Abstract
Influenza B virus (IBV) is one of the two major types of influenza viruses that circulate each year. Unlike influenza A viruses, IBV does not harbor pandemic potential due to its lack of historical circulation in non-human hosts. Many studies and reviews have highlighted important factors for host determination of influenza A viruses. However, much less is known about the factors driving IBV replication in humans. We hypothesize that similar factors influence the host restriction of IBV. Here, we compile and review the current understanding of host factors crucial for the various stages of the IBV viral replication cycle. While we discovered the research in this area of IBV is limited, we review known host factors that may indicate possible host restriction of IBV to humans. These factors include the IBV hemagglutinin (HA) protein, host nuclear factors, and viral immune evasion proteins. Our review frames the current understanding of IBV adaptations to replication in humans. However, this review is limited by the amount of research previously completed on IBV host determinants and would benefit from additional future research in this area.
1. Introduction
Severe influenza infections in humans are typically caused by two influenza virus types: influenza A viruses (IAV) and influenza B viruses (IBV). These infections are suspected to cause up to 5 million severe cases of influenza disease annually [1]. In the 1980s, two antigenically distinct lineages of circulating IBV were identified using reference antiserum neutralization assays [2]. More recently, it was proposed that amino acid changes near the receptor binding site are responsible for the divergence of the two IBV lineages [3]. The lineages are characterized based on their relationship to two reference strains, B/Victoria-like (B/Vic) and B/Yamagata-like (B/Yam). Co-circulation of both lineages necessitated the development of quadrivalent vaccines to better protect against circulating IBV [4,5,6]. Severe disease burden in vulnerable populations has driven a public health pursuit to improve protection against IBV [7,8,9].
Throughout recorded history, IAV has been responsible for four major pandemic outbreaks, in 1918, 1957, 1968, and 2009. These pandemics were caused by the H1N1, H2N2, H3N2, and H1N1 subtypes, respectively [10]. The strains responsible for these pandemics had not previously circulated in humans, resulting in a lack of pre-existing immunity in human populations [11]. The lack of pre-existing immunity subsequently allowed nearly unlimited access to susceptible human hosts and unharnessed human-to-human transmission. Despite sharing many similar proteins to IAV, shown in Figure 1, IBV has not been responsible for pandemic influenza outbreaks. Since its discovery, IBV has been primarily isolated from human hosts. This is a stark contrast to IAV, whose host range consists of birds, swine, and other mammals [12]. Infectious IBV has been isolated from harbor seals, but phylogenetic similarity to human strains circulating at the same location and time suggests the seals were infected with human IBV strains [13]. Therefore, it is likely that humans remain the sole reservoir driving IBV circulation. Consistent IBV circulation in humans likely maintains a level of pre-existing immunity in the population and may contribute to the lack of an IBV-caused pandemic.
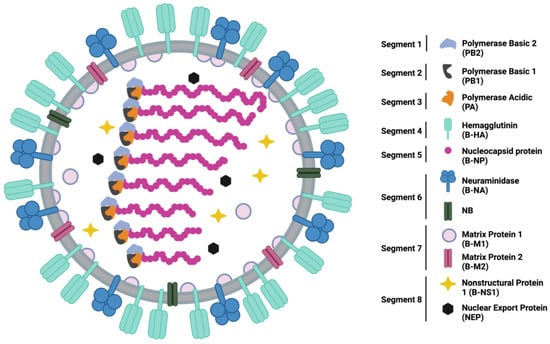
Figure 1.
Schematic representation of the structure and organization of an IBV virion and gene segments with all 11 proteins represented. Schematic created in BioRender.
Due to shared structural and replicative properties with IAV, we hypothesized that the distinct difference in host range between IBV and IAV is unlikely to be due to chance alone. Viral and host factors important to IAV host determination have been reviewed previously [12,14]. However, to the best of our knowledge, no reports have reviewed possible IBV adaptations to human hosts. Because of this, we chose to review the relevant findings suggesting IBV circulation in humans is due to a restriction of host range rather than by chance. We focused on three main areas most likely to contain host restriction factors due to their crucial role in the viral replication cycle: hemagglutinin-receptor interaction, nuclear replication factors, and immune evasion mechanisms. While some evidence to support this hypothesis has been reported across various stages of the viral replication cycle, including receptor binding, we found that research into this area was more limited than we expected. Therefore, this review focuses on the factors that could suggest IBV host range restriction to humans. We discuss how factors identified as host determinants of other influenza viruses could be investigated for a potential role in IBV host restriction. By conducting this literature review, we hope to provide the field of IBV research a framed view of various factors that may restrict IBV replication to humans and prevent circulation in other common hosts of influenza viruses.
2. Hemagglutinin-Specific Adaptations
In the initial stages of infection, IBV must bind to sialic acid residues on the surface of the cells lining the respiratory tract. Receptor binding is facilitated by the IBV hemagglutinin protein (B-HA). This receptor-binding interaction is crucial for successful viral infection and plays a major role in host specificity of IAV [15,16,17]. The receptor binding site of the B-HA is formed through contact of the 140-loop, 160-loop, 190-helix, and 240-loop. The tertiary receptor binding site then interacts with the sialic acid linkages on the surface of respiratory cells [18]. Human influenza viruses, including both lineages of IBV, bind preferentially to α2,6 linked sialic acid residues [19,20,21,22,23,24]. Avian IAV strains preferentially bind α2,3 linked-sialic acids [12,25]. The high abundance of α2,6 linked sialic acid residues in the human upper respiratory tract [16,26,27] provides ample access to susceptible host cells during viral infection. Among IBV, only the B/Victoria-like lineage has been linked with binding α2,3 linked sialic acid residues [20]. Interestingly, it was recently proposed that younger individuals have an increased abundance of α2,3 linked sialic acid residues in the lower respiratory tract compared to adults [28]. This difference could be associated with reports of more common and severe Victoria-like IBV infections in younger individuals than Yamagata-like infections [7,9,29,30]. Currently it is unknown whether the amino acid mutations responsible for the lineage divergence play a role in the difference of sialic acid receptor binding. The bias of B-HA binding α2,6 linked sialic acids may block transmission into hosts whose upper respiratory tracts contain more α2,3 linked sialic acids.
To facilitate membrane fusion between the viral envelope and the host endosome after receptor interaction, B-HA must have been enzymatically activated [31]. While other viruses commonly encode their own proteases [32], influenza viruses rely on host proteases to activate their HA proteins. The transmembrane serine protease 2 (TMPRSS2) is a host protease responsible for enzymatic activation of influenza viruses [33,34,35], paramyxoviruses [36], rotaviruses [37], and coronaviruses [38]. IAV is thought to be highly reliant on TMPRSS2 for efficient viral infection [33,39]. This was also observed when low pathogenic avian IAV strains were used to infect mouse and human airway epithelial cells [40]. Activation by TMPRSS2 and other trypsin-like proteases highly represented in the airways of humans and the gastrointestinal tract of waterfowl, may support the wide host range of IAV [41]. In comparison to IAV, IBV can be activated by a much broader range of human proteases [34,35,42]. Many of these additional proteases, including the kallikrein-family (KLK) proteases [35], are found in abundance in the human respiratory epithelium [34]. This abundance of proteases at the site of infection may point to an adaptation towards human airway infection rather than non-human hosts.
Outside of receptor binding and enzymatic activation, Laporte, et al. has shown that the physical environment of the human upper respiratory tract plays a key role in facilitating IBV infection. Interestingly, the pH required for efficient B-HA activation and membrane fusion was lower for IBV strains than for avian IAV (5.4–5.6 for IBV compared to 5.7–5.8 for avian IAV) [34]. Additionally, the optimal growth temperature of representative IBV strains was shown to be 33 °C, and the viral replicative yield is significantly reduced when the temperature increases up to 39 °C [34]. This is notable because the higher pH levels and temperatures tested in this study mimic the environments of the swine respiratory tract and the avian gastrointestinal tract, the major non-human reservoirs of IAV. Indeed, the avian IAV strains tested had optimal replication conditions that mirrored the avian gastrointestinal tract. Interestingly, the optimal growth conditions of the human IAV strains tested fell in between the IBV and avian IAV optimal growth conditions [34]. Given that the human respiratory tract is at a lower temperature and produces lower endosomal pH, this optimal growth condition observed, suggests an adaptation of IBV to human respiratory infection. Further, IBV replication is likely hindered by environmental factors specific to common non-human influenza hosts.
The receptor abundance in the human respiratory tract, the presence of enzymes capable of B-HA activation, and the optimal physical environment for IBV infection suggest possible host restriction of IBV replication to humans over non-human hosts. A summary of these factors can be seen in Table 1. However, the current lack of IBV research limits the impact of these observations. Overall, there are surprisingly few publications identifying adaptations of B-HA to the human respiratory environment. The few studies characterizing sialic acid binding affinities for B-HA are dated and fail to analyze strains isolated after 2010. Therefore, these studies do not consider the impact of newly emergent clades from both the B/Victoria-like and B/Yamagata-like lineages [43]. Interestingly, the binding affinity of the newest identified B/Victoria-like clade (V1A.3) [44], which contains a 3-amino acid deletion near the receptor-binding site [45,46], has yet to be characterized. This remains a gap in our understanding of possible restriction to human circulation among more recent IBV strains. Similarly, literature investigating B-HA enzymatic activation is based on older reference strains that no longer adequately represent the diversity of currently circulating IBV strains. Finally, little work has pursued the ability of avian- or swine-origin respiratory proteases to activate B-HA. Further work investigating this relationship and pursuing more recently isolated strains would help contribute to our understanding of how B-HA may be acting as a host restriction factor.

Table 1.
Summary of the known characteristics of B-HA and the human respiratory tract that point to possible host restriction to humans.
3. Nuclear Replication Adaptations
Influenza viruses belong to a group of RNA viruses that replicate their genome in the nucleus rather than the cytoplasm [47,48] despite not needing DNA replication machinery. The virus encodes its own RNA-dependent RNA-polymerase (RdRp), which is comprised of three viral protein subunits: PB2, PB1, and PA. These subunits interact to form the IBV polymerase complex (B-Pol). B-Pol lacks the critical function of 5′ mRNA cap production. Neither IAV nor IBV polymerase complexes have the ability to synthesize 5′ caps on their own [48,49]. Addition of a 5′ mRNA cap is required for stable mRNA translation by the host cell and for cap-dependent transcription of a complementary RNA (cRNA) molecule from viral genomic RNA (vRNA) [48,50]. To reach the nucleus, the IBV nucleoprotein (B-NP) contains nuclear localization signals (NLS) to facilitate transport to the nucleus [51]. Once the gene segments reach the nucleus, the virus usurps host factors to cross the nuclear envelope. The host factor importin-α binds three overlapping NLS on the N-terminal tail of B-NP to facilitate nuclear transport [51,52]. Importin-α has been previously described as a species-determinant for IAV [12,53]. However, a study has not been published testing this for IBV replication. Because there has not been a clear connection linking nuclear transport efficiency and replicating virus titers, we can only hypothesize that this may play a role in host restriction of IBV. Further investigation into this mechanism is required for a more definitive connection to IBV host restriction.
Once in the nucleus, B-Pol transcribes the viral RNA to initiate the viral replication cycle. Human acidic nuclear phosphoproteins (ANP32) are crucial for influenza RdRp activity [54,55,56,57]. The most important forms of ANP32 proteins in humans for influenza replication are ANP32A, -B, and -E. These proteins have many functions [58], but one noteworthy function is histone association and interaction with enzymes responsible for epigenetic post-translational modifications. However, influenza viruses are able to hijack these host proteins from their normal function to promote viral polymerase activity [57]. ANP32 proteins stabilize the influenza polymerase complex in a replicase promoting conformation prior to initiation of new RNA production [55]. Similar to IAV, both ANP32A and ANP32B can be utilized for efficient B-Pol activity. However, ANP32E can also support some polymerase activity in IBV but not IAV. Conversely, avian ANP32 proteins are unable to support efficient IBV polymerase activity but support human IAV polymerase activity [54]. Another study investigated the ability of other mammalian ANP32A and -B proteins for their ability to support B-Pol activity. Interestingly, their results showed that swine ANP32 proteins support B-Pol activity to a similar level as human ANP32 proteins. ANP32A and -B from horses, dogs, seals, and bats were only able to support B-Pol activity to a low level [56]. This reliance on mammalian ANP32 proteins may hinder IBV replication in birds but may not be a strong host restriction factor to human circulation alone. Further studies comparing the ability of human and non-human ANP32 proteins to stabilize B-Pol and promote viral replication could further support their role as a host restriction factor for IBV.
Again, few studies describe the interactions of IBV with host nuclear factors for the replication process. As mentioned briefly, the interactions of IBV with human importin-α focus on binding affinities but do not investigate how non-human homologs impact viral replication. These studies also fail to fully establish a mechanism of nuclear transport across the nuclear envelope. To support importin-α acting as a host restriction factor, future studies should assess the ability of different species’ importin-α to support IBV replication. This would provide evidence regarding to what extent IBV can utilize other species’ importin-α for nuclear transport to replicate the viral genome and express the viral proteins. While the importance of human ANP32 proteins was shown more clearly for IBV replication than importin-α, only avian ANP32 proteins were used to compare more than the two primary forms of ANP32 proteins. A major limitation to the studies showing ANP32 proteins supporting B-Pol activity was the absence of a comparison between the ability of host ANP32 proteins to support viral replication. Only B-Pol activity was measured in a minigenome reporter assay [56]. Another limitation is only the B/Florida/4/2006 strain (Yamagata-lineage) was used for this basic analysis, as IBV was not the primary focus of the study. Based on these limitations, we can only conclude that human nuclear factors may serve as possible host restriction factors at this time. Further dedicated studies testing the impact of these and likely other nuclear factors on IBV replication are needed to further support this conclusion.
4. Immune Evasion
A critical relationship between a host and pathogen is immune evasion. Influenza viruses specifically encode proteins to aid in immune evasion and utilize proteins with other viral functions to assist in evasion. A major difference between IAV and IBV in immune evasion is the lack of alternative polymerase protein products encoded in the PB1 and PA gene segments. IAV PB1-F2 [59] and PA-X [60] have immunomodulatory functions that support the replication of IAV [61,62,63], but these proteins are absent in IBV. Host-dependent expression differences in these accessory proteins have been described in IAV [22,64,65], suggesting these alternative products may play a role in the expanded host range. Therefore, there are other mechanisms employed by IBV to avoid the human immune system in the absence of these alternative polymerase factors.
The interferon (IFN) and interferon-stimulated gene (ISG) signaling pathways are a primary defense for the innate immune system in humans. During influenza infection, host cells produce IFN to activate host ISGs and limit total gene expression [66]. This impedes viral protein translation and prevents the production of progeny virions [67]. IBV infection strongly induces IFN expression in both human lung epithelial cell lines [68,69,70] and primary innate immune cells [71,72]. The IBV NS1 (B-NS1) protein has been shown to interfere with human IFN signaling through multiple mechanisms [73,74]. B-NS1 counteracts IFN signaling by preventing activation of the upstream transcription factor, IRF-3 [75]. A major B-NS1 target downstream of IFN signaling is the host IFN-stimulated gene 15 (ISG15) [76]. ISG15 is a ubiquitin-like modification protein that targets host genes to promote antiviral activity, including many that are also activated in response to IFN signaling [77]. B-NS1 specifically binds to human and non-human primate ISG15 but not other mammalian ISG15s [78,79,80]. B-NS1 can further modulate ISG15 activity by sequestering ISG15 target proteins [76,81]. This targeting of human ISG15 activity through multiple mechanisms suggests an important relationship between human ISG15 activity and IBV replication.
In addition to avoiding innate immune detection within the cell, the virus must also evade any adaptive immunity to infected cells. This recognition can come from cytotoxic T lymphocytes (CTLs). One way the virus attempts to avoid CTL recognition is through the downregulation of MHC Class I surface expression in infected cells. After IBV infection, cell lines stably expressing different HLA-alleles showed significantly decreased MHC Class I surface expression compared to IAV infection [82]. Another CTL evasion mechanism of IBV is a loss of an immunodominant epitope within a nuclear export signal (NES) in the IBV M1 (B-M1) protein [83]. This immunodominant epitope is found in the NES of IAV and may represent an adaptive mutation to avoid immune targeting in a conserved functional region [83]. While MHC class I downregulation is not unique to IBV infection [84,85,86], the apparent importance of human CTL evasion for replication may signal an advantage for IBV that could impact its ability to replicate in other hosts. However, little is known about IBV infections in this area. Further testing, comparing the impact of CTL responses on IBV and IAV infection between different hosts could uncover more concrete mechanisms relating immune evasion and host restriction.
We hypothesized that a virus with a limited host range such as IBV will be effective at evading its primary host’s immunity but not other host immunities. This would be due to the absence of viral responses to immune pressures exerted by non-human hosts, limiting virus replication. We also hypothesize that a virus with multiple mechanisms of immune evasion is more likely to have an expanded host range. The literature characterizing the B-NS1 interaction with human ISG15 supports our first hypothesis. As described above, B-NS1 is only able to rescue virus replication in the presence of human ISG15, while ISG15s from non-human species are able to inhibit replication in the presence of B-NS1. This suggests that IBV could be unable to successfully evade an aspect of non-human host immunity that prevents viral replication and spread within that host. We expect this species-specific targeting of ISG15, combined with other immune evasion mechanisms, plays a key role in promoting IBV circulation in humans while also potentially restricting replication in other non-human hosts.
Other immune evasion mechanisms have been characterized for IBV. Protein kinase R (PKR) [87,88], human myxovirus resistance protein 1 (MxA), melanoma differentiation-associated gene 5 (MDA5), and retinoic acid-inducible gene I (RIG-I) [89] have been described as targets of IBV immune evasion. Unfortunately, little has been reported about the ability of IBV to interfere with homologs of these proteins or signaling pathways from other hosts. Without the conclusions of these studies, we cannot adequately conclude what impacts these targets may have on IBV host restriction. If these targets do play a key role in host restriction, we would expect to see a level of species-specificity similar to what has been described with IBV and human ISG15.
5. Summary and Future Research Directions
The lack of an established non-human host presents a major difference between influenza B viruses and influenza A viruses. With the exception of isolated mammalian detection [13], no major animal outbreaks of IBV have been recorded. Despite this, IBV continues to circulate in humans. Here, we compile possible known host restriction factors that optimize IBV circulation in humans. To our surprise, limited studies have pursued this topic. Based on what is known about IBV replication, we then chose to review possible adaptations that may contribute to the lack of a non-human host of IBV. We summarize these possible host-specific adaptations in Figure 2.
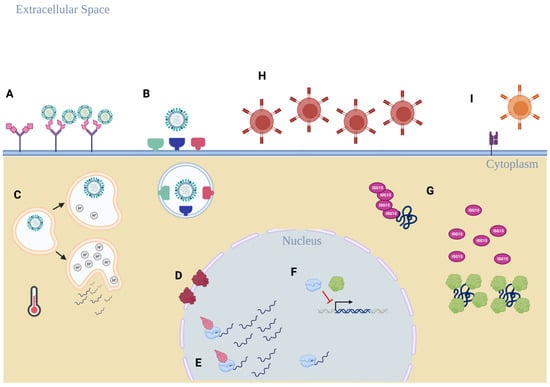
Figure 2.
An overview of the possible host restriction factors that promote IBV circulation in humans. (A) B-HA binding preference for α2,6-linked sialic acid residues on glycoproteins more prevalent in the human respiratory tract. (B) IBV can utilize a broad range of host proteases found in abundance in the human respiratory tract and do not rely on TMPRSS2 for B-HA activation. (A–C) The physical environment in human respiratory epithelial cells and in the extracellular environment favors human replication, including human endosomal pH ranges and upper respiratory tract temperature. (D) IBV replication requires the human importin-α protein to be present to ensure functional replication takes place, but a role as a host restriction factor is currently unclear. (E) Human ANP32 proteins are required for optimal B-Pol efficiency, and a wider range of ANP32 proteins can support B-Pol activity compared to the IAV polymerase complex. (F) B-Pol subunit PB1 and B-NS1 disrupt the host IFN response by preventing expression of anti-viral transcription factors IRF-3 and STAT1, but this interaction has not been evaluated as a possible host restriction factor. (G) B-NS1 selectively prevents ISGylation of host proteins that promote anti-viral responses through sequestration of ISG15 target proteins. (H,I) IBV avoids T-cell recognition of infected cells through surface downregulation of host MHC class I presentation (H) and the lack of a broadly reactive CD8+ T-cell epitope found near the NES of B-M1 (I). Diagram created in BioRender.
The most extensively interrogated, possible IBV host restriction factor is the B-HA protein. The interaction between viral protein and host receptor is a crucial determinant of viral entry into susceptible cells. Unsurprisingly, host receptor characteristics likely shape whether or not that host can be infected. These characteristics are summarized in Table 1. A binding preference of B-HA to host α2,6-glycan structures drives efficient receptor binding to cells of the human respiratory tract. Further, IBV can utilize a wide range of human enzymes highly abundant in the human respiratory tract to activate the B-HA protein. Additionally, the optimal physical environment for IBV replication also seems tailored to the human respiratory tract compared to non-human species. Taken together, these factors suggest IBV adaptation towards cells with receptor structures and environments found in humans rather than non-human hosts.
Possible host restriction factors outside of B-HA are not as well understood. To determine if a host restriction phenomenon has been observed during IBV genome replication, we considered host nuclear factors. The importin-α protein family has been identified as a critical factor for transporting the vRNPs from the cytoplasm into the nucleus [51,52]. However, current literature only highlights the interaction between IBV vRNPs and human importin-α. Currently, it is unclear whether other species’ importin-α family proteins interact with IBV vRNPs to support IBV replication. Additionally, evidence from IAV replication studies suggests that the interactions between host importin-α proteins and the viral polymerase complex may help determine species specificity [90,91,92]. Therefore, it is possible that these proteins may have a similar host restriction role for IBV as well. Another family of nuclear proteins, the ANP32 protein family, has also been shown to be crucial in supporting IBV replication [54,56,57]. Unlike the importin-α protein family, studies have shown that B-Pol activity is optimized with mammalian forms of theANP32 family proteins rather than avian species’ ANP32 [54,56]. These results suggest that this protein family may be acting as a host species limiting factor rather than a host restriction factor.
IBV has also evolved several mechanisms to escape human immune recognition. B-NS1 drives multiple mechanisms that obstruct the human IFN response [73,74]. Targeting of human-specific ISG15 by B-NS1 suggests an integral role between IBV replication and the human IFN response during infection. IBV also encodes mechanisms to avoid long-lasting memory CTL responses by downregulating host MHC Class I molecules on the surface of infected cells [82] and accumulating mutations in the B-M1 protein’s sequence to protect a conserved functional region from an immunodominant CTL epitope [83]. This effort to evade different aspects of the immune system obviously relates to its importance in promoting viral replication. However, more detailed investigations analyzing the ability of IBV to actively evade immune recognition in other hosts would improve our understanding of this as a possible host restriction factor.
A major theme to this research topic is the relative lack of studies investigating the possible IBV host restriction factors. Numerous studies have identified host factors or host–viral interactions critical for efficient IBV viral replication. However, these studies often fail to compare the human factors with non-human homologs, leading to a gap in our knowledge of why IBV only circulates in humans. Table 2 provides an outline of the host factors known to be crucial for IBV replication. The open questions associated with these factors may provide further insights into their role in IBV host restriction.

Table 2.
Summary of questions remaining to be answered about possible IBV host restriction factors.
Another potential avenue to discovery of IBV host restriction factors is considering the restriction factors of other types of influenza viruses. Factors that limit IAV, especially avian IAV, have been comprehensively reviewed [12,14,93,94]. Importantly, while strains of IAV can infect many different species, the host restriction factors identified often limit circulating strains to a particular host. Since these factors limit IAV crossover from one species to another, they may also play a role in host restriction of IBV. In fact, the importin-α and ANP32 protein families are described as host restriction factors for IAV as well [12,93]. However, much more is known about the role of these protein families for IAV host restriction than IBV restriction. As described above, more questions remain to clearly identify how IBV interaction with these proteins may impact host restriction. One host restriction factor well described for IAV with little attention for a role in IBV host restriction is the neuraminidase (NA) protein [14,95,96]. Various subtypes of IAV NA (A-NA) have been described to accumulate deletions in the stalk region when jumping from one host species to another [97,98,99]. This repeated deletion likely represents a critical mutation to overcome a host restriction factor to facilitate transmission in a new host species. Investigating this or other possible IBV host restriction mechanisms mediated by the IBV NA (B-NA) protein is critical to possibly understanding why IBV only replicates in humans.
Two other influenza types have also been discovered and circulate in mammals. Influenza C viruses (ICV) are distantly related to IBV and primarily infect humans [100]. Influenza D viruses (IDV) are closely related to ICV and were characterized as a second ICV subtype [101]. Both virus types differ from IBV and IAV genetically by encoding a hemagglutinin-esterase-fusion (HEF) protein [102] rather than distinct HA and NA proteins. ICV and IDV also bind to a chemically different receptor than IBV or IAV [103,104,105]. Wider natural host ranges for ICV and IDV compared to IBV have been described [106]. Unfortunately, little is known mechanistically about the host restriction factors for ICV or IDV. However, TMPRSS2 activates the ICV HEF protein similar to A-HA activation [107]. Host ANP32A protein has also been connected with ICV RdRp stability [55]. Additionally, ICV NS1 (C-NS1) and IDV NS1 (D-NS1) have been shown to possess IFN antagonistic properties [73,108]. The similarities observed between IBV, ICV, and IDV suggest similar factors may be contributing to host restriction and warrant further investigation. Uncovering these host restriction factors for one virus type may accelerate the discovery of the restriction factors for the others as well.
Despite an established host restriction environment for influenza viruses, host spillover events are detected. The most well-known examples are the pandemic spillover events from past influenza pandemics [10]. However, potential for reverse zoonoses, infections of animals from human circulation, is also possible. One reverse zoonosis pathway that is currently receiving attention is between humans and swine, most often in the H1N1pdm09 subtype [109,110,111]. The risk for reassortment and adaptation of these viruses to pigs remains a concern [112]. The SARS-CoV-2 pandemic has also opened a renewed interest in the impact of reverse zoonoses on global health [113,114]. While a spillover of IBV leading to sustained circulation in swine or other non-human hosts is unlikely, a clearer understanding of IBV host restriction could contribute to understanding barriers that prevent influenza and other viruses from undergoing frequent reverse zoonotic events.
A potential piece of non-molecular evidence supporting strong host restriction factors for IBV circulation is the possible extinction of the Yamagata-like lineage. The 2019–2020 flu season was predominantly characterized by the emergence of the new Victoria-like clade V1A.3 [44,46]. This led to low detection levels of Yamagata-like strains in the US early in the season [44], followed by an impact on influenza circulation due to non-pharmaceutical measures adopted to combat the spread of SARS-CoV-2 [115,116]. Despite influenza circulation rebounding in the more recent seasons [117], no Yamagata lineage detection since 2020 has led the field to consider whether the Yamagata lineage has possibly become extinct [118,119]. For a virus with a non-human reservoir, surveillance of the non-human hosts would likely detect virus even if human circulation was disrupted. However, for a virus that only circulates in humans, a break in human circulation would end the replication cycle and render the virus extinct. While we currently cannot rule out undetected circulation in an isolated population, the continued lack of detection of the Yamagata lineage suggests that the lineage may have become extinct. This lack of detection from humans and non-human hosts further supports the idea of only a single, restricted host population for IBV.
Based on our review of the available literature, there appears to be some evidence of IBV adaptation to human circulation throughout different stages of the replication cycle. Researchers have identified crucial interactions during receptor binding, genome replication, and immune evasion between viral and human factors that permit optimal virus replication to occur. The fact that these interactions occur throughout the virus replication cycle and not just during viral entry suggests that there is likely an important relationship between the human host and IBV replication. Many of these interactions require more comparison studies to investigate the impact that the factors from different host species have on IBV replication. This would help support the role of these factors as IBV host restriction factors. However, logically analyzing the available evidence suggests these adaptations could limit the ability of IBV to circulate in hon-human hosts. A better understanding of these interactions could unlock new information about how influenza viruses are restricted to their hosts. This information could then be leveraged as novel, specific antiviral targets in the future.
Author Contributions
M.J.P. and E.A.W. were both responsible for the conceptualization and writing of this review. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institutes of Health, NIAID grant number R01AI147109. The funders had no role in the manuscript preparation or decision to submit for publication.
Data Availability Statement
No new data was created or statistically analyzed for this study.
Acknowledgments
We would like to acknowledge Erika Petro-Turnquist for her effort in proofreading the manuscript. Figures used in this manuscript were created using BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Influenza (Seasonal). 6 November 2018. Available online: https://www.who.int/en/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 22 May 2023).
- Rota, P.A.; Wallis, T.R.; Harmon, M.W.; Rota, J.S.; Kendal, A.P.; Nerome, K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 1990, 175, 59–68. [Google Scholar] [CrossRef]
- Rosu, M.E.; Lexmond, P.; Bestebroer, T.M.; Hauser, B.M.; Smith, D.J.; Herfst, S.; Fouchier, R.A.M. Substitutions near the HA receptor binding site explain the origin and major antigenic change of the B/Victoria and B/Yamagata lineages. Proc. Natl. Acad. Sci. USA 2022, 119, e2211616119. [Google Scholar] [CrossRef]
- Shaw, M.W.; Xu, X.; Li, Y.; Normand, S.; Ueki, R.T.; Kunimoto, G.Y.; Hall, H.; Klimov, A.; Cox, N.J.; Subbarao, K. Reappearance and Global Spread of Variants of Influenza B/Victoria/2/87 Lineage Viruses in the 2000–2001 and 2001–2002 Seasons. Virology 2002, 303, 1–8. [Google Scholar] [CrossRef]
- Skowronski, D.M.; Masaro, C.; Kwindt, T.L.; Mak, A.; Petric, M.; Li, Y.; Sebastian, R.; Chong, M.; Tam, T.; De Serres, G. Estimating vaccine effectiveness against laboratory-confirmed influenza using a sentinel physician network: Results from the 2005–2006 season of dual A and B vaccine mismatch in Canada. Vaccine 2007, 25, 2842–2851. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.C.; Chuang, J.H.; Kuo, H.W.; Huang, W.T.; Hsu, Y.F.; Liu, M.T.; Chen, C.H.; Huang, H.H.; Chang, C.H.; Chou, J.H.; et al. Surveillance and vaccine effectiveness of an influenza epidemic predominated by vaccine-mismatched influenza B/Yamagata-lineage viruses in Taiwan, 2011–2012 season. PLoS ONE 2013, 8, e58222. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Blanton, L.; Brammer, L.; Olsen, S.J.; Fry, A.M. Influenza-Associated Pediatric Deaths in the United States, 2010–2016. Pediatrics 2018, 141, e20172918. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Vestergaard, L.S.; Richter, L.; Schmid, D.; Bustos, N.; Asikainen, T.; Trebbien, R.; Denissov, G.; Innos, K.; Virtanen, M.J.; et al. European all-cause excess and influenza-attributable mortality in the 2017/18 season: Should the burden of influenza B be reconsidered? Clin. Microbiol. Infect. 2019, 25, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Zaraket, H.; Hurt, A.C.; Clinch, B.; Barr, I.; Lee, N. Burden of influenza B virus infection and considerations for clinical management. Antiviral Res. 2021, 185, 104970. [Google Scholar] [CrossRef]
- Saunders-Hastings, P.R.; Krewski, D. Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission. Pathogens 2016, 5, 66. [Google Scholar] [CrossRef]
- Reperant, L.A.; Moesker, F.M.; Osterhaus, A.D. Influenza: From zoonosis to pandemic. ERJ Open Res. 2016, 2, 00013-2016. [Google Scholar] [CrossRef]
- Long, J.S.; Mistry, B.; Haslam, S.M.; Barclay, W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019, 17, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Osterhaus, A.D.; Rimmelzwaan, G.F.; Martina, B.E.; Bestebroer, T.M.; Fouchier, R.A. Influenza B virus in seals. Science 2000, 288, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Cauldwell, A.V.; Long, J.S.; Moncorgé, O.; Barclay, W.S. Viral determinants of influenza A virus host range. J. Gen. Virol. 2014, 95, 1193–1210. [Google Scholar] [CrossRef]
- Xiong, X.; McCauley, J.W.; Steinhauer, D.A. Receptor binding properties of the influenza virus hemagglutinin as a determinant of host range. Curr. Top Microbiol. Immunol. 2014, 385, 63–91. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Paulson, J.C. Adaptation of influenza viruses to human airway receptors. J. Biol. Chem. 2021, 296, 100017. [Google Scholar] [CrossRef]
- de Graaf, M.; Fouchier, R.A. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. Embo J. 2014, 33, 823–841. [Google Scholar] [CrossRef]
- Velkov, T. The specificity of the influenza B virus hemagglutinin receptor binding pocket: What does it bind to? J. Mol. Recognit. 2013, 26, 439–449. [Google Scholar] [CrossRef]
- Velkov, T.; Thompson, P.E.; El-Kabbani, O.; Lindh, F.; Stambas, J.; Rockman, S. Agel-capture assay for characterizing the sialyl-glycan selectivity of influenza viruses. Acta Virol. 2011, 55, 131–137. [Google Scholar] [CrossRef]
- Wang, Y.F.; Chang, C.F.; Chi, C.Y.; Wang, H.C.; Wang, J.R.; Su, I.J. Characterization of glycan binding specificities of influenza B viruses with correlation with hemagglutinin genotypes and clinical features. J. Med. Virol. 2012, 84, 679–685. [Google Scholar] [CrossRef]
- Ni, F.; Mbawuike, I.N.; Kondrashkina, E.; Wang, Q. The roles of hemagglutinin Phe-95 in receptor binding and pathogenicity of influenza B virus. Virology 2014, 450, 71–83. [Google Scholar] [CrossRef]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Yang, G.; DeBeauchamp, J.; Crumpton, J.C.; Kim, H.; Li, L.; Wan, X.F.; Kercher, L.; Bowman, A.S.; Webster, R.G.; et al. HA stabilization promotes replication and transmission of swine H1N1 gamma influenza viruses in ferrets. Elife 2020, 9, e56236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Pu, J. Influence of Host Sialic Acid Receptors Structure on the Host Specificity of Influenza Viruses. Viruses 2022, 14, 2141. [Google Scholar] [CrossRef] [PubMed]
- Trebbien, R.; Larsen, L.E.; Viuff, B.M. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol. J. 2011, 8, 434. [Google Scholar] [CrossRef]
- Shinya, K.; Ebina, M.; Yamada, S.; Ono, M.; Kasai, N.; Kawaoka, Y. Avian flu: Influenza virus receptors in the human airway. Nature 2006, 440, 435–436. [Google Scholar] [CrossRef]
- Walther, T.; Karamanska, R.; Chan, R.W.; Chan, M.C.; Jia, N.; Air, G.; Hopton, C.; Wong, M.P.; Dell, A.; Malik Peiris, J.S.; et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013, 9, e1003223. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Nelli, R.K.; Gontu, A.; Satyakumar, R.; Surendran Nair, M.; Subbiah, M. Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses 2021, 13, 262. [Google Scholar] [CrossRef]
- Caini, S.; Kusznierz, G.; Garate, V.V.; Wangchuk, S.; Thapa, B.; de Paula Júnior, F.J.; Ferreira de Almeida, W.A.; Njouom, R.; Fasce, R.A.; Bustos, P.; et al. The epidemiological signature of influenza B virus and its B/Victoria and B/Yamagata lineages in the 21st century. PLoS ONE 2019, 14, e0222381. [Google Scholar] [CrossRef]
- Read, J.M.; Zimmer, S.; Vukotich, C., Jr.; Schweizer, M.L.; Galloway, D.; Lingle, C.; Yearwood, G.; Calderone, P.; Noble, E.; Quadelacy, T.; et al. Influenza and other respiratory viral infections associated with absence from school among schoolchildren in Pittsburgh, Pennsylvania, USA: A cohort study. BMC Infect. Dis. 2021, 21, 291. [Google Scholar] [CrossRef]
- Russell, C.J. Acid-induced membrane fusion by the hemagglutinin protein and its role in influenza virus biology. Curr. Top. Microbiol. Immunol. 2014, 385, 93–116. [Google Scholar] [CrossRef]
- Castro, L.K.; Daugherty, M.D. Tripping the wire: Sensing of viral protease activity by CARD8 and NLRP1 inflammasomes. Curr. Opin. Immunol. 2023, 83, 102354. [Google Scholar] [CrossRef] [PubMed]
- Limburg, H.; Harbig, A.; Bestle, D.; Stein, D.A.; Moulton, H.M.; Jaeger, J.; Janga, H.; Hardes, K.; Koepke, J.; Schulte, L.; et al. TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes. J. Virol. 2019, 93, e00649-19. [Google Scholar] [CrossRef] [PubMed]
- Laporte, M.; Stevaert, A.; Raeymaekers, V.; Boogaerts, T.; Nehlmeier, I.; Chiu, W.; Benkheil, M.; Vanaudenaerde, B.; Pöhlmann, S.; Naesens, L. Hemagglutinin Cleavability, Acid Stability, and Temperature Dependence Optimize Influenza B Virus for Replication in Human Airways. J. Virol. 2019, 94, e01430-19. [Google Scholar] [CrossRef]
- Harbig, A.; Mernberger, M.; Bittel, L.; Pleschka, S.; Schughart, K.; Steinmetzer, T.; Stiewe, T.; Nist, A.; Böttcher-Friebertshäuser, E. Transcriptome profiling and protease inhibition experiments identify proteases that activate H3N2 influenza A and influenza B viruses in murine airways. J. Biol. Chem. 2020, 295, 11388–11407. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Tahara, M.; Sakai, K.; Yamaguchi, H.; Kanou, K.; Shirato, K.; Kawase, M.; Noda, M.; Kimura, H.; Matsuyama, S.; et al. TMPRSS2 Is an Activating Protease for Respiratory Parainfluenza Viruses. J. Virol. 2013, 87, 11930–11935. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Itakura, Y.; Kishimoto, M.; Tabata, K.; Uemura, K.; Ito, N.; Sugiyama, M.; Wastika, C.E.; Orba, Y.; Sawa, H. Host serine proteases TMPRSS2 and TMPRSS11D mediate proteolytic activation and trypsin-independent infection in group A rotaviruses. J. Virol. 2021, 95, e00398-21. [Google Scholar] [CrossRef]
- Rahbar Saadat, Y.; Hosseiniyan Khatibi, S.M.; Zununi Vahed, S.; Ardalan, M. Host Serine Proteases: A Potential Targeted Therapy for COVID-19 and Influenza. Front. Mol. Biosci. 2021, 8, 725528. [Google Scholar] [CrossRef]
- Kido, H.; Okumura, Y.; Takahashi, E.; Pan, H.-Y.; Wang, S.; Yao, D.; Yao, M.; Chida, J.; Yano, M. Role of host cellular proteases in the pathogenesis of influenza and influenza-induced multiple organ failure. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2012, 1824, 186–194. [Google Scholar] [CrossRef]
- Bestle, D.; Limburg, H.; Kruhl, D.; Harbig, A.; Stein, D.A.; Moulton, H.; Matrosovich, M.; Abdelwhab, E.M.; Stech, J.; Böttcher-Friebertshäuser, E. Hemagglutinins of Avian Influenza Viruses Are Proteolytically Activated by TMPRSS2 in Human and Murine Airway Cells. J. Virol. 2021, 95, e0090621. [Google Scholar] [CrossRef]
- Bertram, S.; Glowacka, I.; Steffen, I.; Kühl, A.; Pöhlmann, S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol. 2010, 20, 298–310. [Google Scholar] [CrossRef]
- Sakai, K.; Ami, Y.; Nakajima, N.; Nakajima, K.; Kitazawa, M.; Anraku, M.; Takayama, I.; Sangsriratanakul, N.; Komura, M.; Sato, Y.; et al. TMPRSS2 Independency for Haemagglutinin Cleavage In Vivo Differentiates Influenza B Virus from Influenza A Virus. Sci. Rep. 2016, 6, 29430. [Google Scholar] [CrossRef] [PubMed]
- Virk, R.K.; Jayakumar, J.; Mendenhall, I.H.; Moorthy, M.; Lam, P.; Linster, M.; Lim, J.; Lin, C.; Oon, L.L.E.; Lee, H.K.; et al. Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity. Proc. Natl. Acad. Sci. USA 2020, 117, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Borchering, R.K.; Gunning, C.E.; Gokhale, D.V.; Weedop, K.B.; Saeidpour, A.; Brett, T.S.; Rohani, P. Anomalous influenza seasonality in the United States and the emergence of novel influenza B viruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2012327118. [Google Scholar] [CrossRef] [PubMed]
- Kato-Miyashita, S.; Sakai-Tagawa, Y.; Yamashita, M.; Iwatsuki-Horimoto, K.; Ito, M.; Tokita, A.; Hagiwara, H.; Izumida, N.; Nishino, T.; Wada, N.; et al. Antigenic variants of influenza B viruses isolated in Japan during the 2017–2018 and 2018–2019 influenza seasons. Influenza Other Respir. Viruses 2020, 14, 311–319. [Google Scholar] [CrossRef]
- Tenforde, M.W.; Kondor, R.J.G.; Chung, J.R.; Zimmerman, R.K.; Nowalk, M.P.; Jackson, M.L.; Jackson, L.A.; Monto, A.S.; Martin, E.T.; Belongia, E.A.; et al. Effect of Antigenic Drift on Influenza Vaccine Effectiveness in the United States-2019–2020. Clin. Infect. Dis. 2021, 73, e4244–e4250. [Google Scholar] [CrossRef]
- Te Velthuis, A.J.; Fodor, E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef]
- Walker, A.P.; Fodor, E. Interplay between Influenza Virus and the Host RNA Polymerase II Transcriptional Machinery. Trends Microbiol. 2019, 27, 398–407. [Google Scholar] [CrossRef]
- Bouloy, M.; Plotch, S.J.; Krug, R.M. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc. Natl. Acad. Sci. USA 1978, 75, 4886–4890. [Google Scholar] [CrossRef]
- Reich, S.; Guilligay, D.; Cusack, S. An in vitro fluorescence based study of initiation of RNA synthesis by influenza B polymerase. Nucleic Acids Res. 2017, 45, 3353–3368. [Google Scholar] [CrossRef]
- Labaronne, A.; Milles, S.; Donchet, A.; Jensen, M.R.; Blackledge, M.; Bourhis, J.M.; Ruigrok, R.W.H.; Crépin, T. Structural analysis of the complex between influenza B nucleoprotein and human importin-α. Sci. Rep. 2017, 7, 17164. [Google Scholar] [CrossRef]
- Donchet, A.; Vassal-Stermann, E.; Gérard, F.C.A.; Ruigrok, R.W.H.; Crépin, T. Differential Behaviours and Preferential Bindings of Influenza Nucleoproteins on Importins-α. Viruses 2020, 12, 834. [Google Scholar] [CrossRef] [PubMed]
- Resa-Infante, P.; Gabriel, G. The nuclear import machinery is a determinant of influenza virus host adaptation. Bioessays 2013, 35, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, H.; Xu, L.; Guo, X.; Wang, W.; Ji, Y.; Lin, C.; Wang, Y.; Wang, X. Selective usage of ANP32 proteins by influenza B virus polymerase: Implications in determination of host range. PLoS Pathog. 2020, 16, e1008989. [Google Scholar] [CrossRef] [PubMed]
- Carrique, L.; Fan, H.; Walker, A.P.; Keown, J.R.; Sharps, J.; Staller, E.; Barclay, W.S.; Fodor, E.; Grimes, J.M. Host ANP32A mediates the assembly of the influenza virus replicase. Nature 2020, 587, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Swann, O.C.; Salvesen, H.A.; Staller, E.; Leung, P.B.; Goldhill, D.H.; Zhou, H.; Lillico, S.G.; Whitelaw, C.B.A.; Long, J.S.; et al. Swine ANP32A Supports Avian Influenza Virus Polymerase. J. Virol. 2020, 94, e00132-20. [Google Scholar] [CrossRef]
- Yu, M.; Qu, Y.; Zhang, H.; Wang, X. Roles of ANP32 proteins in cell biology and viral replication. Anim. Dis. 2022, 2, 22. [Google Scholar] [CrossRef]
- Reilly, P.T.; Yu, Y.; Hamiche, A.; Wang, L. Cracking the ANP32 whips: Important functions, unequal requirement, and hints at disease implications. BioEssays 2014, 36, 1062–1071. [Google Scholar] [CrossRef]
- Conenello, G.M.; Zamarin, D.; Perrone, L.A.; Tumpey, T.; Palese, P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007, 3, 1414–1421. [Google Scholar] [CrossRef]
- Jagger, B.W.; Wise, H.M.; Kash, J.C.; Walters, K.A.; Wills, N.M.; Xiao, Y.L.; Dunfee, R.L.; Schwartzman, L.M.; Ozinsky, A.; Bell, G.L.; et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 2012, 337, 199–204. [Google Scholar] [CrossRef]
- Hayashi, T.; MacDonald, L.A.; Takimoto, T. Influenza A Virus Protein PA-X Contributes to Viral Growth and Suppression of the Host Antiviral and Immune Responses. J. Virol. 2015, 89, 6442–6452. [Google Scholar] [CrossRef]
- Klemm, C.; Boergeling, Y.; Ludwig, S.; Ehrhardt, C. Immunomodulatory Nonstructural Proteins of Influenza A Viruses. Trends Microbiol. 2018, 26, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Martinez-Sobrido, L.; Topham, D.J.; DeDiego, M.L. Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins. Viruses 2018, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- James, J.; Howard, W.; Iqbal, M.; Nair, V.K.; Barclay, W.S.; Shelton, H. Influenza A virus PB1-F2 protein prolongs viral shedding in chickens lengthening the transmission window. J. Gen. Virol. 2016, 97, 2516–2527. [Google Scholar] [CrossRef] [PubMed]
- Lutz Iv, M.M.; Dunagan, M.M.; Kurebayashi, Y.; Takimoto, T. Key Role of the Influenza A Virus PA Gene Segment in the Emergence of Pandemic Viruses. Viruses 2020, 12, 365. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, S.; Goraya, M.U.; Maarouf, M.; Huang, S.; Chen, J.L. Host Immune Response to Influenza A Virus Infection. Front. Immunol. 2018, 9, 320. [Google Scholar] [CrossRef]
- Iwasaki, A.; Pillai, P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014, 14, 315–328. [Google Scholar] [CrossRef]
- Jiang, J.; Li, J.; Fan, W.; Zheng, W.; Yu, M.; Chen, C.; Sun, L.; Bi, Y.; Ding, C.; Gao, G.F.; et al. Robust Lys63-Linked Ubiquitination of RIG-I Promotes Cytokine Eruption in Early Influenza B Virus Infection. J. Virol. 2016, 90, 6263–6275. [Google Scholar] [CrossRef]
- Jiao, P.; Fan, W.; Cao, Y.; Zhang, H.; Tian, L.; Sun, L.; Luo, T.; Liu, W.; Li, J. Robust induction of interferon and interferon-stimulated gene expression by influenza B/Yamagata lineage virus infection of A549 cells. PLoS ONE 2020, 15, e0231039. [Google Scholar] [CrossRef]
- Dissanayake, T.K.; Schäuble, S.; Mirhakkak, M.H.; Wu, W.L.; Ng, A.C.; Yip, C.C.Y.; López, A.G.; Wolf, T.; Yeung, M.L.; Chan, K.H.; et al. Comparative Transcriptomic Analysis of Rhinovirus and Influenza Virus Infection. Front. Microbiol. 2020, 11, 1580. [Google Scholar] [CrossRef]
- Österlund, P.; Strengell, M.; Sarin, L.P.; Poranen, M.M.; Fagerlund, R.; Melén, K.; Julkunen, I. Incoming Influenza A Virus Evades Early Host Recognition, while Influenza B Virus Induces Interferon Expression Directly upon Entry. J. Virol. 2012, 86, 11183–11193. [Google Scholar] [CrossRef]
- Mäkelä, S.M.; Österlund, P.; Westenius, V.; Latvala, S.; Diamond, M.S.; Gale, M., Jr.; Julkunen, I. RIG-I Signaling Is Essential for Influenza B Virus-Induced Rapid Interferon Gene Expression. J. Virol. 2015, 89, 12014–12025. [Google Scholar] [CrossRef]
- Nogales, A.; Aydillo, T.; Ávila-Pérez, G.; Escalera, A.; Chiem, K.; Cadagan, R.; DeDiego, M.L.; Li, F.; García-Sastre, A.; Martínez-Sobrido, L. Functional Characterization and Direct Comparison of Influenza A, B, C, and D NS1 Proteins in vitro and in vivo. Front. Microbiol. 2019, 10, 2862. [Google Scholar] [CrossRef]
- Hensen, L.; Kedzierska, K.; Koutsakos, M. Innate and adaptive immunity toward influenza B viruses. Future Microbiol. 2020, 15, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Donelan, N.R.; Dauber, B.; Wang, X.; Basler, C.F.; Wolff, T.; García-Sastre, A. The N- and C-terminal domains of the NS1 protein of influenza B virus can independently inhibit IRF-3 and beta interferon promoter activation. J. Virol. 2004, 78, 11574–11582. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Krug, R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. Embo J. 2001, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Denison, C.; Huibregtse, J.M.; Gygi, S.; Krug, R.M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. USA 2005, 102, 10200–10205. [Google Scholar] [CrossRef]
- Versteeg, G.A.; Hale, B.G.; van Boheemen, S.; Wolff, T.; Lenschow, D.J.; García-Sastre, A. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J. Virol. 2010, 84, 5423–5430. [Google Scholar] [CrossRef]
- Sridharan, H.; Zhao, C.; Krug, R.M. Species specificity of the NS1 protein of influenza B virus: NS1 binds only human and non-human primate ubiquitin-like ISG15 proteins. J. Biol. Chem. 2010, 285, 7852–7856. [Google Scholar] [CrossRef]
- Guan, R.; Ma, L.-C.; Leonard, P.G.; Amer, B.R.; Sridharan, H.; Zhao, C.; Krug, R.M.; Montelione, G.T. Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus. Proc. Natl. Acad. Sci. USA 2011, 108, 13468–13473. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sridharan, H.; Chen, R.; Baker, D.P.; Wang, S.; Krug, R.M. Influenza B virus non-structural protein 1 counteracts ISG15 antiviral activity by sequestering ISGylated viral proteins. Nat. Commun. 2016, 7, 12754. [Google Scholar] [CrossRef]
- Koutsakos, M.; McWilliam, H.E.G.; Aktepe, T.E.; Fritzlar, S.; Illing, P.T.; Mifsud, N.A.; Purcell, A.W.; Rockman, S.; Reading, P.C.; Vivian, J.P.; et al. Downregulation of MHC Class I Expression by Influenza A and B Viruses. Front. Immunol. 2019, 10, 1158. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xiao, W.; Wu, Y.; Fan, W.; Li, L.; Yue, C.; Zhang, Q.; Zhang, D.; Yuan, X.; Yao, S.; et al. Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape. J. Immunol. 2023, 210, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Griffin, B.D.; Verweij, M.C.; Wiertz, E.J. Herpesviruses and immunity: The art of evasion. Vet. Microbiol. 2010, 143, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Quinn, L.L.; Williams, L.R.; White, C.; Forrest, C.; Zuo, J.; Rowe, M. The Missing Link in Epstein-Barr Virus Immune Evasion: The BDLF3 Gene Induces Ubiquitination and Downregulation of Major Histocompatibility Complex Class I (MHC-I) and MHC-II. J. Virol. 2016, 90, 356–367. [Google Scholar] [CrossRef]
- Vinjamuri, S.; Li, L.; Bouvier, M. SARS-CoV-2 ORF8: One protein, seemingly one structure, and many functions. Front. Immunol. 2022, 13, 1035559. [Google Scholar] [CrossRef] [PubMed]
- Dauber, B.; Schneider, J.; Wolff, T. Double-stranded RNA binding of influenza B virus nonstructural NS1 protein inhibits protein kinase R but is not essential to antagonize production of alpha/beta interferon. J. Virol. 2006, 80, 11667–11677. [Google Scholar] [CrossRef]
- Yin, C.; Khan, J.A.; Swapna, G.V.; Ertekin, A.; Krug, R.M.; Tong, L.; Montelione, G.T. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J. Biol. Chem. 2007, 282, 20584–20592. [Google Scholar] [CrossRef]
- Schreiber, A.; Liedmann, S.; Brunotte, L.; Anhlan, D.; Ehrhardt, C.; Ludwig, S. Type I interferon antagonistic properties of influenza B virus polymerase proteins. Cell Microbiol. 2020, 22, e13143. [Google Scholar] [CrossRef]
- Gabriel, G.; Herwig, A.; Klenk, H.D. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008, 4, e11. [Google Scholar] [CrossRef]
- Resa-Infante, P.; Jorba, N.; Zamarreño, N.; Fernández, Y.; Juárez, S.; Ortín, J. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS ONE 2008, 3, e3904. [Google Scholar] [CrossRef]
- Gabriel, G.; Klingel, K.; Otte, A.; Thiele, S.; Hudjetz, B.; Arman-Kalcek, G.; Sauter, M.; Shmidt, T.; Rother, F.; Baumgarte, S.; et al. Differential use of importin-α isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2011, 2, 156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, M.; Bai, S.; Zhao, C.; Li, Z.; Xu, J.; Zhang, X. Influenza A Virus-Host Specificity: An Ongoing Cross-Talk Between Viral and Host Factors. Front. Microbiol. 2021, 12, 777885. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, B.; Duncan, M.; Subbarao, K. Role of the viral polymerase during adaptation of influenza A viruses to new hosts. Curr. Opin. Virol. 2023, 62, 101363. [Google Scholar] [CrossRef]
- Bisset, A.T.; Hoyne, G.F. Evolution and Adaptation of the Avian H7N9 Virus into the Human Host. Microorganisms 2020, 8, 778. [Google Scholar] [CrossRef]
- Ciminski, K.; Ran, W.; Gorka, M.; Lee, J.; Malmlov, A.; Schinköthe, J.; Eckley, M.; Murrieta, R.A.; Aboellail, T.A.; Campbell, C.L.; et al. Bat influenza viruses transmit among bats but are poorly adapted to non-bat species. Nat. Microbiol. 2019, 4, 2298–2309. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.; Zhou, N.; Kawaoka, Y.; Webster, R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 1999, 73, 1146–1155. [Google Scholar] [CrossRef]
- Webby, R.J.; Woolcock, P.R.; Krauss, S.L.; Webster, R.G. Reassortment and interspecies transmission of North American H6N2 influenza viruses. Virology 2002, 295, 44–53. [Google Scholar] [CrossRef]
- Li, J.; Zu Dohna, H.; Cardona, C.J.; Miller, J.; Carpenter, T.E. Emergence and genetic variation of neuraminidase stalk deletions in avian influenza viruses. PLoS ONE 2011, 6, e14722. [Google Scholar] [CrossRef]
- Sederdahl, B.K.; Williams, J.V. Epidemiology and Clinical Characteristics of Influenza C Virus. Viruses 2020, 12, 89. [Google Scholar] [CrossRef]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef]
- Herrler, G.; Dürkop, I.; Becht, H.; Klenk, H.D. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J. Gen. Virol. 1988, 69 Pt 4, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.N.; Herrler, G.; Paulson, J.C.; Klenk, H.D. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J. Biol. Chem. 1986, 261, 5947–5951. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Veit, M. Hemagglutinin-esterase-fusion (HEF) protein of influenza C virus. Protein Cell 2016, 7, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Sreenivasan, C.; Yu, H.; Sheng, Z.; Newkirk, S.J.; An, W.; Smith, D.F.; Chen, X.; Wang, D.; Li, F. Influenza D virus diverges from its related influenza C virus in the recognition of 9-O-acetylated N-acetyl- or N-glycolyl-neuraminic acid-containing glycan receptors. Virology 2020, 545, 16–23. [Google Scholar] [CrossRef]
- Sreenivasan, C.C.; Sheng, Z.; Wang, D.; Li, F. Host Range, Biology, and Species Specificity of Seven-Segmented Influenza Viruses-A Comparative Review on Influenza C and D. Pathogens 2021, 10, 1583. [Google Scholar] [CrossRef]
- Sato, K.; Hayashi, H.; Shimotai, Y.; Yamaya, M.; Hongo, S.; Kawakami, K.; Matsuzaki, Y.; Nishimura, H. TMPRSS2 Activates Hemagglutinin-Esterase Glycoprotein of Influenza C Virus. J. Virol. 2021, 95, e0129621. [Google Scholar] [CrossRef]
- Pachler, K.; Vlasak, R. Influenza C virus NS1 protein counteracts RIG-I-mediated IFN signalling. Virol. J. 2011, 8, 48. [Google Scholar] [CrossRef]
- Chastagner, A.; Enouf, V.; Peroz, D.; Hervé, S.; Lucas, P.; Quéguiner, S.; Gorin, S.; Beven, V.; Behillil, S.; Leneveu, P.; et al. Bidirectional Human-Swine Transmission of Seasonal Influenza A(H1N1)pdm09 Virus in Pig Herd, France, 2018. Emerg. Infect. Dis. 2019, 25, 1940–1943. [Google Scholar] [CrossRef]
- Markin, A.; Ciacci Zanella, G.; Arendsee, Z.W.; Zhang, J.; Krueger, K.M.; Gauger, P.C.; Vincent Baker, A.L.; Anderson, T.K. Reverse-zoonoses of 2009 H1N1 pandemic influenza A viruses and evolution in United States swine results in viruses with zoonotic potential. PLoS Pathog. 2023, 19, e1011476. [Google Scholar] [CrossRef]
- Zeller, M.A.; Ma, J.; Wong, F.Y.; Tum, S.; Hidano, A.; Holt, H.; Chhay, T.; Sorn, S.; Koeut, D.; Seng, B.; et al. The genomic landscape of swine influenza A viruses in Southeast Asia. Proc. Natl. Acad. Sci. USA 2023, 120, e2301926120. [Google Scholar] [CrossRef]
- Rajao, D.S.; Vincent, A.L.; Perez, D.R. Adaptation of Human Influenza Viruses to Swine. Front. Vet. Sci. 2018, 5, 347. [Google Scholar] [CrossRef] [PubMed]
- Munir, K.; Ashraf, S.; Munir, I.; Khalid, H.; Muneer, M.A.; Mukhtar, N.; Amin, S.; Ashraf, S.; Imran, M.A.; Chaudhry, U.; et al. Zoonotic and reverse zoonotic events of SARS-CoV-2 and their impact on global health. Emerg. Microbes Infect. 2020, 9, 2222–2235. [Google Scholar] [CrossRef] [PubMed]
- Goraichuk, I.V.; Arefiev, V.; Stegniy, B.T.; Gerilovych, A.P. Zoonotic and Reverse Zoonotic Transmissibility of SARS-CoV-2. Virus Res. 2021, 302, 198473. [Google Scholar] [CrossRef]
- Olsen, S.J.; Azziz-Baumgartner, E.; Budd, A.P.; Brammer, L.; Sullivan, S.; Pineda, R.F.; Cohen, C.; Fry, A.M. Decreased Influenza Activity during the COVID-19 Pandemic—United States, Australia, Chile, and South Africa, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1305–1309. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Shaman, J.; Pei, S. Quantifying the Impact of COVID-19 Nonpharmaceutical Interventions on Influenza Transmission in the United States. J. Infect. Dis. 2021, 224, 1500–1508. [Google Scholar] [CrossRef]
- Takeuchi, H.; Kawashima, R. Disappearance and Re-Emergence of Influenza during the COVID-19 Pandemic: Association with Infection Control Measures. Viruses 2023, 15, 223. [Google Scholar] [CrossRef] [PubMed]
- Koutsakos, M.; Wheatley, A.K.; Laurie, K.; Kent, S.J.; Rockman, S. Influenza lineage extinction during the COVID-19 pandemic? Nat. Rev. Microbiol. 2021, 19, 741–742. [Google Scholar] [CrossRef]
- Paget, J.; Caini, S.; Del Riccio, M.; van Waarden, W.; Meijer, A. Has influenza B/Yamagata become extinct and what implications might this have for quadrivalent influenza vaccines? Eurosurveillance 2022, 27, 2200753. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).