#Stayathome If You Have a Cold: High SARS-CoV-2 Salivary Viral Loads in Pediatric Patients with Nasopharyngeal Symptoms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Demographic and Clinical Data
2.2. Study Procedures
2.3. SARS-CoV-2 RNA Detection and Quantification in Saliva
2.4. Statistical Analysis
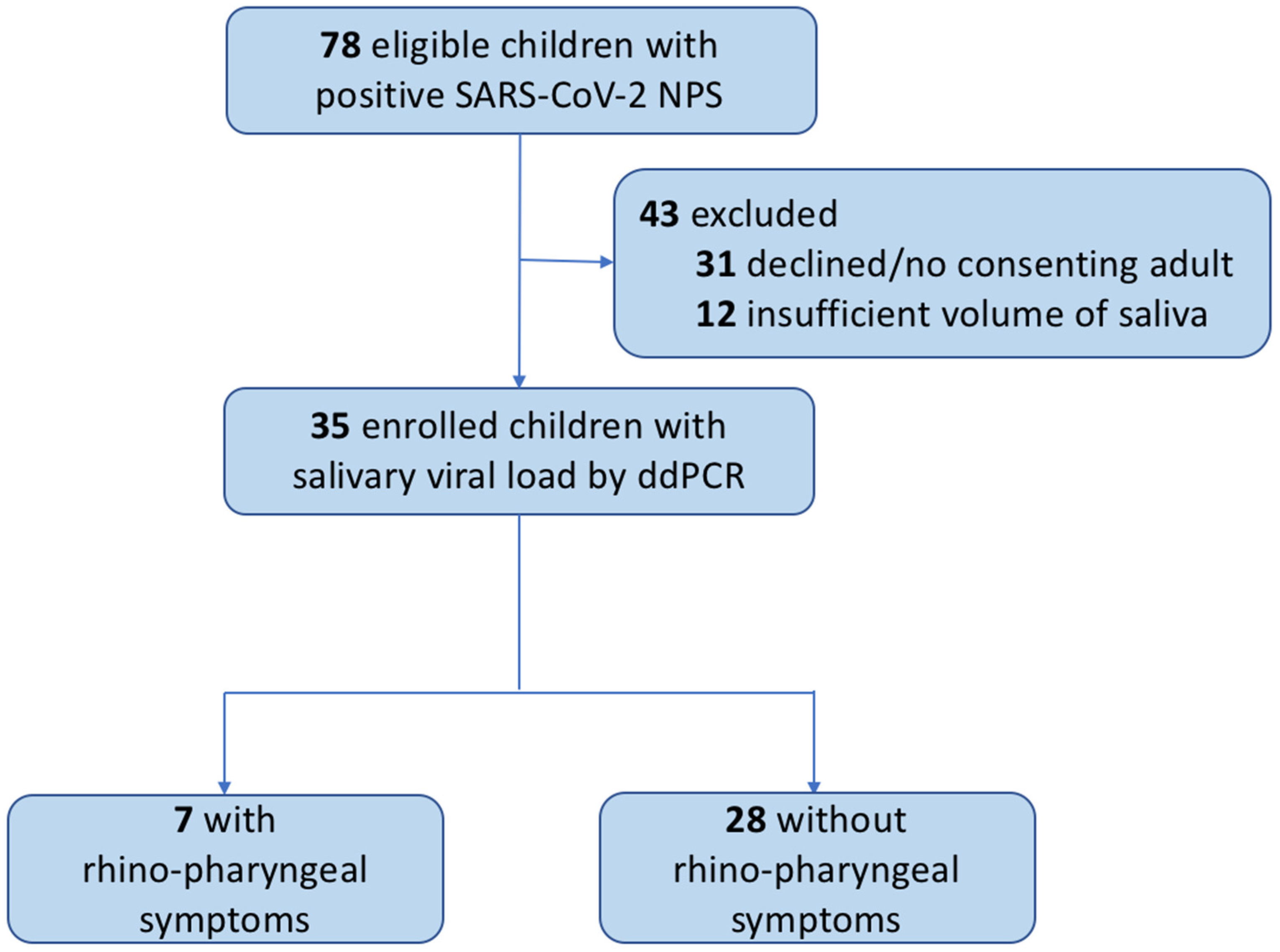
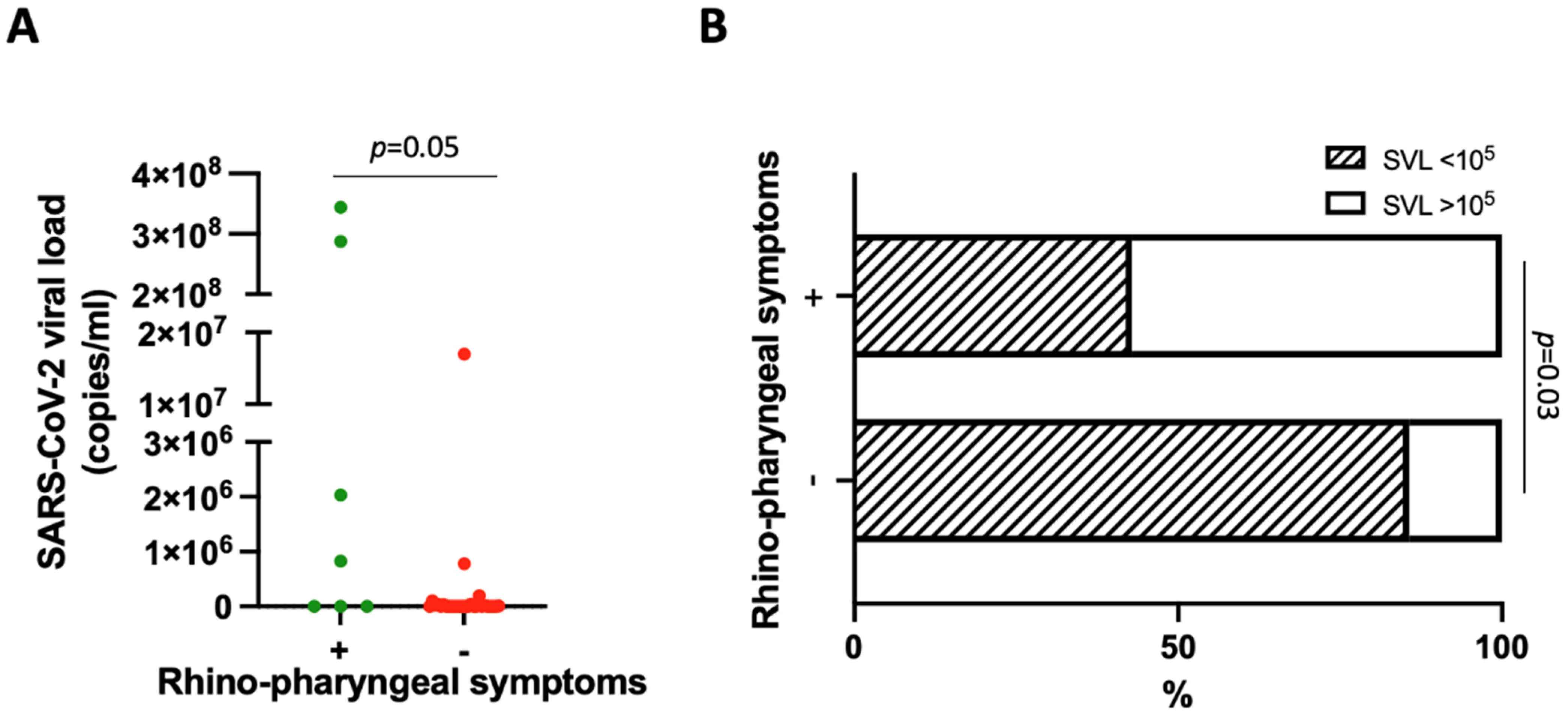
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fougère, Y.; Schwob, J.M.; Miauton, A.; Hoegger, F.; Opota, O.; Jaton, K.; Brouillet, R.; Greub, G.; Genton, B.; Gehri, M.; et al. Performance of RT-PCR on Saliva Specimens Compared With Nasopharyngeal Swabs for the Detection of SARS-CoV-2 in Children: A Prospective Comparative Clinical Trial. Pediatr. Infect. Dis. J. 2021, 40, e300–e304. [Google Scholar] [CrossRef] [PubMed]
- Laura, G.-O.A.; Josué, N.-R.A.; Briceida, L.-M.; Israel, P.-O.; Tania, A.-F.; Nancy, M.-R.; Lourdes, J.-B.; Daniela, D.L.R.-Z.; Fernando, O.-R.; Mauricio, J.-E.C.; et al. Sensitivity of the Molecular Test in Saliva for Detection of COVID-19 in Pediatric Patients With Concurrent Conditions. Front. Pediatr. 2021, 9, 642781. [Google Scholar] [CrossRef] [PubMed]
- Yee, R.; Truong, T.T.; Pannaraj, P.S.; Eubanks, N.; Gai, E.; Jumarang, J.; Turner, L.; Peralta, A.; Lee, Y.; Bard, J.D. Saliva Is a Promising Alternative Specimen for the Detection of SARS-CoV-2 in Children and Adults. J. Clin. Microbiol. 2020, 59, e02686-20. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.A.; Herigon, J.C.; Benedetti, A.; Pollock, N.R.; Denkinger, C.M. Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: A Systematic Review and Meta-analysis. J. Clin. Microbiol. 2021, 59, e02881-20. [Google Scholar] [CrossRef] [PubMed]
- Chua, G.T.; Wong, J.S.C.; To, K.K.W.; Lam, I.C.S.; Yau, F.Y.S.; Chan, W.H.; Ho, P.P.K.; Duque, J.S.R.; Yip, C.C.Y.; Ng, A.C.K.; et al. Saliva viral load better correlates with clinical and immunological profiles in children with coronavirus disease 2019. Emerg. Microbes Infect. 2021, 10, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, L.; Sakthivel, S.K.; Whitaker, B.; Murray, J.; Kamili, S.; Lynch, B.; Malapati, L.; Burke, S.A.; Harcourt, J.; et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1654–1665. [Google Scholar] [CrossRef] [PubMed]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Rashid, F.A.; Sabri, F.S.; Jamil, N.N.; Zain, R.; Hashim, R.; Amran, F.; Kok, H.T.; Samad, M.A.; Ahmad, N. Comparing Nasopharyngeal Swab and Early Morning Saliva for the Identification of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2021, 72, e352–e356. [Google Scholar] [CrossRef] [PubMed]
- Kam, K.Q.; Thoon, K.C.; Maiwald, M.; Chong, C.Y.; Soong, H.Y.; Loo, L.H.; Tan, N.W.H.; Li, J.; Nadua, K.D.; Yung, C.F. SARS-CoV-2 viral RNA load dynamics in the nasopharynx of infected children. Epidemiol. Infect. 2021, 149, e18. [Google Scholar] [CrossRef] [PubMed]
- Salvagno, G.L.; Henry, B.M.; Pighi, L.; De Nitto, S.; Montagnana, M.; Lippi, G. SARS-CoV-2 Omicron infection is associated with high nasopharyngeal viral load. J. Infect. 2022, 84, 834–872. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wang, Z.; Li, J.; Zhang, Y.; Jiang, L.; Fu, Y.; Jin, Y.; Cheng, H.; Li, J.; Chen, Z.; et al. High amounts of SARS-CoV-2 in aerosols exhaled by patients with Omicron variant infection. J. Infect. 2022, 84, e126–e128. [Google Scholar] [CrossRef] [PubMed]
- Granerud, B.K.; Ueland, T.; Lind, A.; Søraas, A.; Fevang, B.; Steffensen, A.K.; Al-Baldawi, H.; Lund-Johansen, F.; Aukrust, P.; Halvorsen, B.; et al. Omicron Variant Generates a Higher and More Sustained Viral Load in Nasopharynx and Saliva Than the Delta Variant of SARS-CoV-2. Viruses 2022, 14, 2420. [Google Scholar] [CrossRef] [PubMed]
| n (%) | ||
|---|---|---|
| Gender | Males | 17 (48.6) |
| Females | 18 (51.4) | |
| Age, median (IQR) | 8 (2–13) | |
| Ethnicity | White | 30 (85.7) |
| Black | 3 (8.6) | |
| Hispanic | 2 (5.7) | |
| Need for hospitalization | Yes | 8 (22.9) |
| No | 27 (77.1) | |
| Prescribed therapy | None | 12 (34.3) |
| Antipyretics | 20 (57.1) | |
| Antibiotics | 14 (40) | |
| Steroids | 9 (25.7) | |
| Referred symptoms at admission | None | 8 (22.9) |
| Fever | 22 (62.9) | |
| Dyspnea | 6 (17.1) | |
| Cough | 5 (14.3) | |
| Diarrhea/abdominal pain | 7 (20) | |
| Vomit | 6 (17.1) | |
| Asthenia | 1 (2.9) | |
| Headache | 3 (8.6) | |
| Rhinitis/pharyngodynia | 7 (20) | |
| Ageusia/anosmia | 0 (0) | |
| Exanthema | 4 (11.4) | |
| Conjunctivitis | 1 (2.9) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monzani, A.; Borgogna, C.; Ferrante, D.; Ciacchini, B.; Felici, E.; Gariglio, M.; Rabbone, I. #Stayathome If You Have a Cold: High SARS-CoV-2 Salivary Viral Loads in Pediatric Patients with Nasopharyngeal Symptoms. Viruses 2023, 15, 81. https://doi.org/10.3390/v15010081
Monzani A, Borgogna C, Ferrante D, Ciacchini B, Felici E, Gariglio M, Rabbone I. #Stayathome If You Have a Cold: High SARS-CoV-2 Salivary Viral Loads in Pediatric Patients with Nasopharyngeal Symptoms. Viruses. 2023; 15(1):81. https://doi.org/10.3390/v15010081
Chicago/Turabian StyleMonzani, Alice, Cinzia Borgogna, Daniela Ferrante, Benedetta Ciacchini, Enrico Felici, Marisa Gariglio, and Ivana Rabbone. 2023. "#Stayathome If You Have a Cold: High SARS-CoV-2 Salivary Viral Loads in Pediatric Patients with Nasopharyngeal Symptoms" Viruses 15, no. 1: 81. https://doi.org/10.3390/v15010081
APA StyleMonzani, A., Borgogna, C., Ferrante, D., Ciacchini, B., Felici, E., Gariglio, M., & Rabbone, I. (2023). #Stayathome If You Have a Cold: High SARS-CoV-2 Salivary Viral Loads in Pediatric Patients with Nasopharyngeal Symptoms. Viruses, 15(1), 81. https://doi.org/10.3390/v15010081









