A Novel Rotavirus Reverse Genetics Platform Supports Flexible Insertion of Exogenous Genes and Enables Rapid Development of a High-Throughput Neutralization Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmid Construction
2.3. Recombinant Rotavirus Rescue
2.4. Virus Infection
2.5. Virus Stock Preparation
2.6. Plaque Assay
2.7. Flow Cytometry and Data Analysis
2.8. Growth Kinetics
2.9. Genetic Stability
2.10. ELISA
2.11. rSA11-GFP Based Micro-Neutralization Assay
2.12. Animal and Human Serum Samples
2.13. Serum Purification
3. Results
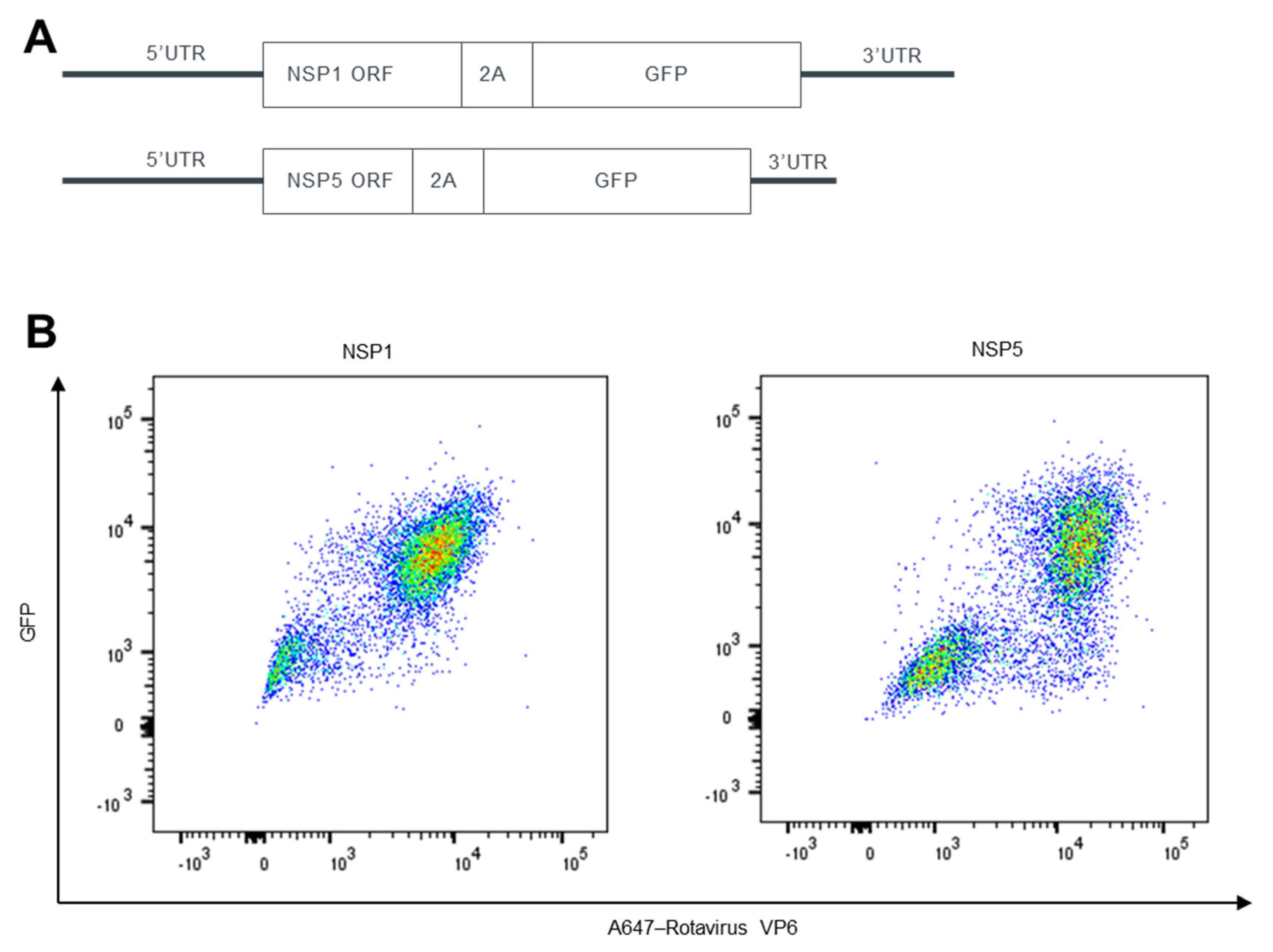
3.1. The Utilization of Three Positions in Rotavirus Genome for Heterologous Gene Expression
3.2. Characteristics of rSA11-GFP
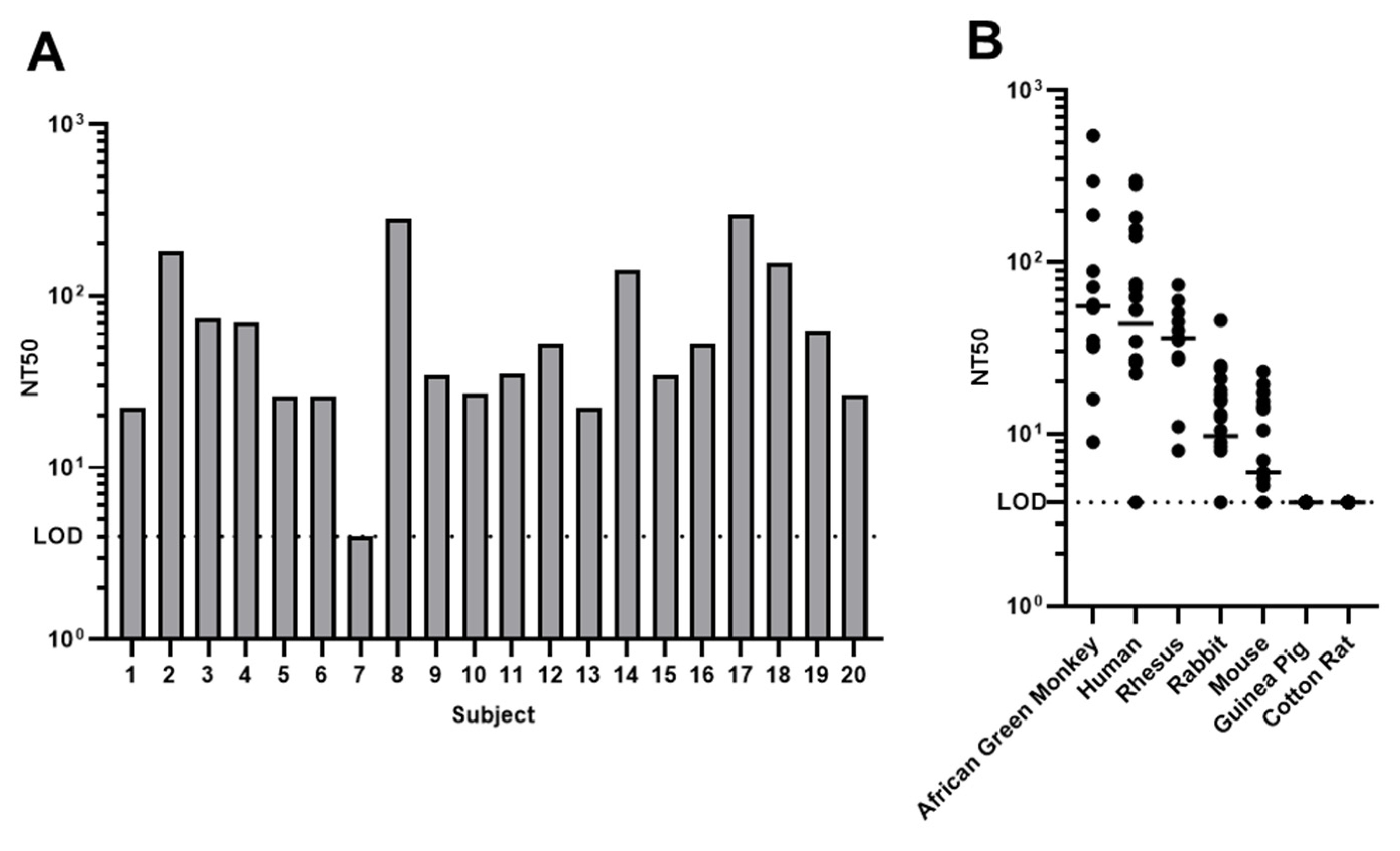
3.3. rSA11-GFP Based Microneutralization Assay
3.4. Pre-Existing Immunity in Human and Other Animal Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burnett, E.; Parashar, U.D.; Tate, J.E. Global Impact of Rotavirus Vaccination on Diarrhea Hospitalizations and Deaths Among Children <5 Years Old: 2006–2019. J. Infect. Dis. 2020, 222, 1731–1739. [Google Scholar] [CrossRef] [PubMed]
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D.; World Health Organization-Coordinated Global Rotavirus Surveillance Network. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clin. Infect. Dis. 2016, 62 (Suppl. S2), S96–S105. [Google Scholar] [CrossRef] [PubMed]
- Varghese, T.; Kang, G.; Steele, A.D. Understanding Rotavirus Vaccine Efficacy and Effectiveness in Countries with High Child Mortality. Vaccines 2022, 10, 346. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Komoto, S.; Kawagishi, T.; Nouda, R.; Nagasawa, N.; Onishi, M.; Matsuura, Y.; Taniguchi, K.; Kobayashi, T. Entirely plasmid-based reverse genetics system for rotaviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 2349–2354. [Google Scholar] [CrossRef]
- Sanchez-Tacuba, L.; Feng, N.; Meade, N.J.; Mellits, K.H.; Jais, P.H.; Yasukawa, L.L.; Resch, T.K.; Jiang, B.; Lopez, S.; Ding, S.; et al. An Optimized Reverse Genetics System Suitable for Efficient Recovery of Simian, Human, and Murine-Like Rotaviruses. J. Virol. 2020, 94, e01294-20. [Google Scholar] [CrossRef]
- Kawagishi, T.; Nurdin, J.A.; Onishi, M.; Nouda, R.; Kanai, Y.; Tajima, T.; Ushijima, H.; Kobayashi, T. Reverse Genetics System for a Human Group A Rotavirus. J. Virol. 2020, 94, e00963-19. [Google Scholar] [CrossRef]
- Komoto, S.; Fukuda, S.; Kugita, M.; Hatazawa, R.; Koyama, C.; Katayama, K.; Murata, T.; Taniguchi, K. Generation of Infectious Recombinant Human Rotaviruses from Just 11 Cloned cDNAs Encoding the Rotavirus Genome. J. Virol. 2019, 93, e02207-18. [Google Scholar] [CrossRef] [PubMed]
- Hamajima, R.; Lusiany, T.; Minami, S.; Nouda, R.; Nurdin, J.A.; Yamasaki, M.; Kobayashi, N.; Kanai, Y.; Kobayashi, T. A reverse genetics system for human rotavirus G2P[4]. J. Gen. Virol. 2022, 103, 001816. [Google Scholar] [CrossRef]
- Diebold, O.; Gonzalez, V.; Venditti, L.; Sharp, C.; Blake, R.A.; Tan, W.S.; Stevens, J.; Caddy, S.; Digard, P.; Borodavka, A.; et al. Using Species a Rotavirus Reverse Genetics to Engineer Chimeric Viruses Expressing SARS-CoV-2 Spike Epitopes. J. Virol. 2022, 96, e0048822. [Google Scholar] [CrossRef]
- Kanda, M.; Fukuda, S.; Hamada, N.; Nishiyama, S.; Masatani, T.; Fujii, Y.; Izumi, F.; Okajima, M.; Taniguchi, K.; Sugiyama, M.; et al. Establishment of a reverse genetics system for avian rotavirus A strain PO-13. J. Gen. Virol. 2022, 103, 001760. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Onishi, M.; Kawagishi, T.; Pannacha, P.; Nurdin, J.A.; Nouda, R.; Yamasaki, M.; Lusiany, T.; Khamrin, P.; Okitsu, S.; et al. Reverse Genetics Approach for Developing Rotavirus Vaccine Candidates Carrying VP4 and VP7 Genes Cloned from Clinical Isolates of Human Rotavirus. J. Virol. 2020, 95, e01374-20. [Google Scholar] [CrossRef] [PubMed]
- Criglar, J.M.; Crawford, S.E.; Zhao, B.; Smith, H.G.; Stossi, F.; Estes, M.K. A Genetically Engineered Rotavirus NSP2 Phosphorylation Mutant Impaired in Viroplasm Formation and Replication Shows an Early Interaction between vNSP2 and Cellular Lipid Droplets. J. Virol. 2020, 94, e00972-20. [Google Scholar] [CrossRef] [PubMed]
- Philip, A.A.; Perry, J.L.; Eaton, H.E.; Shmulevitz, M.; Hyser, J.M.; Patton, J.T. Generation of Recombinant Rotavirus Expressing NSP3-UnaG Fusion Protein by a Simplified Reverse Genetics System. J. Virol. 2019, 93, e01616-19. [Google Scholar] [CrossRef] [PubMed]
- Philip, A.A.; Patton, J.T. Rotavirus as an Expression Platform of Domains of the SARS-CoV-2 Spike Protein. Vaccines 2021, 9, 449. [Google Scholar] [CrossRef]
- Kawagishi, T.; Sanchez-Tacuba, L.; Feng, N.; Costantini, V.P.; Tan, M.; Jiang, X.; Green, K.Y.; Vinje, J.; Ding, S.; Greenberg, H.B. Mucosal and systemic neutralizing antibodies to norovirus induced in infant mice orally inoculated with recombinant rotaviruses. Proc. Natl. Acad. Sci. USA 2023, 120, e2214421120. [Google Scholar] [CrossRef]
- Adler, S.P.; Lewis, N.; Conlon, A.; Christiansen, M.P.; Al-Ibrahim, M.; Rupp, R.; Fu, T.M.; Bautista, O.; Tang, H.; Wang, D.; et al. Phase 1 Clinical Trial of a Conditionally Replication-Defective Human Cytomegalovirus (CMV) Vaccine in CMV-Seronegative Subjects. J. Infect. Dis. 2019, 220, 411–419. [Google Scholar] [CrossRef]
- McIntyre, M.; Rosenbaum, V.; Rappold, W.; Desselberger, M.; Wood, D.; Desselberger, U. Biophysical characterization of rotavirus particles containing rearranged genomes. J. Gen. Virol. 1987, 68 Pt. 11, 2961–2966. [Google Scholar] [CrossRef]
- Troupin, C.; Schnuriger, A.; Duponchel, S.; Deback, C.; Schnepf, N.; Dehee, A.; Garbarg-Chenon, A. Rotavirus rearranged genomic RNA segments are preferentially packaged into viruses despite not conferring selective growth advantage to viruses. PLoS ONE 2011, 6, e20080. [Google Scholar] [CrossRef]
- Jiang, B.; McClure, H.M.; Fankhauser, R.L.; Monroe, S.S.; Glass, R.I. Prevalence of rotavirus and norovirus antibodies in non-human primates. J. Med. Primatol. 2004, 33, 30–33. [Google Scholar] [CrossRef]
- RCWC. Available online: https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg (accessed on 29 September 2023).
- Velazquez, F.R.; Matson, D.O.; Calva, J.J.; Guerrero, L.; Morrow, A.L.; Carter-Campbell, S.; Glass, R.I.; Estes, M.K.; Pickering, L.K.; Ruiz-Palacios, G.M. Rotavirus infection in infants as protection against subsequent infections. N. Engl. J. Med. 1996, 335, 1022–1028. [Google Scholar] [CrossRef]
- Fields, B.N.; Knipe, D.M. Fields Virology; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2022; Volume 3. [Google Scholar]
- Nair, N.; Feng, N.; Blum, L.K.; Sanyal, M.; Ding, S.; Jiang, B.; Sen, A.; Morton, J.M.; He, X.S.; Robinson, W.H.; et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci. Transl. Med. 2017, 9, eaam5434. [Google Scholar] [CrossRef]
- Mackow, E.R.; Shaw, R.D.; Matsui, S.M.; Vo, P.T.; Benfield, D.A.; Greenberg, H.B. Characterization of homotypic and heterotypic VP7 neutralization sites of rhesus rotavirus. Virology 1988, 165, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Matsui, S.M.; Offit, P.A.; Vo, P.T.; Mackow, E.R.; Benfield, D.A.; Shaw, R.D.; Padilla-Noriega, L.; Greenberg, H.B. Passive protection against rotavirus-induced diarrhea by monoclonal antibodies to the heterotypic neutralization domain of VP7 and the VP8 fragment of VP4. J. Clin. Microbiol. 1989, 27, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Hoshino, Y.; Nishikawa, K.; Green, K.Y.; Maloy, W.L.; Morita, Y.; Urasawa, S.; Kapikian, A.Z.; Chanock, R.M.; Gorziglia, M. Cross-reactive and serotype-specific neutralization epitopes on VP7 of human rotavirus: Nucleotide sequence analysis of antigenic mutants selected with monoclonal antibodies. J. Virol. 1988, 62, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.; Zeng, Q.; Matthijnssens, J.; Greenberg, H.B.; Ding, S. Rotavirus NSP1 Contributes to Intestinal Viral Replication, Pathogenesis, and Transmission. mBio 2021, 12, e0320821. [Google Scholar] [CrossRef] [PubMed]
- Shambaugh, C.; Azshirvani, S.; Yu, L.; Pache, J.; Lambert, S.L.; Zuo, F.; Esser, M.T. Development of a High-Throughput Respiratory Syncytial Virus Fluorescent Focus-Based Microneutralization Assay. Clin. Vaccine Immunol. 2017, 24, e00225-17. [Google Scholar] [CrossRef]
- Wang, Z.; Mo, C.; Kemble, G.; Duke, G. Development of an efficient fluorescence-based microneutralization assay using recombinant human cytomegalovirus strains expressing green fluorescent protein. J. Virol. Methods 2004, 120, 207–215. [Google Scholar] [CrossRef]
- Biacchesi, S.; Skiadopoulos, M.H.; Yang, L.; Murphy, B.R.; Collins, P.L.; Buchholz, U.J. Rapid human metapneumovirus microneutralization assay based on green fluorescent protein expression. J. Virol. Methods 2005, 128, 192–197. [Google Scholar] [CrossRef]
- Caddy, S.L.; Vaysburd, M.; Wing, M.; Foss, S.; Andersen, J.T.; O’Connell, K.; Mayes, K.; Higginson, K.; Iturriza-Gomara, M.; Desselberger, U.; et al. Intracellular neutralisation of rotavirus by VP6-specific IgG. PLoS Pathog. 2020, 16, e1008732. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, J.; Radcliffe, S.; Pirrone, A.; Lu, M.; Li, Y.; Cassaday, J.; Newhard, W.; Heidecker, G.J.; Rose II, W.A.; He, X.; et al. A Novel Rotavirus Reverse Genetics Platform Supports Flexible Insertion of Exogenous Genes and Enables Rapid Development of a High-Throughput Neutralization Assay. Viruses 2023, 15, 2034. https://doi.org/10.3390/v15102034
Wei J, Radcliffe S, Pirrone A, Lu M, Li Y, Cassaday J, Newhard W, Heidecker GJ, Rose II WA, He X, et al. A Novel Rotavirus Reverse Genetics Platform Supports Flexible Insertion of Exogenous Genes and Enables Rapid Development of a High-Throughput Neutralization Assay. Viruses. 2023; 15(10):2034. https://doi.org/10.3390/v15102034
Chicago/Turabian StyleWei, Jiajie, Scott Radcliffe, Amanda Pirrone, Meiqing Lu, Yuan Li, Jason Cassaday, William Newhard, Gwendolyn J. Heidecker, William A. Rose II, Xi He, and et al. 2023. "A Novel Rotavirus Reverse Genetics Platform Supports Flexible Insertion of Exogenous Genes and Enables Rapid Development of a High-Throughput Neutralization Assay" Viruses 15, no. 10: 2034. https://doi.org/10.3390/v15102034
APA StyleWei, J., Radcliffe, S., Pirrone, A., Lu, M., Li, Y., Cassaday, J., Newhard, W., Heidecker, G. J., Rose II, W. A., He, X., Freed, D., Citron, M., Espeseth, A., & Wang, D. (2023). A Novel Rotavirus Reverse Genetics Platform Supports Flexible Insertion of Exogenous Genes and Enables Rapid Development of a High-Throughput Neutralization Assay. Viruses, 15(10), 2034. https://doi.org/10.3390/v15102034





