A Bioreactor-Based Yellow Fever Virus-like Particle Production Process with Integrated Process Analytical Technology Based on Transient Transfection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Production
2.1.1. pDNA Production under Standard Conditions
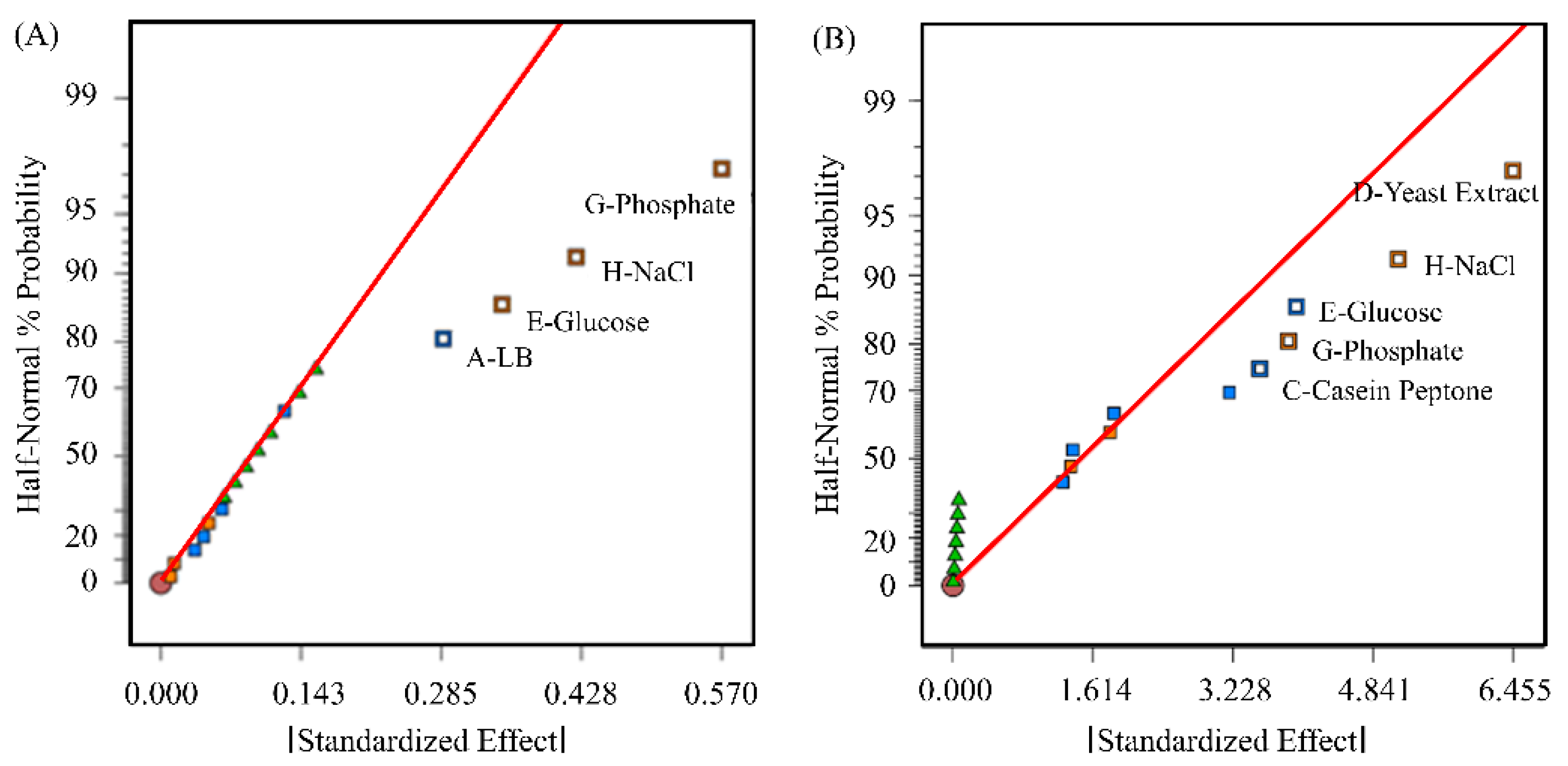
2.1.2. Optimization of pDNA Production Using a Plackett–Burman Design
2.1.3. pDNA Purification
2.2. eGFP or YF-VLP Production
2.2.1. Cell Line and Medium
2.2.2. Transient Transfection in Shaking Flasks
2.2.3. Transient Transfection in a Stirred-Tank Bioreactor
2.3. Analysis
2.3.1. pDNA Analysis
2.3.2. Analysis of E. coli Growth
2.3.3. Offline Determination of Cell Concentration
2.3.4. Metabolite Analysis
2.3.5. SDS-PAGE
2.3.6. Western Blot
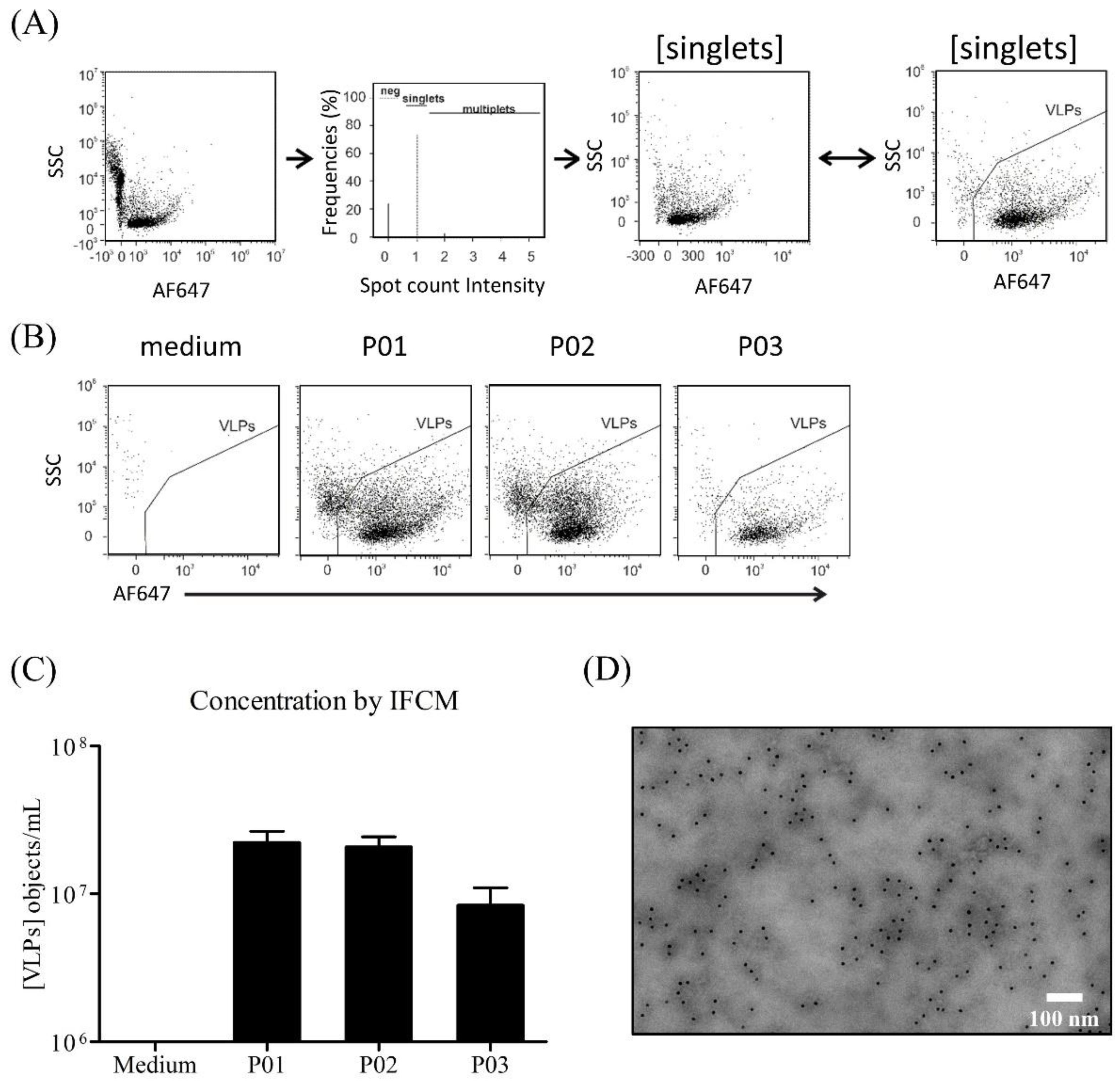
2.3.7. Imaging Flow Cytometry Measurement (IFCM)
2.3.8. Transmission Electron Microscopy (TEM)
2.3.9. Statistical Analysis
3. Results
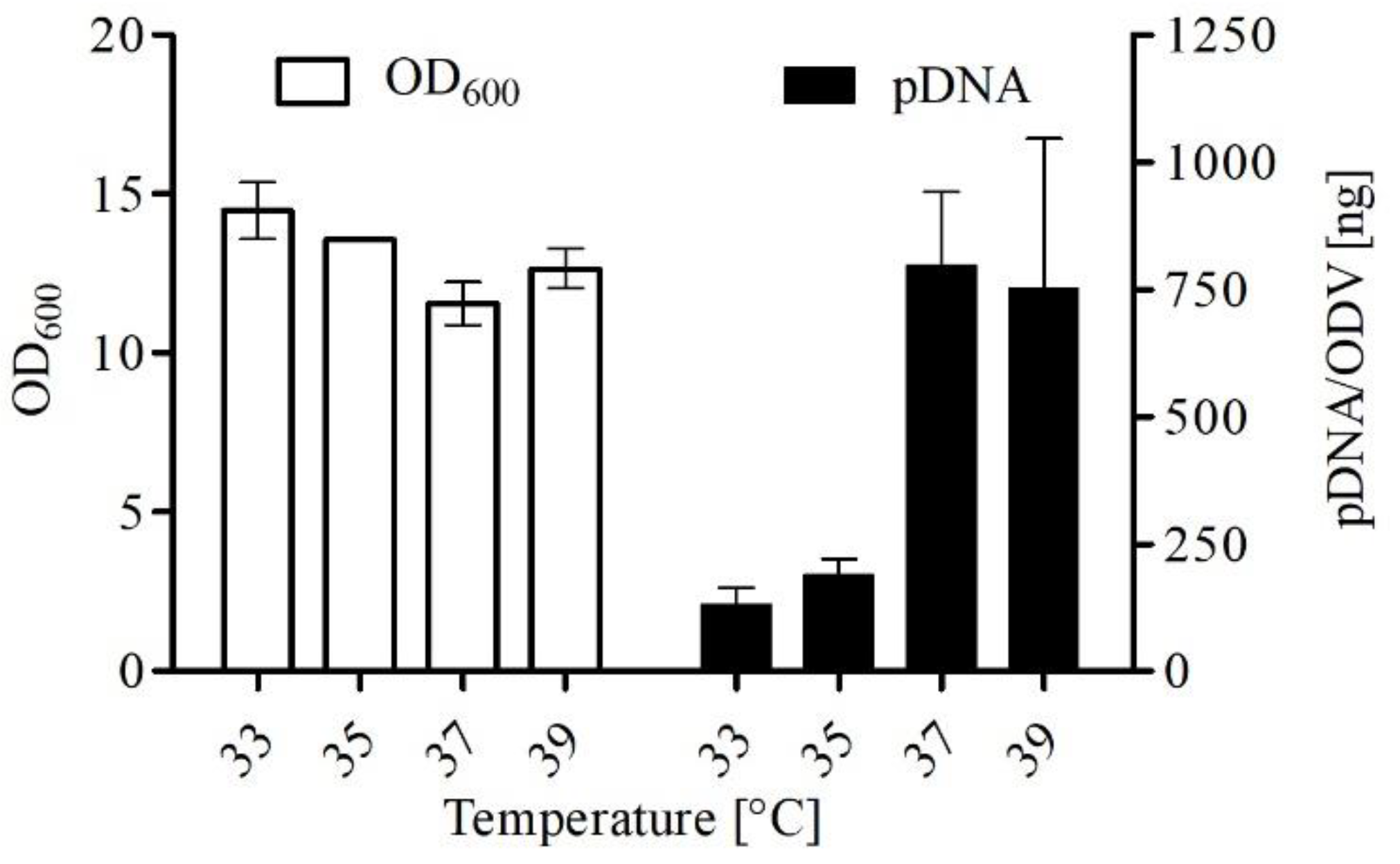
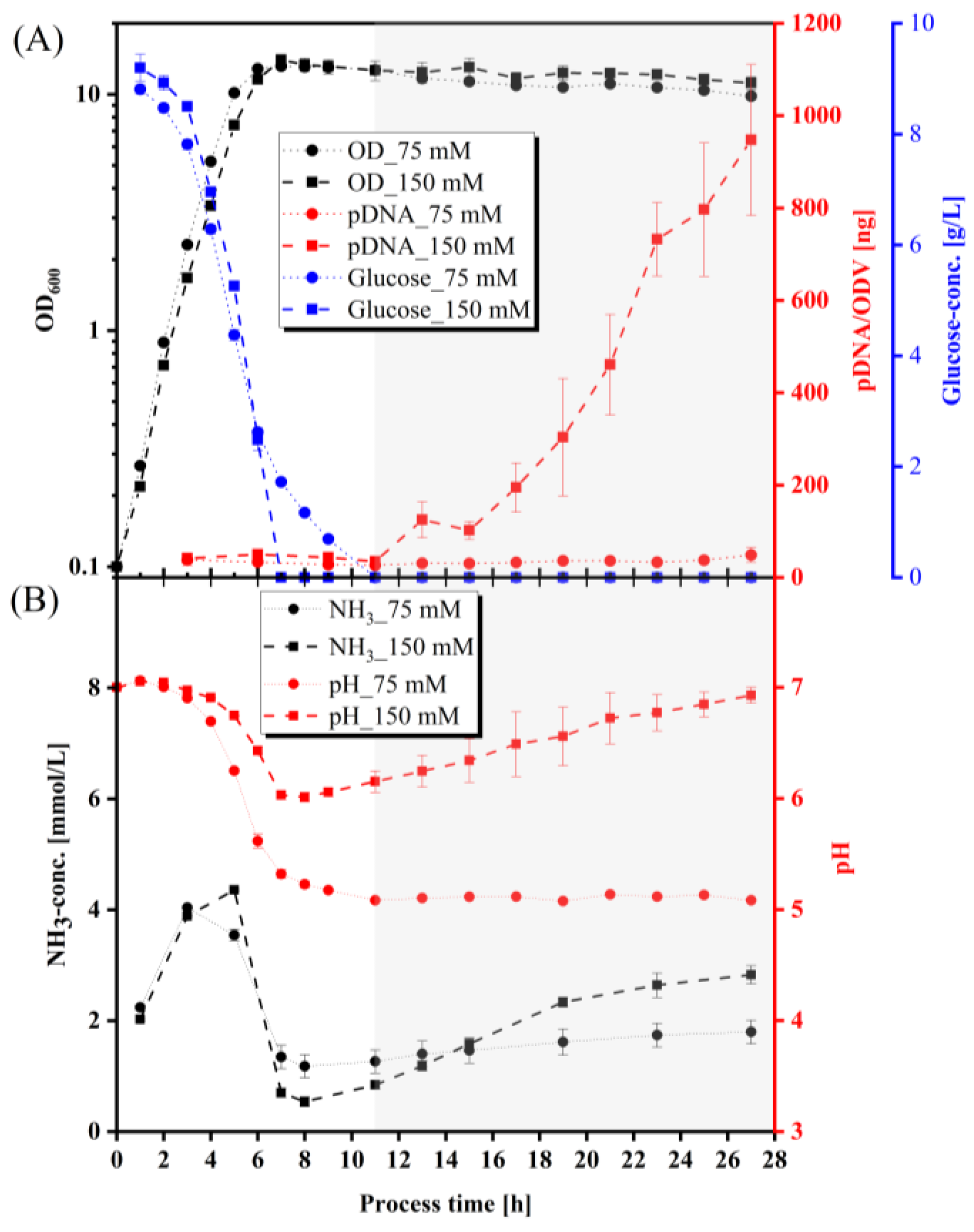
3.1. Optimization of Transfection-Grade pDNA Production
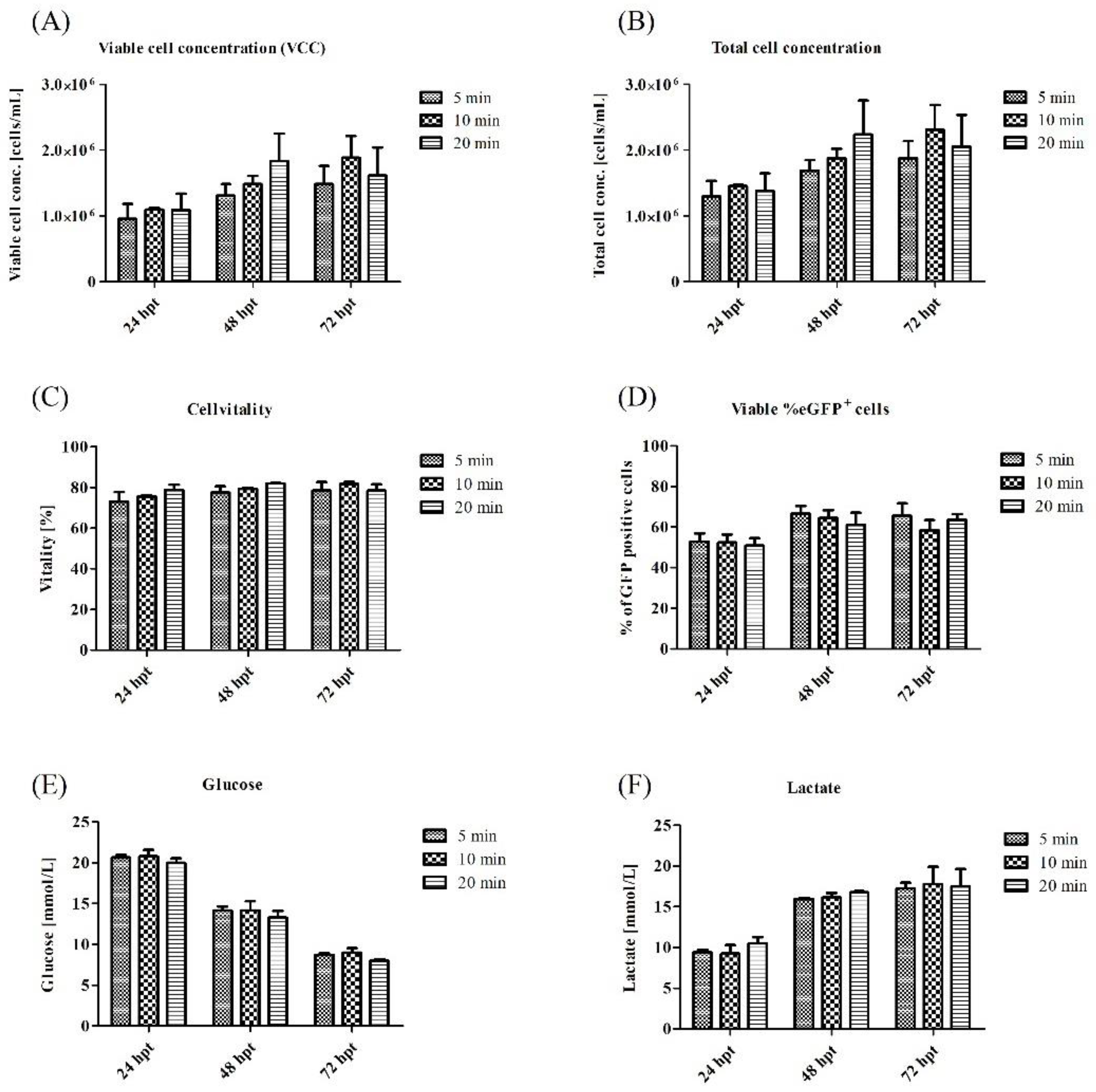
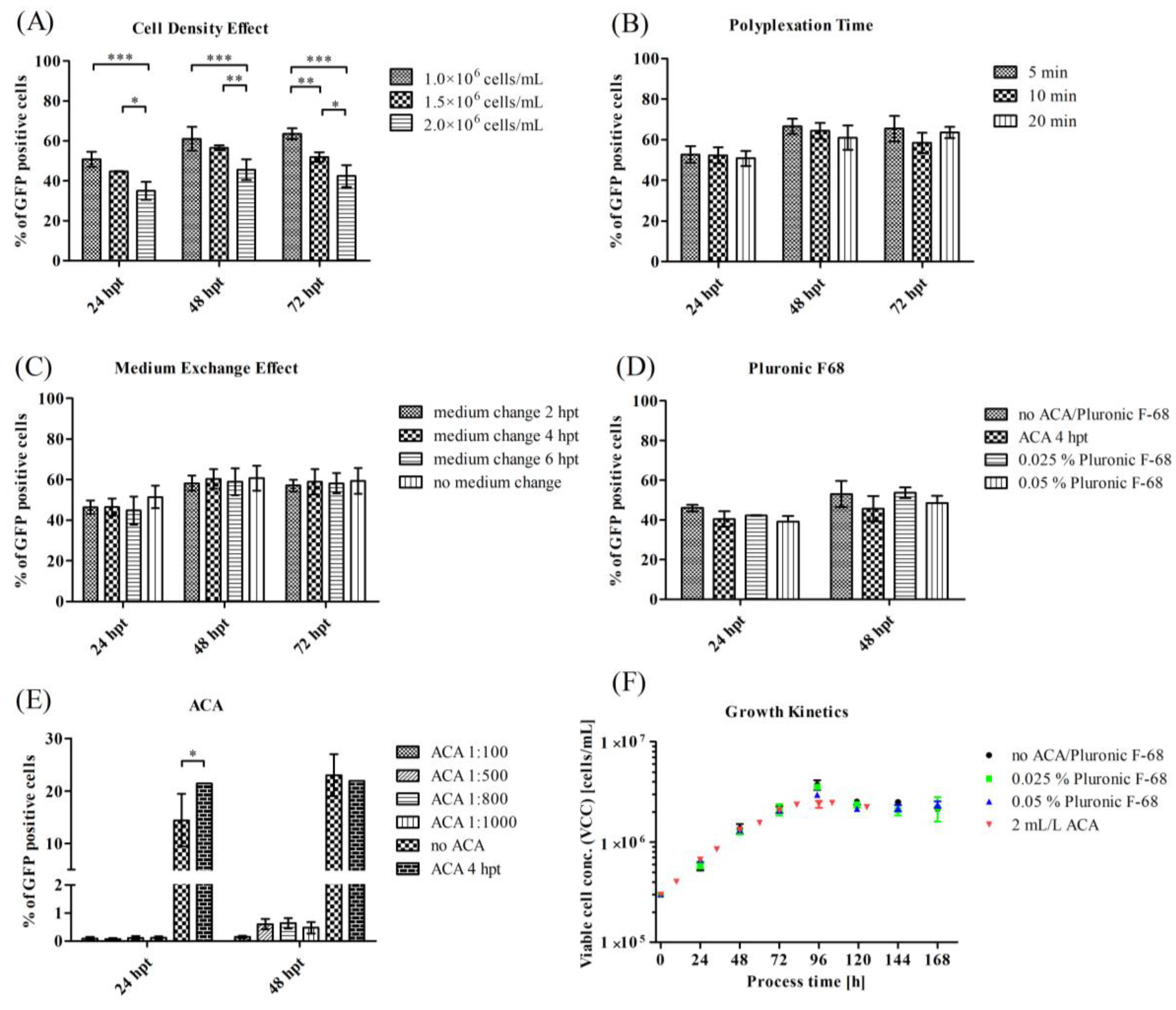
3.2. Optimization of Process Parameters for a Transient VLP Production in a Stirred-Tank Bioreactor
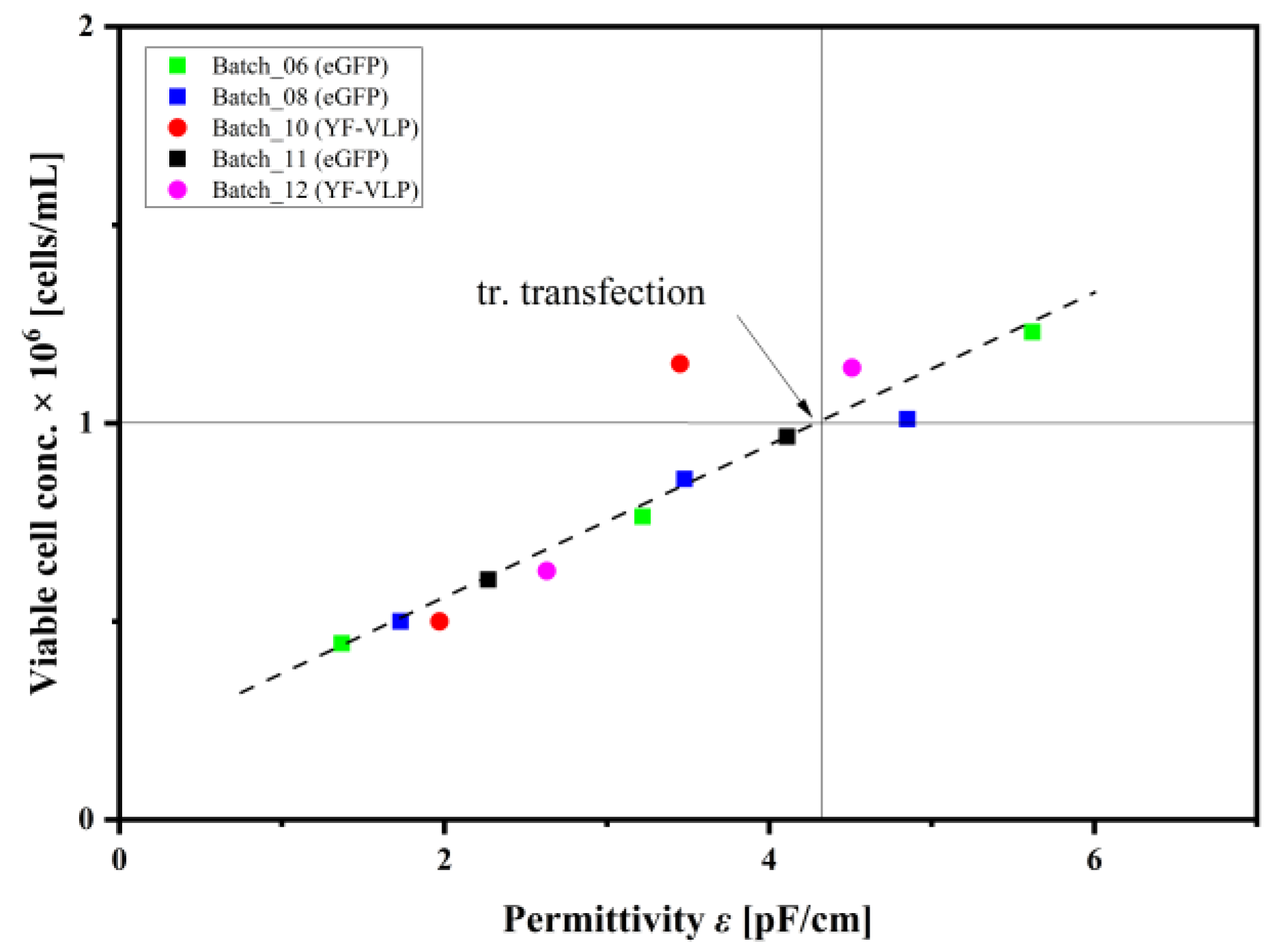
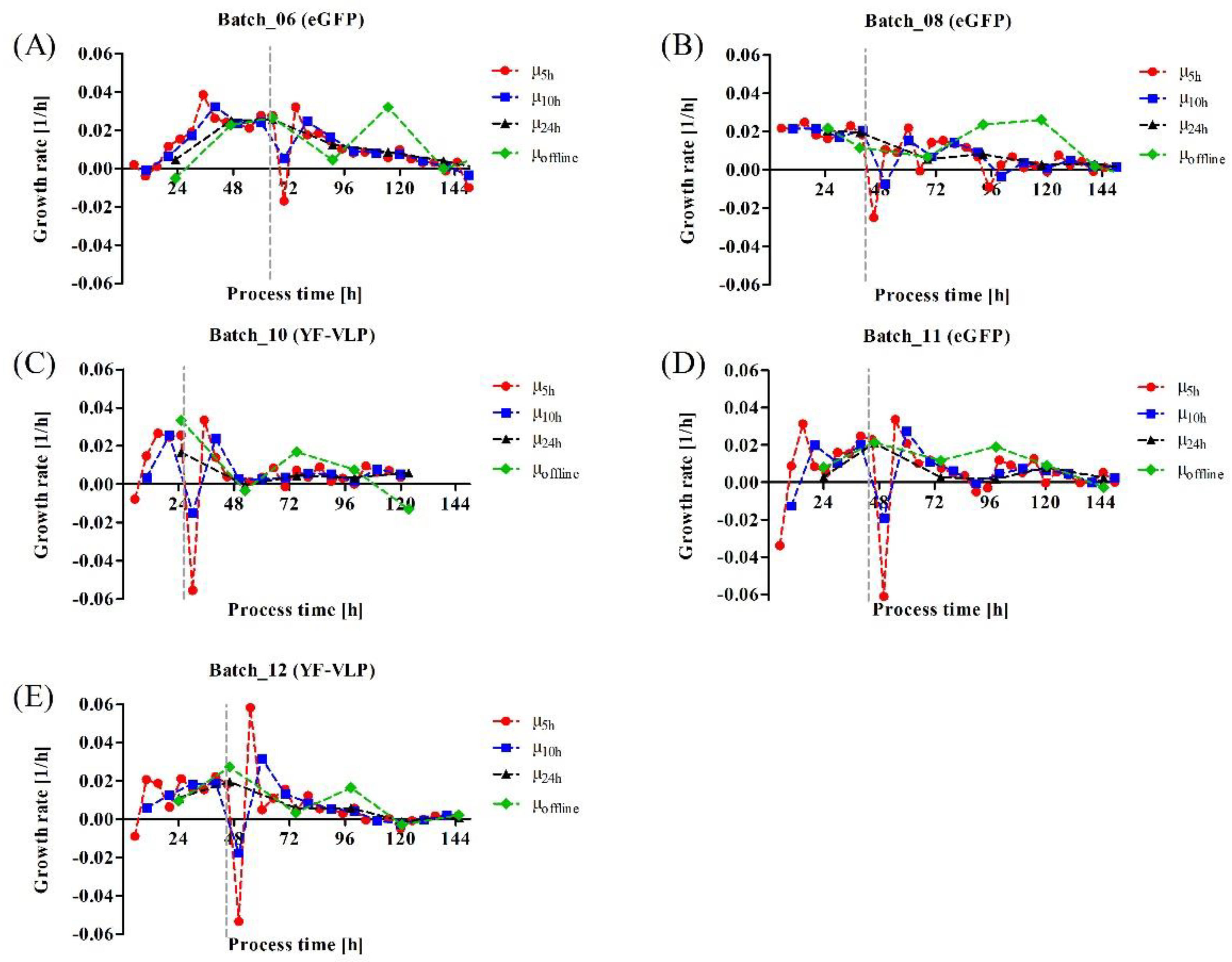
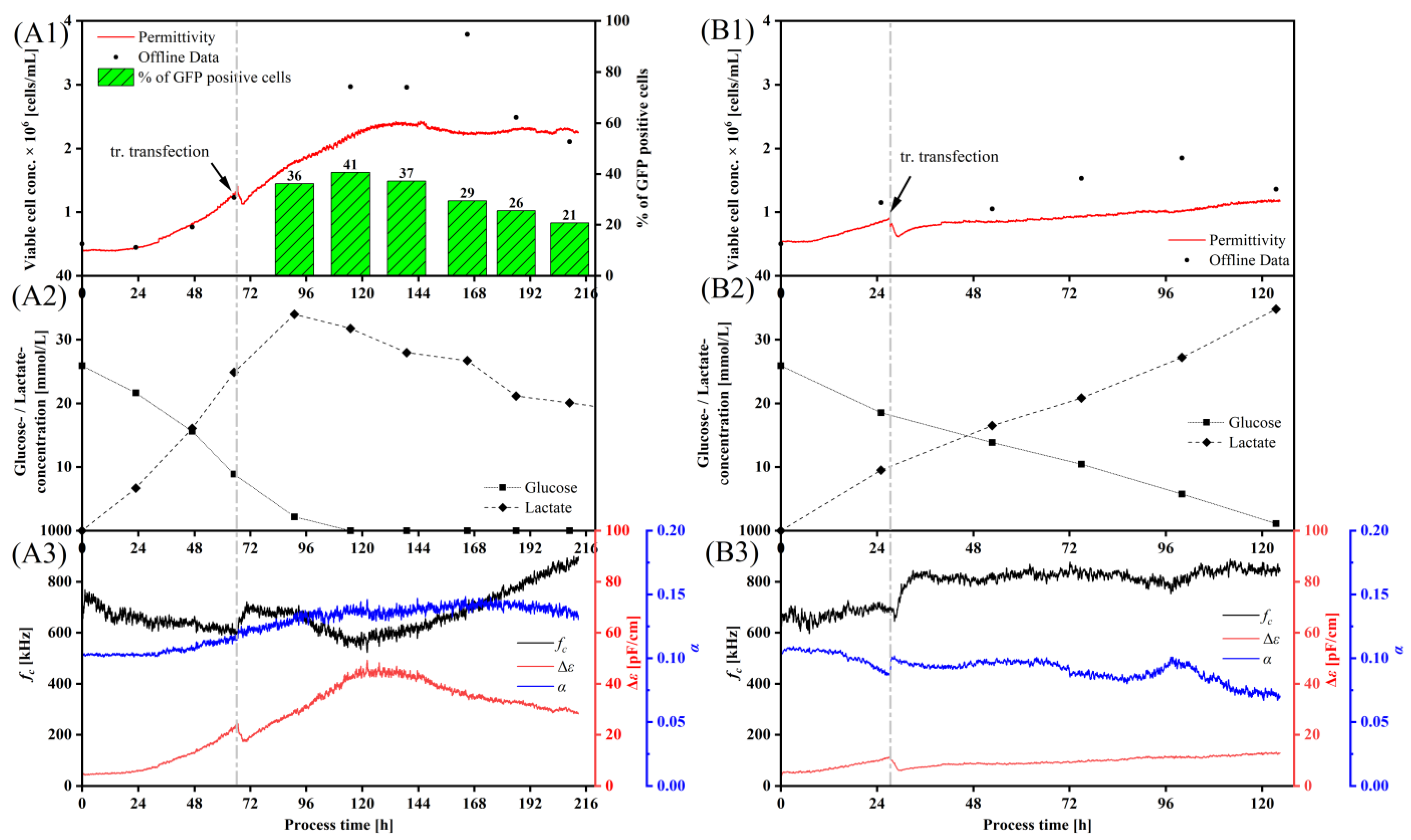
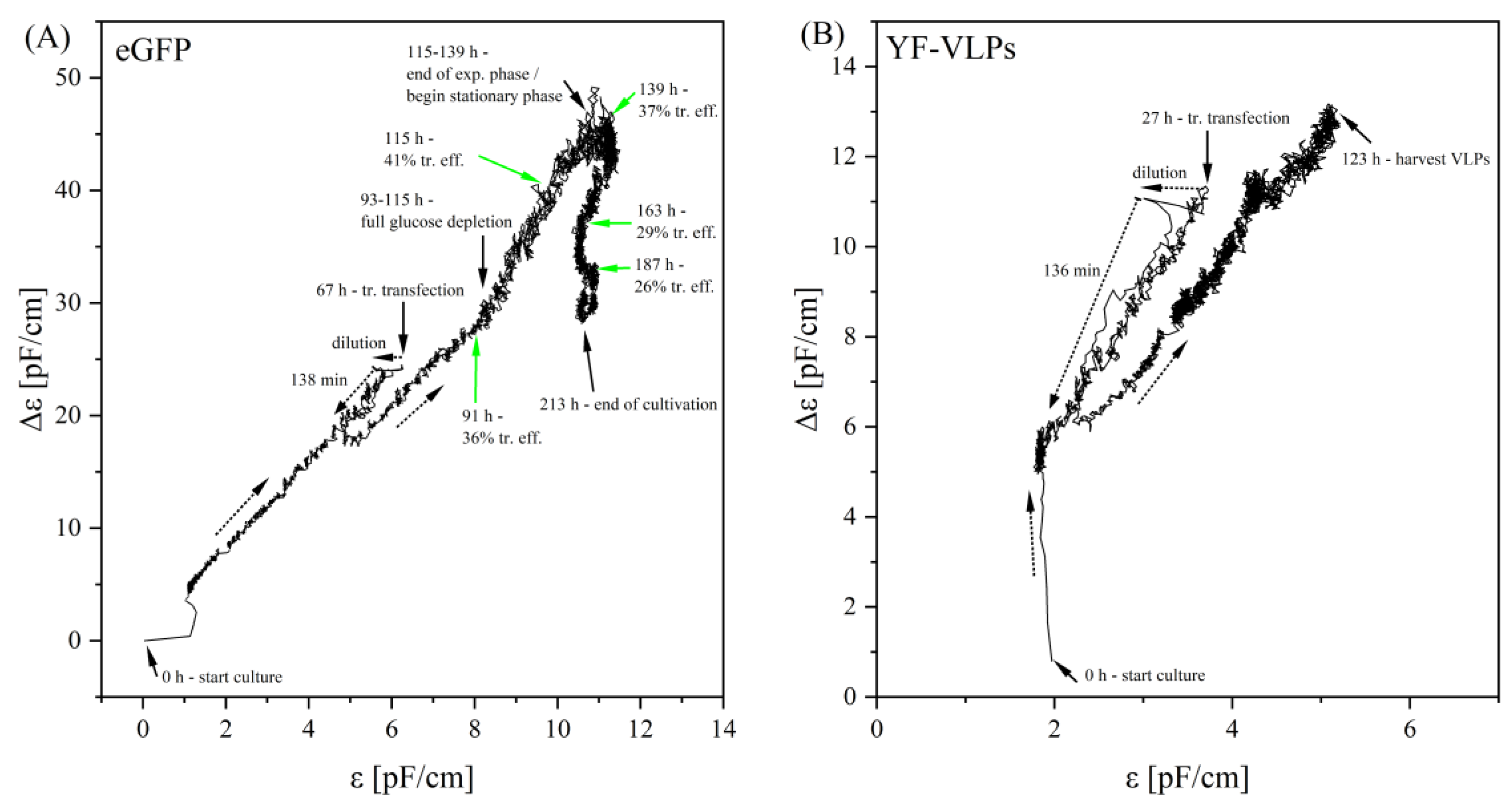
3.3. Monitoring a Transient VLP Production in a Stirred-Tank Bioreactor Using Dielectric Spectroscopy
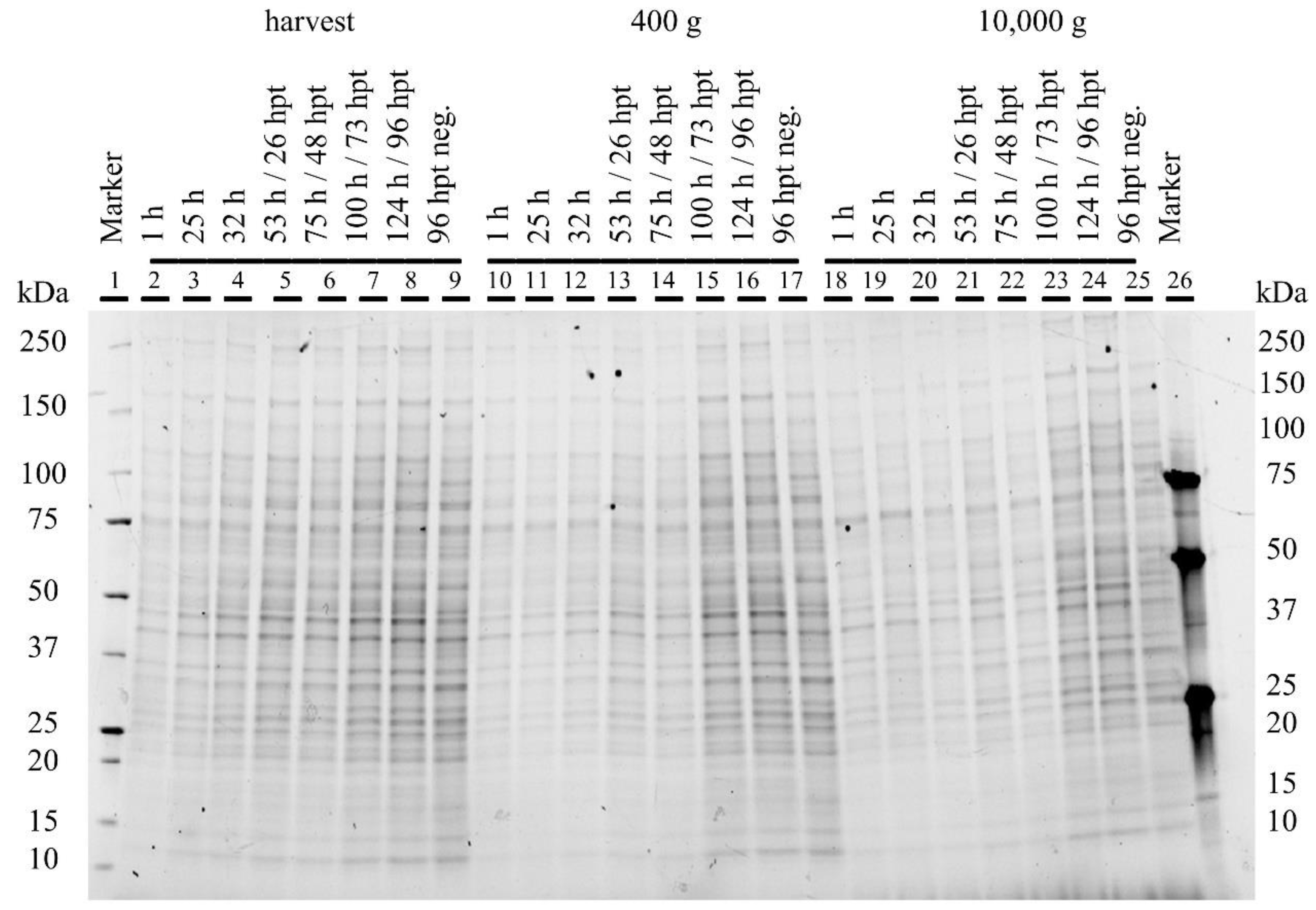
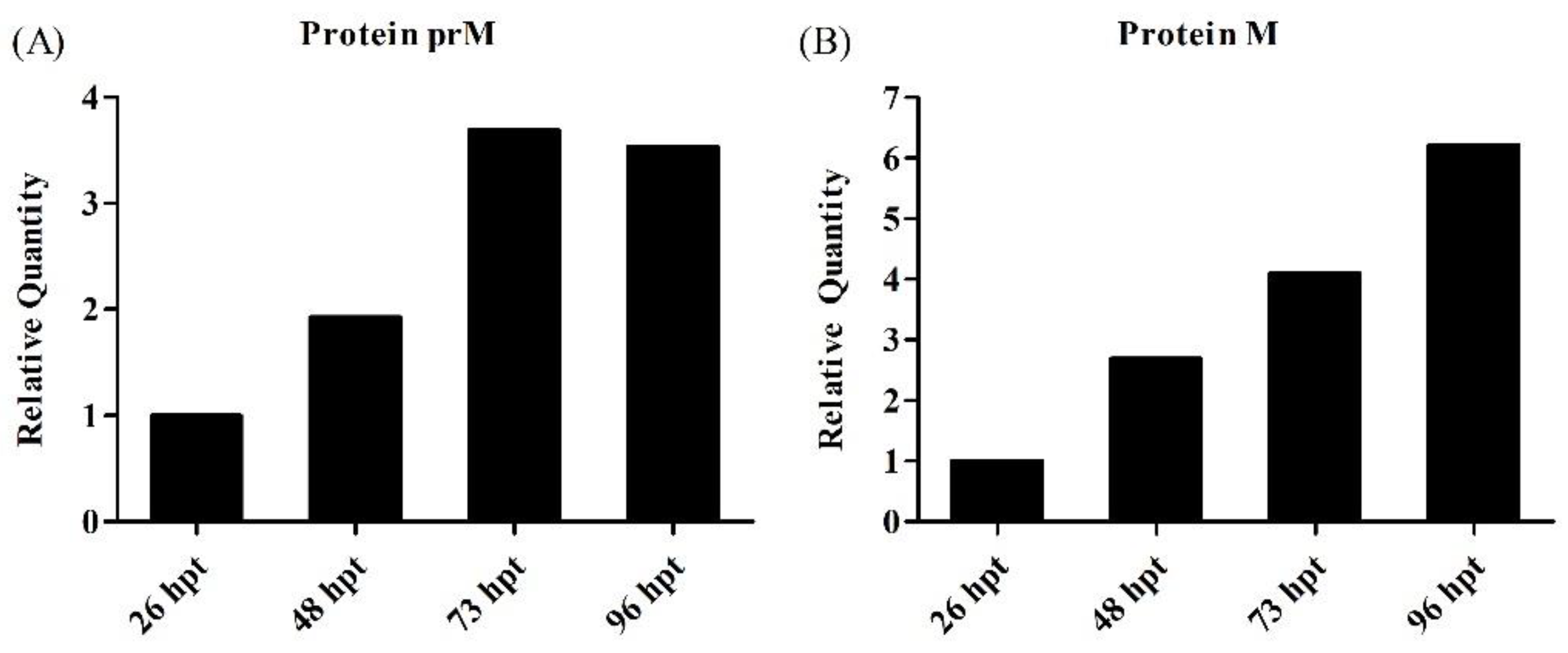
3.4. Characterization of Produced YF-VLPs
4. Discussion
4.1. Impact of Various Parameters on pDNA Production in E. coli
4.2. Influence of Different Process Parameters on Transient Transfection
4.3. Implementation of Dielectric Spectroscopy into a Transient YF-VLP Production Process
4.4. Characterization of the YF-VLP
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



References
- Garske, T.; Van Kerkhove, M.D.; Yactayo, S.; Ronveaux, O.; Lewis, R.F.; Staples, J.E.; Perea, W.; Ferguson, N.M.; Yellow Fever Expert Committee. Yellow Fever in Africa: Estimating the burden of disease and impact of mass vaccination from outbreak and serological data. PLoS Med. 2014, 11, e1001638. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P. Yellow fever vaccine. Expert. Rev. Vaccines 2005, 4, 553–574. [Google Scholar] [CrossRef]
- Barrett, A.D.T.; Higgs, S. Yellow fever: A disease that has yet to be conquered. Annu. Rev. Entomol. 2007, 52, 209–229. [Google Scholar] [CrossRef] [PubMed]
- Frierson, J.G. The Yellow Fever Vaccine: A History. Yale J. Biol. Med. 2020, 83, 77–85. [Google Scholar]
- Grobbelaar, A.A.; Weyer, J.; Moolla, N.; van Vuren, P.J.; Moises, F.; Paweska, J.T. Resurgence of Yellow Fever in Angola, 2015–2016. Emerg. Infect. Dis. 2016, 22, 1854–1855. [Google Scholar] [CrossRef] [PubMed]
- Goldani, L.Z. Yellow fever outbreak in Brazil, 2017. Braz. J. Infect. Dis. 2017, 21, 123–124. [Google Scholar] [CrossRef] [PubMed]
- Noad, R.; Roy, P. Virus-like particles as immunogens. Trends Microbiol. 2013, 11, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Gheysen, D.; Jacobs, E.; Foresta, F.d.; Thiriart, C.; Francotte, M.; Thines, D.; De Wilde, M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 1989, 59, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Delchambre, M.; Gheysen, D.; Thines, D.; Thiriart, C.; Jacobs, E.; Verdin, E.; Horth, M.; Burny, A.; Bex, F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989, 8, 2653–2660. [Google Scholar] [CrossRef]
- Zeltins, A. Construction and characterization of virus-like particles: A review. Mol. Biotechnol. 2013, 53, 92–107. [Google Scholar] [CrossRef]
- Lothert, K.; Dekevic, G.; Loewe, D.; Salzig, D.; Czermak, P.; Wolff, M.W. Upstream and Downstream Processes for Viral Nanoplexes as Vaccines. Methods Mol. Biol. 2021, 2183, 217–248. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, H.; Segawa, M.; Nakamura, M.; Katsuda, T.; Kuwahara, M.; Konishi, E. Production of Japanese encephalitis virus-like particles using the baculovirus-insect cell system. J. Biosci. Bioeng. 2012, 114, 657–662. [Google Scholar] [CrossRef]
- Hirsch, J.; Faber, B.W.; Crowe, J.E.; Verstrepen, B.; Cornelissen, G.E. coli production process yields stable dengue 1 virus-sized particles (VSPs). Vaccine 2020, 38, 3305–3312. [Google Scholar] [CrossRef] [PubMed]
- Ponndorf, D.; Meshcheriakova, Y.; Thuenemann, E.C.; Dobon Alonso, A.; Overman, R.; Holton, N.; Dowall, S.; Kennedy, E.; Stocks, M.; Lomonossoff, G.P.; et al. Plant-made dengue virus-like particles produced by co-expression of structural and non-structural proteins induce a humoral immune response in mice. Plant Biotechnol. J. 2021, 19, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.; Li, Y.; Ahlemeyer, B.; Krieglstein, J.; Kissel, T. In vitro cytotoxicity testing of polycations: Influence of polymerstructure on cell viability an dhemolysis. Biomaterials 2003, 24, 1121–1131. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Symonds, P.; Murray, J.C.; Hunter, A.C.; Debska, G.; Szewczyk, A. A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Mol. Ther. 2005, 11, 990–995. [Google Scholar] [CrossRef]
- Puckette, M.; Primavera, V.; Martel, E.; Barrera, J.; Hurtle, W.; Clark, B.; Kamicker, B.; Zurita, M.; Brake, D.; Neilan, J. Transiently Transfected Mammalian Cell Cultures: An Adaptable and Effective Platform for Virus-like Particle-Based Vaccines against Foot-and-Mouth Disease Virus. Viruses 2022, 14, 989. [Google Scholar] [CrossRef]
- Cervera, L.; Gutiérrez-Granados, S.; Martínez, M.; Blanco, J.; Gòdia, F.; Segura, M.M. Generation of HIV-1 Gag VLPs by transient transfection of HEK 293 suspension cell cultures using an optimized animal-derived component free medium. J. Biotechnol. 2013, 166, 152–165. [Google Scholar] [CrossRef]
- González-Domínguez, I.; Lorenzo, E.; Bernier, A.; Cervera, L.; Gòdia, F.; Kamen, A. A Four-Step Purification Process for Gag VLPs: From Culture Supernatant to High-Purity Lyophilized Particles. Vaccines 2021, 9, 1154. [Google Scholar] [CrossRef]
- Lavado-García, J.; Jorge, I.; Cervera, L.; Vazquez, J.; Gòdia, F. Multiplexed Quantitative Proteomic Analysis of HEK293 Provides Insights into Molecular Changes Associated with the Cell Density Effect, Transient Transfection, and Virus-Like Particle Production. J. Proteome Res. 2020, 19, 1085–1099. [Google Scholar] [CrossRef]
- Alvim, R.G.F.; Lima, T.M.; Silva, J.L.; Silva, J.L.; de Oliveira, G.A.; Castilho, L.R. Process intensification for the production of yellow fever virus-like particles as potential recombinant vaccine antigen. Biotechnol. Bioeng. 2021, 118, 3581–3592. [Google Scholar] [CrossRef]
- Alvim, R.G.F.; Itabaiana, I.; Castilho, L.R. Zika virus-like particles (VLPs): Stable cell lines and continuous perfusion processes as a new potential vaccine manufacturing platform. Vaccine 2019, 37, 6970–6977. [Google Scholar] [CrossRef] [PubMed]
- Druzinec, D.; Weiss, K.; Elseberg, C.; Salzig, D.; Kraume, M.; Pörtner, R.; Czermak, P. Process analytical technology (PAT) in insect and mammalian cell culture processes: Dielectric spectroscopy and focused beam reflectance measurement (FBRM). Methods Mol. Biol. 2014, 1104, 313–341. [Google Scholar] [CrossRef]
- Markx, G.H.; Davey, C.L. The dielectric properties of biological cells at radiofrequencies: Applications in biotechnology. Enzym. Microb. Technol. 1999, 25, 161–171. [Google Scholar] [CrossRef]
- Negrete, A.; Esteban, G.; Kotin, R.M. Process optimization of large-scale production of recombinant adeno-associated vectors using dielectric spectroscopy. Appl. Microbiol. Biotechnol. 2007, 76, 761–772. [Google Scholar] [CrossRef]
- Flores-Cosío, G.; Herrera-López, E.J.; Arellano-Plaza, M.; Gschaedler-Mathis, A.; Kirchmayr, M.; Amaya-Delgado, L. Application of dielectric spectroscopy to unravel the physiological state of microorganisms: Current state, prospects and limits. Appl. Microbiol. Biotechnol. 2020, 104, 6101–6113. [Google Scholar] [CrossRef] [PubMed]
- Schwan, H.P. Electrical properties of tissue and cell suspensions. Adv. Biol. Med. Phys. 1957, 5, 147–209. [Google Scholar] [CrossRef]
- Cole, K.S.; Cole, R.H. Dispersion and Absorption in Dielectrics, I. Alternating Current Characteristics. J. Chem. Phys. 1041, 9, 341–351. [Google Scholar] [CrossRef]
- Kiviharju, K.; Salonen, K.; Moilanen, U.; Meskanen, E.; Leisola, M.; Eerikäinen, T. On-line biomass measurements in bioreactor cultivations: Comparison study of two on-line probes. J. Ind. Microbiol. Biotechnol. 2007, 34, 561–566. [Google Scholar] [CrossRef]
- Olsson, L.; Nielsen, J. On-line and in situ monitoring of biomass in submerged cultivations. Trends Biotechnol. 1997, 15, 517–522. [Google Scholar] [CrossRef]
- Sonnleitner, B.; Locher, G.; Fiechter, A. Biomass determination. J. Biotechnol. 1997, 25, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Carvell, J.P.; Dowd, J.E. On-line Measurements and Control of Viable Cell Density in Cell Culture Manufacturing Processes using Radio-frequency Impedance. Cytotechnology 2006, 50, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Konstantinov, K.; Chuppa, S.; Sajan, E.; Tsai, Y.; Yoon, S.; Golini, F. Real-time biomass-concentration monitoring in animal-cell cultures. Trends Biotechnol. 1994, 12, 324–333. [Google Scholar] [CrossRef]
- Brunner, S.; Sauer, T.; Carotta, S.; Cotten, M.; Saltik, M.; Wagner, E. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000, 7, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Kiviharju, K.; Salonen, K.; Moilanen, U.; Eerikäinen, T. Biomass measurement online: The performance of in situ measurements and software sensors. J. Ind. Microbiol. Biotechnol. 2008, 35, 657–665. [Google Scholar] [CrossRef]
- Justice, C.; Brix, A.; Freimark, D.; Kraume, M.; Pfromm, P.; Eichenmueller, B.; Czermak, P. Process control in cell culture technology using dielectric spectroscopy. Biotechnol. Adv. 2011, 29, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Zitzmann, J.; Weidner, T.; Eichner, G.; Salzig, D.; Czermak, P. Dielectric Spectroscopy and Optical Density Measurement for the Online Monitoring and Control of Recombinant Protein Production in Stably Transformed Drosophila melanogaster S2 Cells. Sensors 2018, 18, 900. [Google Scholar] [CrossRef]
- Grein, T.A.; Loewe, D.; Dieken, H.; Salzig, D.; Weidner, T.; Czermak, P. High titer oncolytic measles virus production process by integration of dielectric spectroscopy as online monitoring system. Biotechnol. Bioeng. 2018, 115, 1186–1194. [Google Scholar] [CrossRef]
- Dekevic, G.; Tasto, L.; Czermak, P.; Salzig, D. Statistical experimental designs to optimize the transient transfection of HEK 293T cells and determine a transfer criterion from adherent cells to larger-scale cell suspension cultures. J. Biotechnol. 2022, 346, 23–34. [Google Scholar] [CrossRef]
- Welsh, J.A.; Van Der Pol, E.; Arkesteijn, G.J.A.; Bremer, M.; Brisson, A.; Coumans, F.; Dignat-George, F.; Duggan, E.; Ghiran, I.; Giebel, B.; et al. MIFlowCyt-EV: A framework for standardized reporting of extracellular vesicle flow cytometry experiments. J. Extracell. Vesicles 2020, 9, 1713526. [Google Scholar] [CrossRef]
- Tertel, T.; Bremer, M.; Maire, C.; Lamszus, K.; Peine, S.; Jawad, R.; Andaloussi, S.E.L.; Giebel, B.; Ricklefs, F.L.; Görgens, A. High-Resolution Imaging Flow Cytometry Reveals Impact of Incubation Temperature on Labeling of Extracellular Vesicles with Antibodies. Cytom. A 2020, 97, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Görgens, A.; Bremer, M.; Ferrer-Tur, R.; Murke, F.; Tertel, T.; Horn, P.A.; Thalmann, S.; Welsh, J.A.; Probst, C.; Guerin, C.; et al. Optimisation of imaging flow cytometry for the analysis of single extracellular vesicles by using fluorescence-tagged vesicles as biological reference material. J. Extracell. Vesicles 2019, 8, 1587567. [Google Scholar] [CrossRef] [PubMed]
- Tertel, T.; Görgens, A.; Giebel, B. Analysis of individual extracellular vesicles by imaging flow cytometry. Methods Enzymol. 2020, 645, 55–78. [Google Scholar] [CrossRef]
- Wang, C.; Börger, V.; Sardari, M.; Murke, F.; Skuljec, J.; Pul, R.; Hagemann, N.; Dzyubenko, E.; Dittrich, R.; Gregorius, J.; et al. Mesenchymal Stromal Cell-Derived Small Extracellular Vesicles Induce Ischemic Neuroprotection by Modulating Leukocytes and Specifically Neutrophils. Stroke 2020, 51, 1825–1834. [Google Scholar] [CrossRef] [PubMed]
- Siebertz, K.; van Bebber, D.; Hochkirchen, T. (Eds.) Statistische Versuchsplanung: Design of Experiments (DoE); Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Williams, J.A.; Luke, J.; Langtry, S.; Meskanen, E.; Leisola, M.; Eerikäinen, T. Generic plasmid DNA production platform incorporating low metabolic burden seed-stock and fed-batch fermentation processes. Biotechnol. Bioeng. 2009, 103, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Geisse, S.; Henke, M. Large-scale transient transfection of mammalian cells: A newly emerging attractive option for recombinant protein production. J. Struct. Funct. Genom. 2005, 6, 165–170. [Google Scholar] [CrossRef]
- Pear, W. Transient transfection methods for preparation of high-titer retroviral supernatants. Curr. Protoc. Mol. Biol. 2001, 9, mb0911s36. [Google Scholar] [CrossRef]
- Tom, R.; Bisson, L.; Durocher, Y. Transfection of HEK293-EBNA1 Cells in Suspension with Linear PEI for Production of Recombinant Proteins. CSH Protoc. 2008, 3, 1–4. [Google Scholar] [CrossRef][Green Version]
- Droste, M.; Tertel, T.; Jeruschke, S.; Dittrich, R.; Kontopoulou, E.; Walkenfort, B.; Börger, V.; Hoyer, P.F.; Büscher, A.K.; Thakur, B.K.; et al. Single Extracellular Vesicle Analysis Performed by Imaging Flow Cytometry and Nanoparticle Tracking Analysis Evaluate the Accuracy of Urinary Extracellular Vesicle Preparation Techniques Differently. Int. J. Mol. Sci. 2021, 22, 12436. [Google Scholar] [CrossRef]
- Miller, A.; Sitter, R.R. Using the Folded-Over 12-Run Plackett—Burman Design to Consider Interactions. Technometrics 2001, 43, 44–55. [Google Scholar] [CrossRef]
- Yu, X.; Hallett, S.G.; Sheppard, J.; Watson, A.K. Application of the Plackett-Burman experimental design to evaluate nutritional requirements for the production of Colletotrichum coccodes spores. Appl. Microbiol. Biotechnol. 1997, 47, 301–305. [Google Scholar] [CrossRef]
- Stanbury, P.F.; Whitaker, A.; Hall, S.J. Principles of Fermentation Technology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Kepes, A.; Meury, J.; Robin, A.; Jimeno, J. Some Ion Transport Systems in E. coli (Transport of Potassium and of Anionic Sugars). In Biochemistry of Membrane Transport; Semenza, G., Carafoli, E., Eds.; Springer: Berlin/Heidelberg, Germany, 1977; pp. 633–647. [Google Scholar]
- Pinhal, S.; Ropers, D.; Geiselmann, J.; De Jong, H. Acetate Metabolism and the Inhibition of Bacterial Growth by Acetate. J. Bacteriol. 2019, 201, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Luli, G.W.; Strohl, W.R. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 1990, 56, 1004–1011. [Google Scholar] [CrossRef]
- Ongkudon, C.M.; Pickering, R.; Webster, D.; Danquah, M.K. Cultivation of E. coli carrying a plasmid-based Measles vaccine construct (4.2 kbp pcDNA3F) employing medium optimisation and pH-temperature induction techniques. Microb. Cell Fact. 2011, 10, 16. [Google Scholar] [CrossRef]
- Wang, Z.; Le, G.; Shi, Y.; Węgrzyn, G. Medium design for plasmid DNA production based on stoichiometric model. Process Biochem. 2001, 36, 1085–1093. [Google Scholar] [CrossRef]
- Carnes, A.E. Fermentation Design for the Manufacture of Therapeutic Plasmid DNA; BioProcess International: Boston, MI, USA, 2005; pp. 36–44. [Google Scholar]
- Lahijani, R.; Hulley, G.; Soriano, G.; Horn, N.A.; Marquet, M. High-yield production of pBR322-derived plasmids intended for human gene therapy by employing a temperature-controllable point mutation. Hum. Gene Ther. 1996, 7, 1971–1980. [Google Scholar] [CrossRef]
- Wong, E.M.; Muesing, M.A.; Polisky, B. Temperature-sensitive copy number mutants of CoIE1 are located in an untranslated region of the plasmid genome. Proc. Natl. Acad. Sci. USA 1982, 79, 3570–3574. [Google Scholar] [CrossRef]
- Lin-Chao, S.; Chen, W.T.; Wong, T.T. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol. Microbiol. 1992, 6, 3385–3393. [Google Scholar] [CrossRef]
- Carnes, A.E.; Hodgson, C.P.; Williams, J.A. Inducible Escherichia coli fermentation for increased plasmid DNA production. Biotechnol. Appl. Biochem. 2006, 45, 155–166. [Google Scholar] [CrossRef]
- Durland, R.H.; Eastman, E.M. Manufacturing and quality control of plasmid-based gene expression systems. Adv. Drug Deliv. Rev. 1998, 30, 33–48. [Google Scholar] [CrossRef]
- Satyagal, V.N.; Agrawal, P. A generalized model of plasmid replication. Biotechnol. Bioeng. 1989, 33, 1135–1144. [Google Scholar] [CrossRef]
- Seo, J.H.; Bailey, J.E. Effects of recombinant plasmid content on growth properties and cloned gene product formation in Escherichia coli. Biotechnol. Bioeng. 1985, 27, 1668–1674. [Google Scholar] [CrossRef]
- Chen, W.; Graham, C.; Ciccarelli, R.B. Automated fed-batch fermentation with feed-back controls based on dissolved oxygen (DO) and pH for production of DNA vaccines. J. Ind. Microbiol. Biotechnol. 1997, 18, 43–48. [Google Scholar] [CrossRef]
- Kim, B.G.; Shuler, M.L. Analysis of pBR322 replication kinetics and its dependency on growth rate. Biotechnol. Bioeng. 1990, 36, 233–242. [Google Scholar] [CrossRef]
- Reinikainen, P.; Korpela, K.; Nissinen, V.; Olkku, J.; Söderlund, H.; Markkanen, P. Escherichia coli plasmid production in fermenter. Biotechnol. Bioeng. 1989, 33, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Namdev, P.K.; Irwin, N.; Thompson, B.G.; Gray, M.R. Effect of oxygen fluctuations on recombinant Escherichia coli fermentation. Biotechnol. Bioeng. 1993, 41, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Prather, K.J.; Sagar, S.; Murphy, J.; Chartrain, M. Industrial scale production of plasmid DNA for vaccine and gene therapy: Plasmid design, production, and purification. Enzym. Microb. Technol. 2003, 33, 865–883. [Google Scholar] [CrossRef]
- Follmann, M.; Ochrombel, I.; Krämer, R.; Trötschel, C.; Poetsch, A.; Rückert, C.; Hüser, A.; Persicke, M.; Seiferling, D.; Kalinowski, J.; et al. Functional genomics of pH homeostasis in Corynebacterium glutamicum revealed novel links between pH response, oxidative stress, iron homeostasis and methionine synthesis. BMC Genom. 2009, 10, 621. [Google Scholar] [CrossRef] [PubMed]
- Cortés, J.T.; Flores, N.; Bolívar, F.; Lara, A.R.; Ramírez, O.T. Physiological effects of pH gradients on Escherichia coli during plasmid DNA production. Biotechnol. Bioeng. 2016, 113, 598–611. [Google Scholar] [CrossRef]
- O’Mahony, K.; Freitag, R.; Hilbrig, F.; Müller, P.; Schumacher, I. Strategies for high titre plasmid DNA production in Escherichia coli DH5α. Process Biochem. 2007, 42, 1039–1049. [Google Scholar] [CrossRef]
- Derouazi, M.; Girard, P.; van Tilborgh, F.; Iglesias, K.; Muller, N.; Bertschinger, M.; Wurm, F.M. Serum-free large-scale transient transfection of CHO cells. Biotechnol. Bioeng. 2004, 87, 537–545. [Google Scholar] [CrossRef]
- Yang, S.; Shi, H.; Chu, X.; Zhou, X.; Sun, P. A rapid and efficient polyethylenimine-based transfection method to prepare lentiviral or retroviral vectors: Useful for making iPS cells and transduction of primary cells. Biotechnol. Lett. 2016, 38, 1631–1641. [Google Scholar] [CrossRef]
- Backliwal, G.; Hildinger, M.; Hasija, V.; Wurm, F.M. High-density transfection with HEK-293 cells allows doubling of transient titers and removes need for a priori DNA complex formation with PEI. Biotechnol. Bioeng. 2008, 99, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Schlaeger, E.J.; Christensen, K. Transient gene expression in mammalian cells grown in serum-free suspension culture. Cytotechnology 1999, 30, 71–83. [Google Scholar] [CrossRef]
- Nadeau, I.; Kamen, A. Production of adenovirus vector for gene therapy. Biotechnol. Adv. 2003, 20, 475–489. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H.T.; Tran, T.T.B.; Chan, L.C.L.; Nielsen, L.K.; Reid, S. Effect of the peak cell density of recombinant AcMNPV-infected Hi5 cells on baculovirus yields. Appl. Microbiol. Biotechnol. 2015, 99, 1687–1700. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Dalby, B.; Chen, W.; Kilzer, J.M.; Chiou, H.C. Transient transfection factors for high-level recombinant protein production in suspension cultured mammalian cells. Mol. Biotechnol. 2008, 39, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.; Bieber, T.; Li, Y.; Elsässer, H.P.; Kissel, T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: Effect of molecular weight on transfection efficiency and cytotoxicity. Pharm. Res. 1999, 16, 1273–1279. [Google Scholar] [CrossRef]
- Ducommun, P.; Kadouri, A.; Stockar, U.; Marison, I.W. On-line determination of animal cell concentration in two industrial high-density culture processes by dielectric spectroscopy. Biotechnol. Bioeng. 2002, 77, 316–323. [Google Scholar] [CrossRef]
- Ducommun, P.; Bolzonella, I.; Rhiel, M.; Pugeaud, P.; Von Stockar, U.; Marison, I.W. On-line determination of animal cell concentration. Biotechnol. Bioeng. 2001, 72, 515–522. [Google Scholar] [CrossRef]
- Choosakoonkriang, S.; Lobo, B.A.; Koe, G.S.; Koe, J.G.; Middaugh, C.R. Biophysical characterization of PEI/DNA complexes. J. Pharm. Sci. 2003, 92, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Kircheis, R.; Schüller, S.; Brunner, S.; Ogris, M.; Heider, K.H.; Zauner, W.; Wagner, E. Polycation-based DNA complexes for tumor-targeted gene deliveryin vivo. J. Gene Med. 1999, 1, 111–120. [Google Scholar] [CrossRef]
- Ogris, M.; Brunner, S.; Schüller, S.; Kircheis, R.; Wagner, E. PEGylated DNA/transferrin-PEI complexes: Reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999, 6, 595–605. [Google Scholar] [CrossRef]
- Ansorge, S.; Lanthier, S.; Transfiguracion, J.; Henry, O.; Kamen, A. Monitoring lentiviral vector production kinetics using online permittivity measurements. Biochem. Eng. J. 2011, 54, 16–25. [Google Scholar] [CrossRef]
- Patel, P.; Markx, G.H. Dielectric measurement of cell death. Enzym. Microb. Technol. 2008, 43, 463–470. [Google Scholar] [CrossRef]
- Ron, A.; Fishelson, N.; Croitoriu, N.; Benayahu, D.; Shacham-Diamand, Y. Theoretical examination of aggregation effect on the dielectric characteristics of spherical cellular suspension. Biophys. Chem. 2009, 140, 39–50. [Google Scholar] [CrossRef]
- Ansorge, S.; Esteban, G.; Schmid, G. Multifrequency permittivity measurements enable on-line monitoring of changes in intracellular conductivity due to nutrient limitations during batch cultivations of CHO cells. Biotechnol. Prog. 2010, 26, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Ansorge, S.; Esteban, G.; Schmid, G. On-line monitoring of responses to nutrient feed additions by multi-frequency permittivity measurements in fed-batch cultivations of CHO cells. Cytotechnology 2010, 62, 121–132. [Google Scholar] [CrossRef]
- Liste-Calleja, L.; Lecina, M.; Lopez-Repullo, J.; Albiol, J.; Solà, C.; Cairó, J.J. Lactate and glucose concomitant consumption as a self-regulated pH detoxification mechanism in HEK293 cell cultures. Appl. Microbiol. Biotechnol. 2015, 99, 9951–9960. [Google Scholar] [CrossRef]
- Petiot, E.; El-Wajgali, A.; Esteban, G.; Gény, C.; Pinton, H.; Marc, A. Real-time monitoring of adherent Vero cell density and apoptosis in bioreactor processes. Cytotechnology 2012, 64, 429–441. [Google Scholar] [CrossRef]
- Ma, F.; Zhang, A.; Chang, D.; Velev, O.D.; Wiltberger, K.; Kshirsagar, R. Real-time monitoring and control of CHO cell apoptosis by in situ multifrequency scanning dielectric spectroscopy. Proc. Biochem. 2019, 80, 138–145. [Google Scholar] [CrossRef]
- González-Correa, C.A.; Colina-Gallo, E.; Miranda-Mercado, D.A. The alpha parameter of the Cole-Cole model as an indicator of fibromyalgia. J. Phys. Conf. Ser. 2019, 1272, 12003. [Google Scholar] [CrossRef]
- Boigard, H.; Alimova, A.; Martin, G.R.; Katz, A.; Gottlieb, P.; Galarza, J.M. Zika virus-like particle (VLP) based vaccine. PLoS Negl. Trop. Dis. 2017, 11, e0005608. [Google Scholar] [CrossRef]
- Kümmerer, B.M.; Rice, C.M. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 2002, 76, 4773–4784. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, M.C.; Sequeira, P.C.; Galler, R. The yellow fever 17D virus as a platform for new live attenuated vaccines. Hum. Vaccines Immunother. 2014, 10, 1256–1265. [Google Scholar] [CrossRef]
- Beeck, A.; Molenkamp, R.; Caron, M.; Ben Younes, A.; Bredenbeek, P.; Dubuisson, J. Role of the transmembrane domains of prM and E proteins in the formation of yellow fever virus envelope. J. Virol. 2003, 77, 813–820. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wu, L.; Chen, L.; Qin, Y.; Pan, Z.; Chen, M. Vesicular stomatitis virus-based vaccines expressing EV71 virus-like particles elicit strong immune responses and protect newborn mice from lethal challenges. Vaccine 2016, 34, 4196–4204. [Google Scholar] [CrossRef]
- Steppert, P.; Burgstaller, D.; Klausberger, M.; Berger, E.; Pereira Aguilar, P.; Schneider, T.A.; Kramberger, P.; Tover, A.; Nöbauer, K.; Razzazi-Fazeli, E.; et al. Purification of HIV-1 gag virus-like particles and separation of other extracellular particles. J. Chromatogr. A 2016, 1455, 93–101. [Google Scholar] [CrossRef]
- Lin, S.Y.; Chiu, H.Y.; Chiang, B.L.; Hu, Y.C. Development of EV71 virus-like particle purification processes. Vaccine 2015, 33, 5966–5973. [Google Scholar] [CrossRef]
- Negrete, A.; Pai, A.; Shiloach, J. Use of hollow fiber tangential flow filtration for the recovery and concentration of HIV virus-like particles produced in insect cells. J. Virol. Methods 2014, 195, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Staubach, S.; Bauer, F.N.; Tertel, T.; Börger, V.; Stambouli, O.; Salzig, D.; Giebel, B. Scaled preparation of extracellular vesicles from conditioned media. Adv. Drug Deliv. Rev. 2021, 177, 113940. [Google Scholar] [CrossRef] [PubMed]
- Garg, H.; Mehmetoglu-Gurbuz, T.; Joshi, A. Virus Like Particles (VLP) as multivalent vaccine candidate against Chikungunya, Japanese Encephalitis, Yellow Fever and Zika Virus. Sci. Rep. 2020, 10, 4017. [Google Scholar] [CrossRef] [PubMed]
- Krol, E.; Brzuska, G.; Szewczyk, B. Production and Biomedical Application of Flavivirus-like Particles. Trends Biotechnol. 2019, 37, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Hausig-Punke, F.; Dekevic, G.; Sobotta, F.H.; Solomun, J.I.; Richter, F.; Salzig, D.; Traeger, A.; Brendel, J.C. Efficient Transfection via an Unexpected Mechanism by Near Neutral Polypiperazines with Tailored Response to Endosomal pH. Macromol. Biosci. 2023, 23, e2200517. [Google Scholar] [CrossRef]

| Standard | 8 | 6 | 5 | 1 | 7 | 3 | 4 | 12 | 11 | 9 | 10 | 2 | Center Point | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| LB | 1 | −1 | −1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Meat-peptone | 1 | −1 | −1 | 1 | −1 | −1 | 1 | −1 | −1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Casein-peptone | −1 | −1 | 1 | −1 | −1 | 1 | −1 | −1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Yeast extract | −1 | 1 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Glucose | −1 | −1 | 1 | 1 | 1 | −1 | 1 | −1 | 1 | −1 | −1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Glycerol | 1 | 1 | 1 | 1 | −1 | 1 | −1 | −1 | −1 | −1 | −1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Phosphate * | −1 | 1 | −1 | −1 | 1 | 1 | 1 | −1 | −1 | 1 | −1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NaCl | 1 | −1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MgSO4 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Dummy 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | 1 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Dummy 2 | 1 | 1 | −1 | −1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dekevic, G.; Tertel, T.; Tasto, L.; Schmidt, D.; Giebel, B.; Czermak, P.; Salzig, D. A Bioreactor-Based Yellow Fever Virus-like Particle Production Process with Integrated Process Analytical Technology Based on Transient Transfection. Viruses 2023, 15, 2013. https://doi.org/10.3390/v15102013
Dekevic G, Tertel T, Tasto L, Schmidt D, Giebel B, Czermak P, Salzig D. A Bioreactor-Based Yellow Fever Virus-like Particle Production Process with Integrated Process Analytical Technology Based on Transient Transfection. Viruses. 2023; 15(10):2013. https://doi.org/10.3390/v15102013
Chicago/Turabian StyleDekevic, Gregor, Tobias Tertel, Lars Tasto, Deborah Schmidt, Bernd Giebel, Peter Czermak, and Denise Salzig. 2023. "A Bioreactor-Based Yellow Fever Virus-like Particle Production Process with Integrated Process Analytical Technology Based on Transient Transfection" Viruses 15, no. 10: 2013. https://doi.org/10.3390/v15102013
APA StyleDekevic, G., Tertel, T., Tasto, L., Schmidt, D., Giebel, B., Czermak, P., & Salzig, D. (2023). A Bioreactor-Based Yellow Fever Virus-like Particle Production Process with Integrated Process Analytical Technology Based on Transient Transfection. Viruses, 15(10), 2013. https://doi.org/10.3390/v15102013







