmon, M.A. Pleckstrin Homology (PH) Domains and Phosphoinositides. Biochem. Soc. Symp. 2007, 74, 81–93. [Google Scholar] [CrossRef]
- Lemmon, M.A. Pleckstrin Homology (PH) Domains. Handb. Cell Signal. Second. Ed. 2009, 2, 1093–1101. [Google Scholar] [CrossRef]
- Harlan, J.E.; Hajduk, P.J.; Yoon, H.S.; Fesik, S.W. Pleckstrin Homology Domains Bind to Phosphatidylinositol-4,5-Bisphosphate. Nature 1994, 371, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Szentpetery, Z.; Balla, A.; Kim, Y.J.; Lemmon, M.A.; Balla, T. Live Cell Imaging with Protein Domains Capable of Recognizing Phosphatidylinositol 4,5-Bisphosphate; a Comparative Study. BMC Cell Biol. 2009, 10, 67. [Google Scholar] [CrossRef]
- Hammond, G.R.V.; Balla, T. Polyphosphoinositide Binding Domains: Key to Inositol Lipid Biology. Biochim. Biophys. Acta 2015, 1851, 746–758. [Google Scholar] [CrossRef]
- Pemberton, J.G.; Balla, T. Polyphosphoinositide-Binding Domains: Insights from Peripheral Membrane and Lipid-Transfer Proteins. Adv. Exp. Med. Biol. 2019, 1111, 77–137. [Google Scholar] [CrossRef]
- Groux-Degroote, S.; van Dijk, S.M.; Wolthoorn, J.; Neumann, S.; Theos, A.C.; De Mazière, A.M.; Klumperman, J.; van Meer, G.; Sprong, H. Glycolipid-Dependent Sorting of Melanosomal from Lysosomal Membrane Proteins by Lumenal Determinants. Traffic 2008, 9, 951–963. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, L.; Stipkovits, L.; Szathmary, S.; Li, X.; Liu, Y. The Strategy of Picornavirus Evading Host Antiviral Responses: Nonstructural Proteins Suppress the Production of IFNs. Front. Microbiol. 2018, 9, 2943. [Google Scholar] [CrossRef]
- Flather, D.; Semler, B.L. Picornaviruses and Nuclear Functions: Targeting a Cellular Compartment Distinct from the Replication Site of a Positive-Strand RNA Virus. Front. Microbiol. 2015, 6, 594. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Chen, T.-C.; Weng, K.-F.; Chang, S.-C.; Chen, L.-L.; Shih, S.-R. Viral and Host Proteins Involved in Picornavirus Life Cycle. J. Biomed. Sci. 2009, 16, 103. [Google Scholar] [CrossRef]
- Martínez-Salas, E.; Francisco-Velilla, R.; Fernandez-Chamorro, J.; Lozano, G.; Diaz-Toledano, R. Picornavirus IRES Elements: RNA Structure and Host Protein Interactions. Virus Res. 2015, 206, 62–73. [Google Scholar] [CrossRef]
- Kim, H.; Aponte-Diaz, D.; Sotoudegan, M.S.; Shengjuler, D.; Arnold, J.J.; Cameron, C.E. The Enterovirus Genome Can Be Translated in an IRES-Independent Manner That Requires the Initiation Factors EIF2A/EIF2D. PLoS Biol. 2023, 21, e3001693. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Guo, Y.; Wang, D.; Quan, R.; Wang, J.; Liu, J. Seneca Valley Virus 3C pro Antagonizes Type I Interferon Response by Targeting STAT1-STAT2-IRF9 and KPNA1 Signals. J. Virol. 2023, 97, e0072723. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Quan, R.; Wang, D.; Liu, J. Seneca Valley Virus 3Cpro Cleaves Heterogeneous Nuclear Ribonucleoprotein K to Facilitate Viral Replication. Front. Microbiol. 2022, 13, 945443. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, J.D.; Tsai, W.-C.; Lloyd, R.E. Multiple Poliovirus Proteins Repress Cytoplasmic RNA Granules. Viruses 2015, 7, 6127–6140. [Google Scholar] [CrossRef]
- Fitzgerald, K.D.; Chase, A.J.; Cathcart, A.L.; Tran, G.P.; Semler, B.L. Viral Proteinase Requirements for the Nucleocytoplasmic Relocalization of Cellular Splicing Factor SRp20 during Picornavirus Infections. J. Virol. 2013, 87, 2390–2400. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jackson, T.; Belsham, G.J. Picornaviruses: A View from 3A. Viruses 2021, 13, 456. [Google Scholar] [CrossRef]
- White, J.P.; Cardenas, A.M.; Marissen, W.E.; Lloyd, R.E. Inhibition of Cytoplasmic MRNA Stress Granule Formation by a Viral Proteinase. Cell Host Microbe 2007, 2, 295–305. [Google Scholar] [CrossRef]
- Yi, S.-J.; Kim, K. Histone Tail Cleavage as a Novel Epigenetic Regulatory Mechanism for Gene Expression. BMB Rep. 2018, 51, 211–218. [Google Scholar] [CrossRef]
- Das, C.; Tyler, J.K. Histone Exchange and Histone Modifications during Transcription and Aging. Biochim. Biophys. Acta 2013, 1819, 332–342. [Google Scholar] [CrossRef]
- Werbeck, N.D.; Shukla, V.K.; Kunze, M.B.A.; Yalinca, H.; Pritchard, R.B.; Siemons, L.; Mondal, S.; Greenwood, S.O.R.; Kirkpatrick, J.; Marson, C.M.; et al. A Distal Regulatory Region of a Class I Human Histone Deacetylase. Nat. Commun. 2020, 11, 3841. [Google Scholar] [CrossRef]
- Zhou, P.; Wu, E.; Alam, H.B.; Li, Y. Histone Cleavage as a Mechanism for Epigenetic Regulation: Current Insights and Perspectives. Curr. Mol. Med. 2014, 14, 1164–1172. [Google Scholar] [CrossRef]
- Shi, J.; Perryman, J.M.; Yang, X.; Liu, X.; Musser, D.M.; Boehr, A.K.; Moustafa, I.M.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Rational Control of Poliovirus RNA-Dependent RNA Polymerase Fidelity by Modulating Motif-D Loop Conformational Dynamics. Biochemistry 2019, 58, 3735–3743. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; Liu, X.; Yang, X. Targeting Structural Dynamics of the RNA-Dependent RNA Polymerase for Anti-Viral Strategies. Curr. Opin. Virol. 2014, 9, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Gohara, D.W.; Crotty, S.; Arnold, J.J.; Yoder, J.D.; Andino, R.; Cameron, C.E. Poliovirus RNA-Dependent RNA Polymerase (3Dpol): Structural, Biochemical, and Biological Analysis of Conserved Structural Motifs A and B. J. Biol. Chem. 2000, 275, 25523–25532. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, X.; Lee, C.A.; Moustafa, I.M.; Smidansky, E.D.; Lum, D.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Vaccine-Derived Mutation in Motif D of Poliovirus RNA-Dependent RNA Polymerase Lowers Nucleotide Incorporation Fidelity*. J. Biol. Chem. 2013, 288, 32753–32765. [Google Scholar] [CrossRef]
- Sholders, A.J.; Peersen, O.B. Distinct Conformations of a Putative Translocation Element in Poliovirus Polymerase. J. Mol. Biol. 2014, 426, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Kundu, P.; Raychaudhuri, S.; Tsai, W.; Dasgupta, A. Shutoff of RNA Polymerase II Transcription by Poliovirus Involves 3C Protease-Mediated Cleavage of the TATA-Binding Protein at an Alternative Site: Incomplete Shutoff of Transcription Interferes with Efficient Viral Replication. J. Virol. 2005, 79, 9702–9713. [Google Scholar] [CrossRef]
- Weng, K.F.; Li, M.L.; Hung, C.T.; Shih, S.R. Enterovirus 71 3C Protease Cleaves a Novel Target CstF-64 and Inhibits Cellular Polyadenylation. PLoS Pathog. 2009, 5, e1000593. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, H.; Zhu, Z.; Yang, F.; Ma, L.; Cai, X.; Xue, Q.; Zheng, H. Seneca Valley Virus 3Cpro Abrogates the IRF3- and IRF7-Mediated Innate Immune Response by Degrading IRF3 and IRF7. Virology 2018, 518, 1–7. [Google Scholar] [CrossRef]
- Wen, W.; Yin, M.; Zhang, H.; Liu, T.; Chen, H.; Qian, P.; Hu, J.; Li, X. Seneca Valley Virus 2C and 3C Inhibit Type I Interferon Production by Inducing the Degradation of RIG-I. Virology 2019, 535, 122–129. [Google Scholar] [CrossRef]
- Martínez-Salas, E. The Impact of RNA Structure on Picornavirus IRES Activity. Trends Microbiol. 2008, 16, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Yalamanchili, P.; Weidman, K.; Dasgupta, A. Cleavage of Transcriptional Activator Oct-1 by Poliovirus Encoded Protease 3C Pro. Virology 1997, 239, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Maggioli, M.F.; Fernandes, M.H.V.; Joshi, L.R.; Sharma, B.; Tweet, M.M.; Noll, J.C.G.; Bauermann, F.V.; Diel, D.G. Persistent Infection and Transmission of Senecavirus A from Carrier Sows to Contact Piglets. J. Virol. 2019, 93, e00819-19. [Google Scholar] [CrossRef] [PubMed]
- Galicia-Vázquez, G.; Cencic, R.; Robert, F.; Agenor, A.Q.; Pelletier, J. A Cellular Response Linking EIF4AI Activity to EIF4AII Transcription. RNA 2012, 18, 1373–1384. [Google Scholar] [CrossRef]
- Kim, Y.; Lovell, S.; Tiew, K.-C.; Mandadapu, S.R.; Alliston, K.R.; Battaile, K.P.; Groutas, W.C.; Chang, K.-O. Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses. J. Virol. 2012, 86, 11754–11762. [Google Scholar] [CrossRef]
- Novoa, I.; Carrasco, L. Cleavage of Eukaryotic Translation Initiation Factor 4G by Exogenously Added Hybrid Proteins Containing Poliovirus 2Apro in HeLa Cells: Effects on Gene Expression. Mol. Cell Biol. 1999, 19, 2445–2454. [Google Scholar] [CrossRef]
- Kafasla, P.; Lin, H.; Curry, S.; Jackson, R.J. Activation of Picornaviral IRESs by PTB Shows Differential Dependence on Each PTB RNA-Binding Domain. RNA 2011, 17, 1120–1131. [Google Scholar] [CrossRef]
- Gradi, A.; Foeger, N.; Strong, R.; Svitkin, Y.V.; Sonenberg, N.; Skern, T.; Belsham, G.J. Cleavage of Eukaryotic Translation Initiation Factor 4GII within Foot-and-Mouth Disease Virus-Infected Cells: Identification of the L-Protease Cleavage Site In Vitro. J. Virol. 2004, 78, 3271–3278. [Google Scholar] [CrossRef]
- Huot, M.-É.; Brown, C.M.; Lamarche-Vane, N.; Richard, S. An Adaptor Role for Cytoplasmic Sam68 in Modulating Src Activity during Cell Polarization. Mol. Cell Biol. 2009, 29, 1933–1943. [Google Scholar] [CrossRef]
- Ullmer, W.; Semler, B.L. Diverse Strategies Used by Picornaviruses to Escape Host RNA Decay Pathways. Viruses 2016, 8, 335. [Google Scholar] [CrossRef]
- De Jesús-González, L.A.; Palacios-Rápalo, S.; Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Gutiérrez-Escolano, A.L.; Del Ángel, R.M. The Nuclear Pore Complex Is a Key Target of Viral Proteases to Promote Viral Replication. Viruses 2021, 13, 706. [Google Scholar] [CrossRef]
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T.; et al. The Molecular Architecture of the Nuclear Pore Complex. Nature 2007, 450, 695–701. [Google Scholar] [CrossRef]
- Grossman, E.; Medalia, O.; Zwerger, M. Functional Architecture of the Nuclear Pore Complex. Annu. Rev. Biophys. 2012, 41, 557–584. [Google Scholar] [CrossRef]
- Lizcano-Perret, B.; Michiels, T. Nucleocytoplasmic Trafficking Perturbation Induced by Picornaviruses. Viruses 2021, 13, 1210. [Google Scholar] [CrossRef]
- Sun, D.; Wen, X.; Wang, M.; Mao, S.; Cheng, A.; Yang, X.; Jia, R.; Chen, S.; Yang, Q.; Wu, Y.; et al. Apoptosis and Autophagy in Picornavirus Infection. Front. Microbiol. 2019, 10, 2032. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Wang, S.; Meng, X.; Song, F.; Huo, W.; Zhang, S.; Chang, J.; Li, J.; Zheng, B.; et al. Coxsackievirus A6 Induces Cell Cycle Arrest in G0/G1 Phase for Viral Production. Front. Cell Infect. Microbiol. 2018, 8, 279. [Google Scholar] [CrossRef] [PubMed]
- Shubin, A.V.; Demidyuk, I.V.; Lunina, N.A.; Komissarov, A.A.; Roschina, M.P.; Leonova, O.G.; Kostrov, S. V Protease 3C of Hepatitis A Virus Induces Vacuolization of Lysosomal/Endosomal Organelles and Caspase-Independent Cell Death. BMC Cell Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Calandria, C.; Irurzun, A.; Barco, Á.; Carrasco, L. Individual Expression of Poliovirus 2Apro and 3Cpro Induces Activation of Caspase-3 and PARP Cleavage in HeLa Cells. Virus Res. 2004, 104, 39–49. [Google Scholar] [CrossRef]
- Odell, A.F.; Mannion, A.J.; Jones, P.F.; Cook, G.P. Negative Regulation of P53 by the Poliovirus Receptor PVR Is a Target of a Human Cytomegalovirus Immune Evasion Molecule. bioRxiv 2022. [Google Scholar] [CrossRef]
- Autret, A.; Martin-Latil, S.; Mousson, L.; Wirotius, A.; Petit, F.; Arnoult, D.; Colbère-Garapin, F.; Estaquier, J.; Blondel, B. Poliovirus Induces Bax-Dependent Cell Death Mediated by c-Jun NH2-Terminal Kinase. J. Virol. 2007, 81, 7504–7516. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pampin, M.; Simonin, Y.; Blondel, B.; Percherancier, Y.; Chelbi-Alix, M.K. Cross Talk between PML and P53 during Poliovirus Infection: Implications for Antiviral Defense. J. Virol. 2006, 80, 8582–8592. [Google Scholar] [CrossRef]
- Belov, G.A.; Romanova, L.I.; Tolskaya, E.A.; Kolesnikova, M.S.; Lazebnik, Y.A.; Agol, V.I. The Major Apoptotic Pathway Activated and Suppressed by Poliovirus. J. Virol. 2003, 77, 45–56. [Google Scholar] [CrossRef]
- Li, M.-L.; Lin, J.-Y.; Chen, B.-S.; Weng, K.-F.; Shih, S.-R.; Calderon, J.D.; Tolbert, B.S.; Brewer, G. EV71 3C Protease Induces Apoptosis by Cleavage of HnRNP A1 to Promote Apaf-1 Translation. PLoS ONE 2019, 14, e0221048. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, Y.; Wu, C.; Zhou, D.; Gong, J.; Zhao, T.; Jin, Z. Development of FRET and Stress Granule Dual-Based System to Screen for Viral 3C Protease Inhibitors. Molecules 2023, 28, 3020. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Su, J.; Wang, H.; Chang, J.; Wang, S.; Zheng, W.; Cai, Y.; Wei, W.; Gordy, J.T.; Markham, R.; et al. Disruption of MDA5-Mediated Innate Immune Responses by the 3C Proteins of Coxsackievirus A16, Coxsackievirus A6, and Enterovirus D68. J. Virol. 2017, 91, 10-1128. [Google Scholar] [CrossRef]
- Song, J.; Quan, R.; Wang, D.; Liu, J. Seneca Valley Virus 3C pro Mediates Cleavage and Redistribution of Nucleolin To Facilitate Viral Replication. Microbiol. Spectr. 2022, 10, e0030422. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liu, Q.; Zhang, L.; Zhang, Q.; Hu, L.; Li, C.; Wang, S.; Li, J.; Zhang, Y.; Yu, H.; et al. Encephalomyocarditis Virus 3C Protease Relieves TRAF Family Member-Associated NF-KB Activator (TANK) Inhibitory Effect on TRAF6-Mediated NF-KB Signaling through Cleavage of TANK. J. Biol. Chem. 2015, 290, 27618–27632. [Google Scholar] [CrossRef]
- Hung, H.-C.; Wang, H.-C.; Shih, S.-R.; Teng, I.-F.; Tseng, C.-P.; Hsu, J.T.-A. Synergistic Inhibition of Enterovirus 71 Replication by Interferon and Rupintrivir. J. Infect. Dis. 2011, 203, 1784–1790. [Google Scholar] [CrossRef]
- Lei, X.; Liu, X.; Ma, Y.; Sun, Z.; Yang, Y.; Jin, Q.; He, B.; Wang, J. The 3C Protein of Enterovirus 71 Inhibits Retinoid Acid-Inducible Gene I-Mediated Interferon Regulatory Factor 3 Activation and Type I Interferon Responses. J. Virol. 2010, 84, 8051–8061. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morosky, S.A.; Delorme-Axford, E.; Dybdahl-Sissoko, N.; Oberste, M.S.; Wang, T.; Coyne, C.B. The Coxsackievirus B 3Cpro Protease Cleaves MAVS and TRIF to Attenuate Host Type I Interferon and Apoptotic Signaling. PLoS Pathog. 2011, 7, e1001311. [Google Scholar] [CrossRef]
- Zhao, T.; Yang, L.; Sun, Q.; Arguello, M.; Ballard, D.W.; Hiscott, J.; Lin, R. The NEMO Adaptor Bridges the Nuclear Factor-ΚB and Interferon Regulatory Factor Signaling Pathways. Nat. Immunol. 2007, 8, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liang, Y.; Qu, L.; Chen, Z.; Yi, M.; Li, K.; Lemon, S.M. Disruption of Innate Immunity Due to Mitochondrial Targeting of a Picornaviral Protease Precursor. Proc. Natl. Acad. Sci. USA 2007, 104, 7253–7258. [Google Scholar] [CrossRef]
- Bruurs, L.J.M.; Müller, M.; Schipper, J.G.; Rabouw, H.H.; Boersma, S.; van Kuppeveld, F.J.M.; Tanenbaum, M.E. Antiviral Responses Are Shaped by Heterogeneity in Viral Replication Dynamics. Nat. Microbiol. 2023, 8, 2115–2129. [Google Scholar] [CrossRef] [PubMed]
- Dias Junior, A.G.; Sampaio, N.G.; Rehwinkel, J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol. 2019, 27, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Andreas, P.; Oliver, S.; Choon-Ping, T.; Jan, R.; Hiroki, K.; Osamu, T.; Shizuo, A.; Michael, W.; Giampietro, S.; Caetano, R.e.S. Activation of MDA5 Requires Higher-Order RNA Structures Generated during Virus Infection. J. Virol. 2009, 83, 10761–10769. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like Receptors: Their Regulation and Roles in RNA Sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Savitsky, D.; Tamura, T.; Yanai, H.; Taniguchi, T. Regulation of Immunity and Oncogenesis by the IRF Transcription Factor Family. Cancer Immunol. Immunother. 2010, 59, 489–510. [Google Scholar] [CrossRef]
- Andersen, J.; VanScoy, S.; Cheng, T.-F.; Gomez, D.; Reich, N.C. IRF-3-Dependent and Augmented Target Genes during Viral Infection. Genes. Immun. 2008, 9, 168–175. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Andino, R. Switch from Translation to RNA Replication in a Positive-Stranded RNA Virus. Minerva Anestesiol. 1998, 12, 2293–2304. [Google Scholar] [CrossRef]
- Lloyd, R.E. Translational Control by Viral Proteinases. Virus Res. 2006, 119, 76–88. [Google Scholar] [CrossRef]
- Amero, C.D.; Arnold, J.J.; Moustafa, I.M.; Cameron, C.E.; Foster, M.P. Identification of the OriI-Binding Site of Poliovirus 3C Protein by Nuclear Magnetic Resonance Spectroscopy. J. Virol. 2008, 82, 4363–4370. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Gauss-Müller, V.; Cordes, S.; Tamura, R.; Okitsu, K.; Shuang, W.; Nakamoto, S.; Fujiwara, K.; Imazeki, F.; Yokosuka, O. Hepatitis A Virus (HAV) Proteinase 3C Inhibits HAV IRES-Dependent Translation and Cleaves the Polypyrimidine Tract-Binding Protein. J. Viral Hepat. 2010, 17, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Hoon, B.S.; Ki, K.Y.; Jae, K.W.; Sungchan, C.; Rang, O.H.; Jung-Eun, K.; Key, J.S. Translation of Polioviral MRNA Is Inhibited by Cleavage of Polypyrimidine Tract-Binding Proteins Executed by Polioviral 3Cpro. J. Virol. 2002, 76, 2529–2542. [Google Scholar] [CrossRef]
- Valesano, A.L.; Taniuchi, M.; Fitzsimmons, W.J.; Islam, M.O.; Ahmed, T.; Zaman, K.; Haque, R.; Wong, W.; Famulare, M.; Lauring, A.S. The Early Evolution of Oral Poliovirus Vaccine Is Shaped by Strong Positive Selection and Tight Transmission Bottlenecks. Cell Host Microbe 2021, 29, 32–43.e4. [Google Scholar] [CrossRef] [PubMed]
- Reineke, L.C.; Lloyd, R.E. The Stress Granule Protein G3BP1 Recruits Protein Kinase R to Promote Multiple Innate Immune Antiviral Responses. J. Virol. 2015, 89, 2575–2589. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.; Kanagavelu, S.; Fukata, M. A Unique Host Defense Pathway: TRIF Mediates Both Antiviral and Antibacterial Immune Responses. Microbes Infect. 2013, 15, 1–10. [Google Scholar] [CrossRef][Green Version]
- Lei, X.; Xiao, X.; Xue, Q.; Jin, Q.; He, B.; Wang, J. Cleavage of Interferon Regulatory Factor 7 by Enterovirus 71 3C Suppresses Cellular Responses. J. Virol. 2013, 87, 1690–1698. [Google Scholar] [CrossRef]
- Andino, R.; Rieckhof, G.E.; Achacoso, P.L.; Baltimore, D. Poliovirus RNA Synthesis Utilizes an RNP Complex Formed around the 5’-End of Viral RNA. EMBO J. 1993, 12, 3587–3598. [Google Scholar] [CrossRef]
- Fass, D.M.; Reis, S.A.; Ghosh, B.; Hennig, K.M.; Joseph, N.F.; Zhao, W.N.; Nieland, T.J.; Guan, J.S.; Kuhnle, C.E.G.; Tang, W.; et al. Crebinostat: A novel cognitive enhancer that inhibits histone deacetylase activity and modulates chromatin-mediated neuroplasticity. Neuropharmacology 2013, 64, 81–96. [Google Scholar] [CrossRef]
- Paul, A.V.; Wimmer, E. Initiation of Protein-Primed Picornavirus RNA Synthesis. Virus Res. 2015, 206, 12–26. [Google Scholar] [CrossRef]
- Herold, J.; Andino, R. Poliovirus RNA Replication Requires Genome Circularization through a Protein–Protein Bridge. Mol. Cell 2001, 7, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Parsley, T.B.; Towner, J.S.; Blyn, L.B.; Ehrenfeld, E.; Semler, B.L. Poly (RC) Binding Protein 2 Forms a Ternary Complex with the 5’-Terminal Sequences of Poliovirus RNA and the Viral 3CD Proteinase. RNA 1997, 3, 1124–1134. [Google Scholar] [PubMed]
- Harris, K.S.; Xiang, W.; Alexander, L.; Lane, W.S.; Paul, A.V.; Wimmer, E. Interaction of Poliovirus Polypeptide 3CDpro with the 5′ and 3′ Termini of the Poliovirus Genome. Identification of Viral and Cellular Cofactors Needed for Efficient Binding. J. Biol. Chem. 1994, 269, 27004–27014. [Google Scholar] [CrossRef] [PubMed]
- Xiang, W.; Harris, K.S.; Alexander, L.; Wimmer, E. Interaction between the 5’-Terminal Cloverleaf and 3AB/3CDpro of Poliovirus Is Essential for RNA Replication. J. Virol. 1995, 69, 3658–3667. [Google Scholar] [CrossRef]
- Yang, Y.; Rijnbrand, R.; Watowich, S.; Lemon, S.M. Genetic Evidence for an Interaction between a Picornaviral Cis-Acting RNA Replication Element and 3CD Protein*. J. Biol. Chem. 2004, 279, 12659–12667. [Google Scholar] [CrossRef]
- Shen, M.; Wang, Q.; Yang, Y.; Pathak, H.B.; Arnold, J.J.; Castro, C.; Lemon, S.M.; Cameron, C.E. Human Rhinovirus Type 14 Gain-of-Function Mutants for OriI Utilization Define Residues of 3C(D) and 3Dpol That Contribute to Assembly and Stability of the Picornavirus VPg Uridylylation Complex. J. Virol. 2007, 81, 12485–12495. [Google Scholar] [CrossRef]
- Kusov, Y.Y.; Gauss-Müller, V. In Vitro RNA Binding of the Hepatitis A Virus Proteinase 3C (HAV 3Cpro) to Secondary Structure Elements within the 5’ Terminus of the HAV Genome. RNA 1997, 3, 291–302. [Google Scholar]
- Peters, H.; Kusov, Y.Y.; Meyer, S.; Benie, A.J.; Bäuml, E.; Wolff, M.; Rademacher, C.; Peters, T.; Gauss-Müller, V. Hepatitis A Virus Proteinase 3C Binding to Viral RNA: Correlation with Substrate Binding and Enzyme Dimerization. Biochem. J. 2005, 385, 363–370. [Google Scholar] [CrossRef]
- Kusov, Y.Y.; Morace, G.; Probst, C.; Gauss-Müller, V. Interaction of Hepatitis A Virus (HAV) Precursor Proteins 3AB and 3ABC with the 5’ and 3’ Termini of the HAV RNA. Virus Res. 1997, 51, 151–157. [Google Scholar] [CrossRef]
- Walker, P.A.; Leong, L.E.-C.; Porter, A.G. Sequence and Structural Determinants of the Interaction between the 5′-Noncoding Region of Picornavirus RNA and Rhinovirus Protease 3C*. J. Biol. Chem. 1995, 270, 14510–14516. [Google Scholar] [CrossRef]
- Leong, L.E.; Walker, P.A.; Porter, A.G. Human Rhinovirus-14 Protease 3C (3Cpro) Binds Specifically to the 5′-Noncoding Region of the Viral RNA. Evidence That 3Cpro Has Different Domains for the RNA Binding and Proteolytic Activities. J. Biol. Chem. 1993, 268, 25735–25739. [Google Scholar] [CrossRef] [PubMed]
- Nayak, A.; Goodfellow, I.G.; Woolaway, K.E.; Birtley, J.; Curry, S.; Belsham, G.J. Role of RNA Structure and RNA Binding Activity of Foot-and-Mouth Disease Virus 3C Protein in VPg Uridylylation and Virus Replication. J. Virol. 2006, 80, 9865–9875. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Molla, A.; Harris, K.S.; Paul, A.V.; Shin, S.H.; Mugavero, J.; Wimmer, E. Stimulation of Poliovirus Proteinase 3Cpro-Related Proteolysis by the Genome-Linked Protein VPg and Its Precursor 3AB. J. Biol. Chem. 1994, 269, 27015–27020. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Welch, J.L.; Arnold, J.J.; Boehr, D.D. Long-Range Interaction Networks in the Function and Fidelity of Poliovirus RNA-Dependent RNA Polymerase Studied by Nuclear Magnetic Resonance. Biochemistry 2010, 49, 9361–9371. [Google Scholar] [CrossRef] [PubMed]
- Plotch, S.J.; Palant, O. Poliovirus Protein 3AB Forms a Complex with and Stimulates the Activity of the Viral RNA Polymerase, 3Dpol. J. Virol. 1995, 69, 7169–7179. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, K.; Bhat, R.; Rao, V.U.B.; Nain, A.; Rallapalli, K.L.; Gangopadhyay, S.; Singh, R.P.; Banerjee, M.; Jayaram, B. Toward Development of Generic Inhibitors against the 3C Proteases of Picornaviruses. FEBS J. 2019, 286, 765–787. [Google Scholar] [CrossRef] [PubMed]
- Gouvea, I.E.; Santos, J.A.N.; Burlandy, F.M.; Tersariol, I.L.S.; da Silva, E.E.; Juliano, M.A.; Juliano, L.; Cunha, R.L.O.R. Poliovirus 3C Proteinase Inhibition by Organotelluranes. Biol. Chem. 2011, 392, 587–591. [Google Scholar] [CrossRef]
- McKinlay, M.A.; Collett, M.S.; Hincks, J.R.; Oberste, M.S.; Pallansch, M.A.; Okayasu, H.; Sutter, R.W.; Modlin, J.F.; Dowdle, W.R. Progress in the Development of Poliovirus Antiviral Agents and Their Essential Role in Reducing Risks That Threaten Eradication. J. Infect. Dis. 2014, 210, S447–S453. [Google Scholar] [CrossRef]
- Rhoden, E.; Liu, H.-M.; Wang-Chern, S.; Oberste, M.S. Anti-Poliovirus Activity of Protease Inhibitor AG-7404, and Assessment of In Vitro Activity in Combination with Antiviral Capsid Inhibitor Compounds. Antivir. Res. 2013, 98, 186–191. [Google Scholar] [CrossRef]
- Patick, A.K.; Brothers, M.A.; Maldonado, F.; Binford, S.; Maldonado, O.; Fuhrman, S.; Petersen, A.; Smith, G.J.; Zalman, L.S.; Burns-Naas, L.A.; et al. In Vitro Antiviral Activity and Single-Dose Pharmacokinetics in Humans of a Novel, Orally Bioavailable Inhibitor of Human Rhinovirus 3C Protease. Antimicrob. Agents Chemother. 2005, 49, 2267–2275. [Google Scholar] [CrossRef]
- Groaz, E.; De Clercq, E.; Herdewijn, P. Anno 2021: Which Antivirals for the Coming Decade? Annu. Rep. Med. Chem. 2021, 57, 49–107. [Google Scholar] [CrossRef] [PubMed]
- Jagdeo, J.M.; Dufour, A.; Klein, T.; Solis, N.; Kleifeld, O.; Kizhakkedathu, J.; Luo, H.; Overall, C.M.; Jan, E. N-Terminomics TAILS Identifies Host Cell Substrates of Poliovirus and Coxsackievirus B3 3C Proteinases That Modulate Virus Infection. J. Virol. 2018, 92, e02211-17. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, A. Molecular Aspects of Poliovirus Pathogenesis. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2007, 83, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Kafasla, P.; Morgner, N.; Robinson, C.V.; Jackson, R.J. Polypyrimidine Tract-Binding Protein Stimulates the Poliovirus IRES by Modulating EIF4G Binding. EMBO J. 2010, 29, 3710–3722. [Google Scholar] [CrossRef]
- Chase, A.J.; Daijogo, S.; Semler, B.L. Inhibition of Poliovirus-Induced Cleavage of Cellular Protein PCBP2 Reduces the Levels of Viral RNA Replication. J. Virol. 2014, 88, 3192–3201. [Google Scholar] [CrossRef] [PubMed]
- Kuyumcu-Martinez, N.M.; Joachims, M.; Lloyd, R.E. Efficient Cleavage of Ribosome-Associated Poly(A)-Binding Protein by Enterovirus 3C Protease. J. Virol. 2002, 76, 2062–2074. [Google Scholar] [CrossRef] [PubMed]
- Kuyumcu-Martinez, M.; Belliot, G.; Sosnovtsev, S.V.; Chang, K.-O.; Green, K.Y.; Lloyd, R.E. Calicivirus 3C-like Proteinase Inhibits Cellular Translation by Cleavage of Poly(A)-Binding Protein. J. Virol. 2004, 78, 8172–8182. [Google Scholar] [CrossRef]
- Wan, Q.; Song, D.; Li, H.; He, M. Stress Proteins: The Biological Functions in Virus Infection, Present and Challenges for Target-Based Antiviral Drug Development. Signal Transduct. Target. Ther. 2020, 5, 125. [Google Scholar] [CrossRef]
- Gorman, S.D.; D’Amico, R.N.; Winston, D.S.; Boehr, D.D. Engineering Allostery into Proteins BT—Protein Allostery in Drug Discovery; Zhang, J., Nussinov, R., Eds.; Springer: Singapore, 2019; pp. 359–384. ISBN 978-981-13-8719-7. [Google Scholar]
- Ma, Y.; Li, L.; He, S.; Shang, C.; Sun, Y.; Liu, N.; Meek, T.D.; Wang, Y.; Shang, L. Application of Dually Activated Michael Acceptor to the Rational Design of Reversible Covalent Inhibitor for Enterovirus 71 3C Protease. J. Med. Chem. 2019, 62, 6146–6162. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, S.; Huang, Y.; Chen, F.; Chen, L.; Ding, D.; Zheng, Y.; Li, H.; Xiao, J.; Feng, J.; et al. Virus-like Particle Vaccines for Poliovirus Types 1, 2, and 3 with Enhanced Thermostability Expressed in Insect Cells. Vaccine 2019, 37, 2340–2347. [Google Scholar] [CrossRef]
- Kassem, A.F.; Batran, R.Z.; Abbas, E.M.H.; Elseginy, S.A.; Shaheen, M.N.F.; Elmahdy, E.M. New 4-Phenylcoumarin Derivatives as Potent 3C Protease Inhibitors: Design, Synthesis, Anti-HAV Effect and Molecular Modeling. Eur. J. Med. Chem. 2019, 168, 447–460. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).