Identification, Sequencing, and Molecular Analysis of RNA2 of Artichoke Italian Latent Virus Isolates from Known Hosts and a New Host Plant Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source of Plant Material
2.2. Extraction of Total Nucleic Acids, cDNA Synthesis, and PCR
2.3. Cloning and Sequencing
2.4. Sequence and Recombination Analyses
3. Results
3.1. Complete Sequence of Genomic RNA2 of AILV Isolates
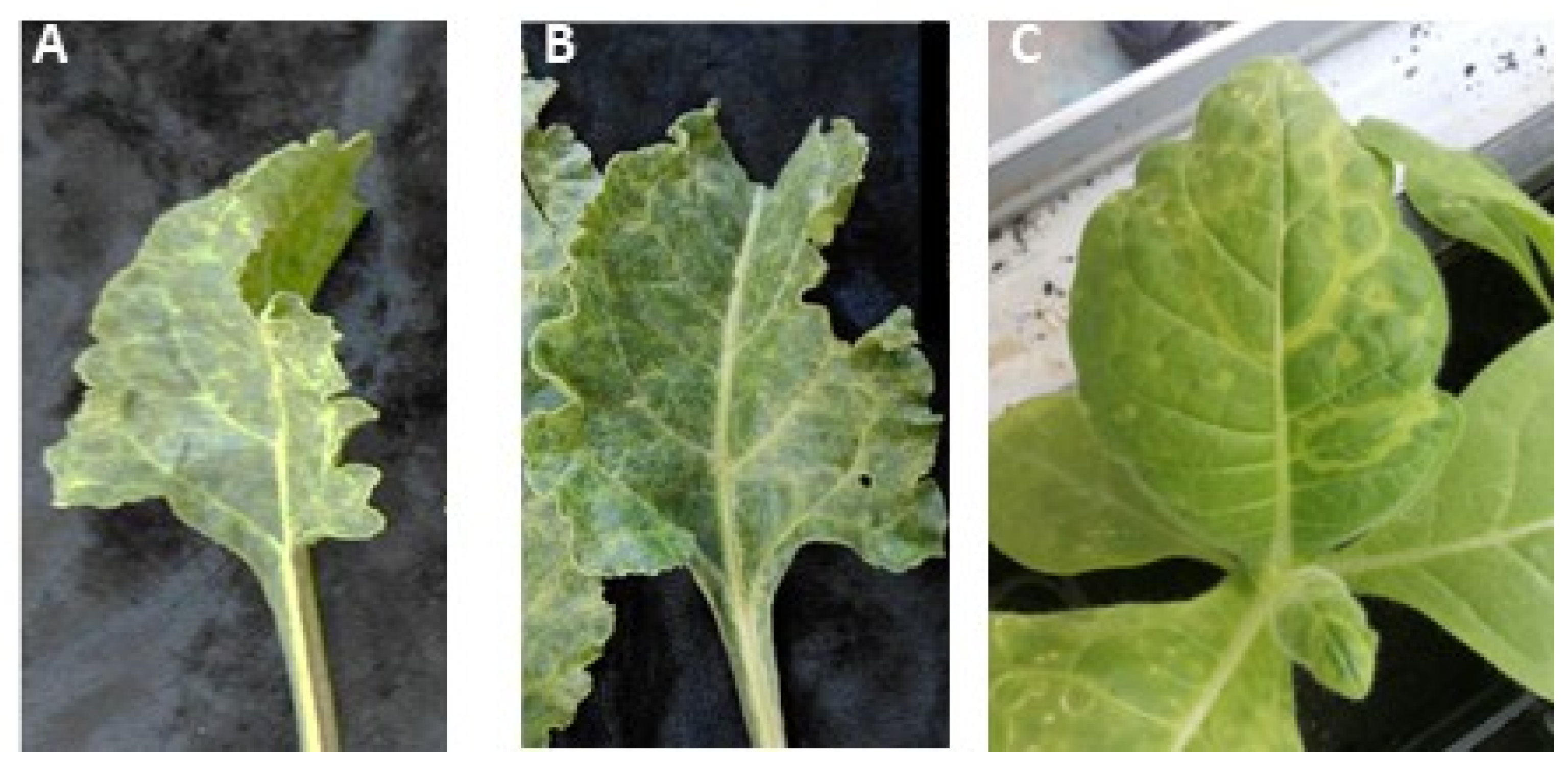
3.2. Identification of AILV in Chard Plants
3.3. Mechanical Transmission of AILV Chard Plants onto Herbaceous Host
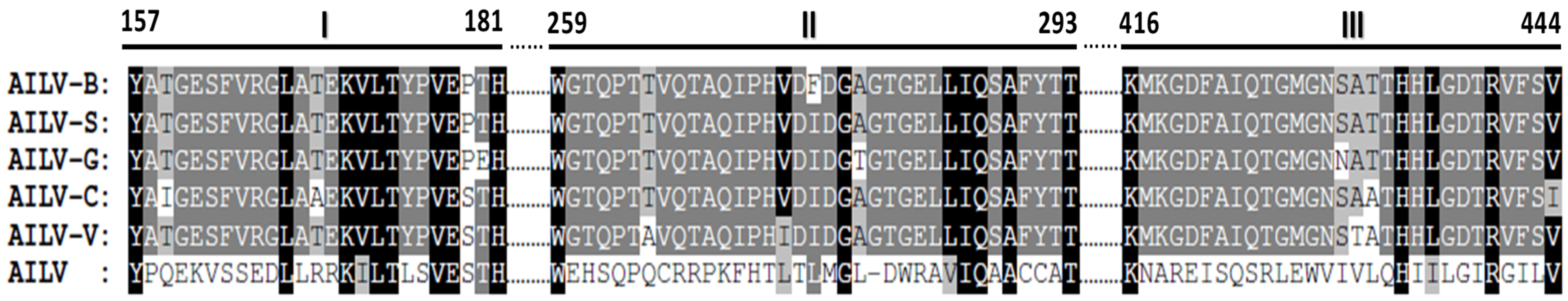
3.4. Genetic Variability of Genomic RNA2 of AILV Isolates
3.5. Phylogenetic Analysis of AILV Isolates
3.6. Recombination Analysis
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cadman, C.H. Biology of Soil-Borne Viruses. Ann. Rev. Phytopathol. 1963, 1, 143–172. [Google Scholar] [CrossRef]
- Harrison, B.D.; Murant, A.F. Nepovirus Group. CMI/AAB. Descriptions of Plant Viruses. Neth. J. Plant Pathol. 1977, 185, 64. [Google Scholar]
- Sanfacon, H. Secoviridae: A Family of Plant Picorna-Like Viruses with Monopartite or Bipartite Genomes. eLS 2015, 1–14. [Google Scholar] [CrossRef]
- ICTV. 2023. Available online: https://ictv.global (accessed on 13 July 2023).
- Majorana, G.; Rana, G.L. Un nuovo virus latente isolato da Carciofo in Puglia/a new latent virus isolated from Artichoke in Apulia. Phytopathol Medit. 1970, 9, 193–196. [Google Scholar]
- Fuchs, M.; Schmitt-Keichinger, C.; Sanfaçon, H. A renaissance in nepovirus research provides new insights into their molecular interface with hosts and vectors. Adv. Virus Res. 2017, 97, 1–105. [Google Scholar]
- Vovlas, C.; Martelli, G.P.; Quacquarelli, A. Le virosi delle piante ortensi in Puglia. VI. II complesso delle maculature anulari della Cicoria. Phytopathol. Medit. 1971, 10, 244–254. [Google Scholar]
- Vovlas, C. Le malformazioni fogliari, una nuova virosi del Geranio. Phytopathol. Medit. 1974, 13, 139–142. [Google Scholar]
- Quacquarelli, A.; Martelli, G.P. Ricerche sull’agente dell’arricciamento maculato del carciofo. I. Ospiti differenziali e proprietà. In Proceedings of the 1st Congr. Unione Fitopatol. Medit, Bari-Napoli, Italy, 26 September–1 October 1966; pp. 168–177. [Google Scholar]
- Savino, V.; Gallitelli, D.; Jankulova, M.; Rana, G.L. A comparison of four isolates of artichoke Italian latent virus (AILV). Phytopathol. Medit. 1977, 16, 41. [Google Scholar]
- Jankulova, M.; Savino, V.; Gallitelli, D.; Quacquarelli, A.; Martelli, G.P. Isolation of artichoke Italian latent virus from the grapevine in Bulgaria. In Proceedings of the 6th ICVG Meeting, Cordoba, Spain, 12–17 September 1976; Monografias INIA, Madrid, Spain. 18, pp. 143–148. [Google Scholar]
- Quacquarelli, A.; Rana, G.L.; Martelli, G.P. Some weeds as host of pathogenic viruses in Apulia. Poljopr. Znan. Smotra 1976, 39, 561. [Google Scholar]
- Elbeaino, T.; Belghacem, I.; Mascia, T.; Gallitelli, D.; Digiaro, M. Next generation sequencing and molecular analysis of artichoke Italian latent virus. Arch. Virol. 2017, 162, 1805–1809. [Google Scholar] [CrossRef]
- Le Gall, O.; Lanneau, M.; Candresse, T.; Dunez, J. The nucleotide sequence of the RNA-2 of an isolate of the English serotype of tomato black ring virus: RNA recombination in the history of nepoviruses. J. General Virol. 1995, 76, 1279–1283. [Google Scholar] [CrossRef]
- Vigne, E.; Marmonier, A.; Fuchs, M. Multiple interspecies recombination events within RNA2 of Grapevine fanleaf virus and Arabis mosaic virus. Arch. Virol. 2008, 153, 1771–1776. [Google Scholar] [CrossRef] [PubMed]
- Elbeaino, T.; Digiaro, M.; Gerbermeskel, S.; Martelli, G.P. Grapevine deformation virus: Complete sequencing and evidence of recombination events derived from Grapevine fanleaf virus and Arabis mosaic virus. Virus Res. 2012, 166, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Digiaro, M.; Yahyaoui, E.; Martelli, G.P.; Elbeaino, T. The sequencing of the complete genome of a Tomato black ring virus (TBRV) and of the RNA2 of three Grapevine chrome mosaic virus (GCMV) isolates from grapevine reveals the possible recombinant origin of GCMV. Virus Genes 2015, 50, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Digiaro, M.; Nehdi, S.; Elbeaino, T. Complete sequence of RNA1 of Grapevine Anatolian ringspot virus. Arch. Virol. 2012, 157, 2013–2016. [Google Scholar] [CrossRef] [PubMed]
- Elbeaino, T.; Digiaro, M.; Fallanaj, F.; Kuzmanovic, S.; Martelli, G.P. Complete nucleotides sequence and genome organization of Grapevine Bulgarian latent virus. Arch. Virol. 2011, 156, 875–879. [Google Scholar] [CrossRef]
- Walker, M.; Chisholm, J.; Wei, T.; Ghoshal, B.; Saeed, H.; Rott, M.; Sanfaçon, H. Complete genome sequence of three tomato ringspot virus isolates: Evidence for reassortment and recombination. Arch. Virol. 2015, 60, 543–547. [Google Scholar] [CrossRef]
- Salleh, W.; Minutillo, S.A.; Spano, R.; Zammouri, S.; Gallitelli, D.; Mnari-Hattab, M. Occurrence of artichoke-infecting viruses in Tunisia. EPPO Bull. 2017, 47, 48–56. [Google Scholar] [CrossRef]
- Minutillo, S.A.; Mascia, T.; Gallitelli, D. A DNA probe mix for the multiplex detection of ten artichoke viruses. Eur. J. Plant Pathol. 2012, 134, 459–465. [Google Scholar] [CrossRef]
- Diener, T.O.; Schneider, I.R. Virus degradation and nucleic acid release in single-phase phenol systems. Arch. Biochem. Biophys. 1968, 124, 401–412. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sanfaçon, H. Re-examination of nepovirus polyprotein cleavage sites highlights the diverse specificities and evolutionary relationships of nepovirus 3C-like protease. Arch. Virol. 2022, 167, 2529–2543. [Google Scholar] [CrossRef]
- Grieco, F.; Saponari, M.; Alkowni, R.; Savino, V.; Garau, R.; Martelli, G.P. Progress in diagnosis of olive viruses. Infor. Fitopatol. 2000, 11, 49–52. [Google Scholar]
- Chen, L.; Brannigan, K.; Clark, R.; Gilbertson, R.L. Characterization of curtoviruses associated with curly top disease of tomato in California and monitoring for these viruses in beet leafhoppers. Plant Dis. 2010, 94, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Nemchinov, L.G.; Hammond, J.; Jordan, R.; Hammond, R.W. The complete nucleotide sequence, genome organization, and specific detection of Beet mosaic virus. Arch. Virol. 2004, 149, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Kundu, K.; Rysánek, P. Detection of beet yellows virus by RT-PCR and immunocapture RT-PCR in Tetragonia expansa and Beta vulgaris. Acta Virol. 2004, 48, 177–182. [Google Scholar] [PubMed]
- Sabokkhiz, M.A.; Jafarpour, B.; Shahriari Ahmadi, F.; Tarighi, S.I. dentification of Turnip mosaic virus isolated from Canola in northeast area of Iran. Afr. J. Biotechnol. 2012, 11, 14553–14560. [Google Scholar]
- Chaouachi, M.; Fortabat, M.N.; Geldreich, A.; Yot, P.; Kerlan, C.; Kebdani, N.; Audeon, C.; Romaniuk, M.; Bertheau, Y. An accurate real-time PCR test for the detection and quantification of cauliflower mosaic virus (CaMV): Applicable in GMO screening. Eur. Food Res. Technol. 2007, 227, 789–798. [Google Scholar] [CrossRef]
- Fuchs, M.; Jean-Michel Hily, J.M.; Petrzik, K.; Sanfaçon, H.; Thompson, J.R.; van der Vlugt, R.; Wetzel, T.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Secoviridae. J. Gen. Virol. 2022, 103, 001807. [Google Scholar] [CrossRef]
- Hily, J.M.; Poulicard, N.; Kubina, J.; Reynard, J.S.; Spilmont, A.S.; Fuchs, M.; Lemaire, O.; and Vigne, E. Metagenomic analysis of nepoviruses: Diversity, evolution and identification of a genome region in members of subgroup A that appears to be important for host range. Arch. Virol. 2021, 166, 2789–2801. [Google Scholar] [CrossRef]

| Virus-Isolate | AILV-B | AILV-S | AILV-C | AILV-G | AILV-V |
|---|---|---|---|---|---|
| AILV-B | ID 1 | 98.9 | 94.6 | 96.2 | 85.2 |
| AILV-S | 98.2 | ID | 95.6 | 97.3 | 86.2 |
| AILV-C | 92.7 | 94.4 | ID | 97.3 | 87.5 |
| AILV-G | 92.7 | 94.4 | 100 | ID | 88.2 |
| AILV-V | 93.0 | 94.7 | 99.6 | 99.6 | ID |
| Virus-Isolate | AILV-B | AILV-S | AILV-C | AILV-G | AILV-V |
|---|---|---|---|---|---|
| AILV-B | ID 1 | 97.8 | 91.6 | 88.7 | 84.7 |
| AILV-S | 99 | ID | 93.7 | 90.7 | 86.4 |
| AILV-C | 88.5 | 89.3 | ID | 90 | 85.9 |
| AILV-G | 86.8 | 87.7 | 86.1 | ID | 90.1 |
| AILV-V | 82.3 | 83.1 | 82.2 | 86.2 | ID |
| Virus-Isolate | AILV-B | AILV-S | AILV-C | AILV-G | AILV-V | AILV-X87254 |
|---|---|---|---|---|---|---|
| AILV-B | ID 1 | 99 | 94.9 | 96.8 | 94.9 | 80.4 |
| AILV-S | 99.6 | ID | 95.8 | 97.8 | 95.7 | 81 |
| AILV-C | 88.1 | 88.4 | ID | 94.9 | 94.5 | 80 |
| AILV-G | 93.1 | 93.5 | 88.4 | ID | 94.7 | 80.2 |
| AILV-V | 88.6 | 88.9 | 87.3 | 89 | ID | 84.3 |
| AILV-X87254 | 85.9 | 86.3 | 84.8 | 86.3 | 94.5 | ID |
| Domain | Isolate | Position (nt) | Parental Isolates (Major × Minor) | RDP4 (p Value) 1 |
|---|---|---|---|---|
| 2A | AILV-C | 251-300 | AILV-V × AILV-B | RGBMC3sS (4.516 × 10−64) |
| 2A | AILV-G | 730-800 | AILV-S × AILV-V | RGBMC3sS (1.462 × 10−67) |
| 2B | AILV-C | 1292-1406 | AILV-V × AILV-C | RGBMC3sS (1.124 × 10−71) |
| 2B | AILV-C | 1304-1408 | AILV-V × AILV-B | RGBMC3sS(4.734 × 10−38) |
| 2C | AILV-G | 2245- 2851 | AILV-B × AILV-V | RGBMC3sS (1.561 × 10−27) |
| 2C | AILV-C | 2762-2961 | AILV-B × AILV-V | RGBMC3sS (1.053 × 10−29) |
| 3′UTR | AILV-G | 4450-4621 | AILV-B × AILV-C | RGBMC3sS (2.573 × 10−45) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbeaino, T.; Ben Slimen, A.; Belgacem, I.; Mnari-Hattab, M.; Spanò, R.; Digiaro, M.; Abdelkhalek, A. Identification, Sequencing, and Molecular Analysis of RNA2 of Artichoke Italian Latent Virus Isolates from Known Hosts and a New Host Plant Species. Viruses 2023, 15, 2170. https://doi.org/10.3390/v15112170
Elbeaino T, Ben Slimen A, Belgacem I, Mnari-Hattab M, Spanò R, Digiaro M, Abdelkhalek A. Identification, Sequencing, and Molecular Analysis of RNA2 of Artichoke Italian Latent Virus Isolates from Known Hosts and a New Host Plant Species. Viruses. 2023; 15(11):2170. https://doi.org/10.3390/v15112170
Chicago/Turabian StyleElbeaino, Toufic, Amani Ben Slimen, Imen Belgacem, Monia Mnari-Hattab, Roberta Spanò, Michele Digiaro, and Ahmed Abdelkhalek. 2023. "Identification, Sequencing, and Molecular Analysis of RNA2 of Artichoke Italian Latent Virus Isolates from Known Hosts and a New Host Plant Species" Viruses 15, no. 11: 2170. https://doi.org/10.3390/v15112170
APA StyleElbeaino, T., Ben Slimen, A., Belgacem, I., Mnari-Hattab, M., Spanò, R., Digiaro, M., & Abdelkhalek, A. (2023). Identification, Sequencing, and Molecular Analysis of RNA2 of Artichoke Italian Latent Virus Isolates from Known Hosts and a New Host Plant Species. Viruses, 15(11), 2170. https://doi.org/10.3390/v15112170










