New RT-PCR Assay for the Detection of Current and Future SARS-CoV-2 Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Lineage Consensus Sequences
2.2. Identification of SARS-CoV-2 Ultra-Conserved Elements and Design of RT-PCR Assays
2.3. In Silico PCR Simulations
2.4. Samples, Sample Processing, and RNA Extraction Procedures
2.5. RT-PCR Testing
2.6. Annealing Temperature Optimization
2.7. Quantification of Viral Load by Droplet Digital RT-PCR (ddRT-PCR)
2.8. Limit of Detection Calculation
3. Results
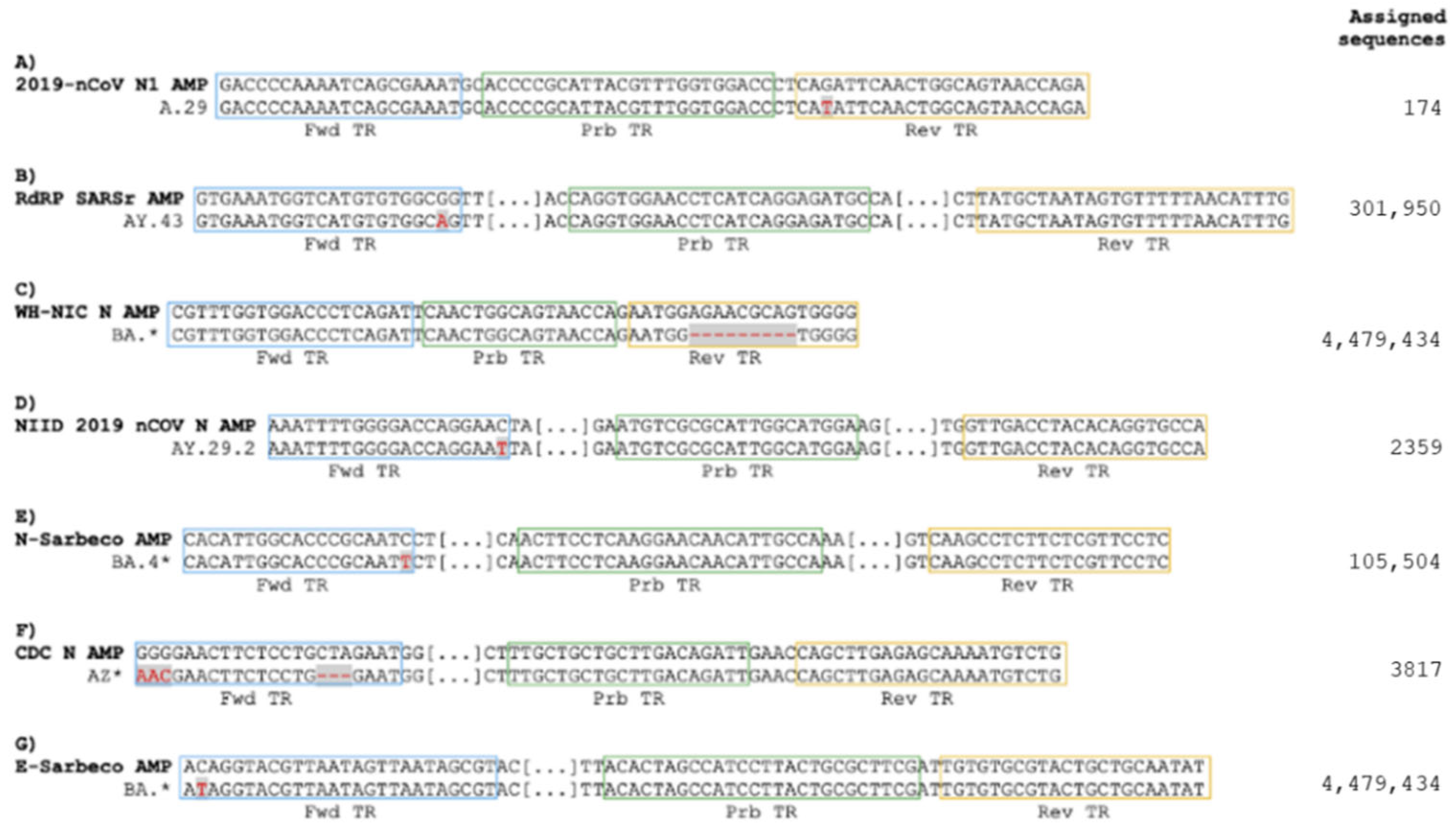
3.1. Omicron Mutations Affect Only One of the Real-Time RT-PCR Assays Recommended by WHO for the Detection of SARS-CoV-2
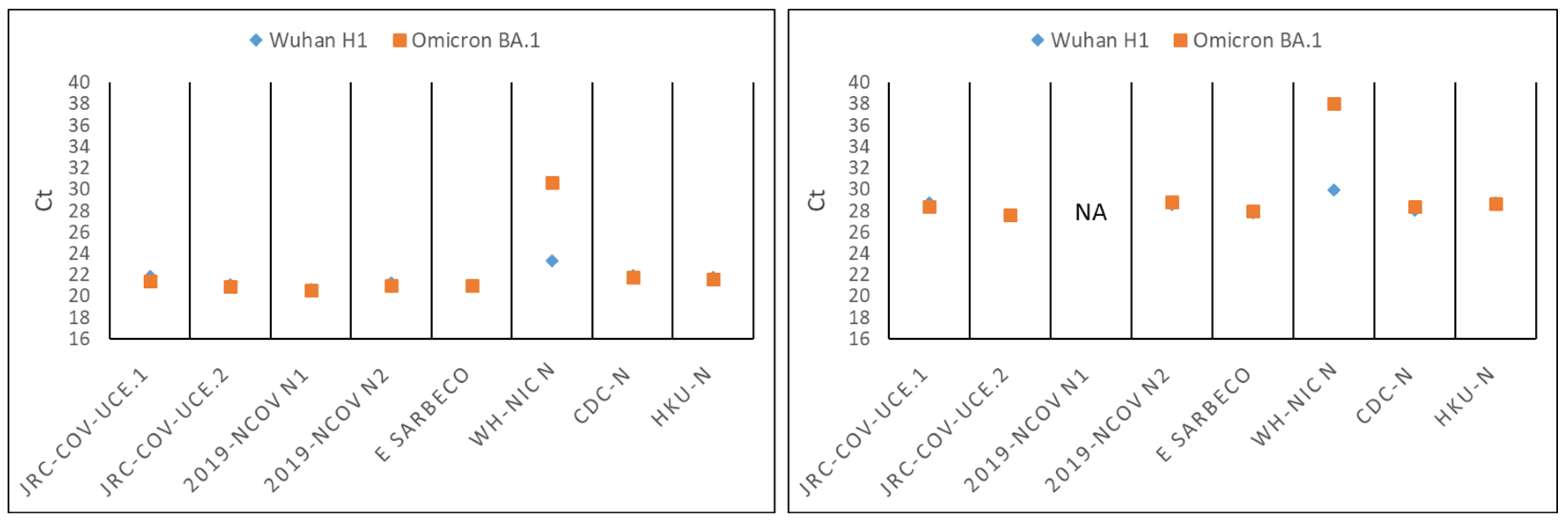
3.2. Development of a Novel Duplex Real-Time (dd)RT-PCR Assay Targeting the Ultra-Conserved Elements of the SARS-CoV-2 Genome
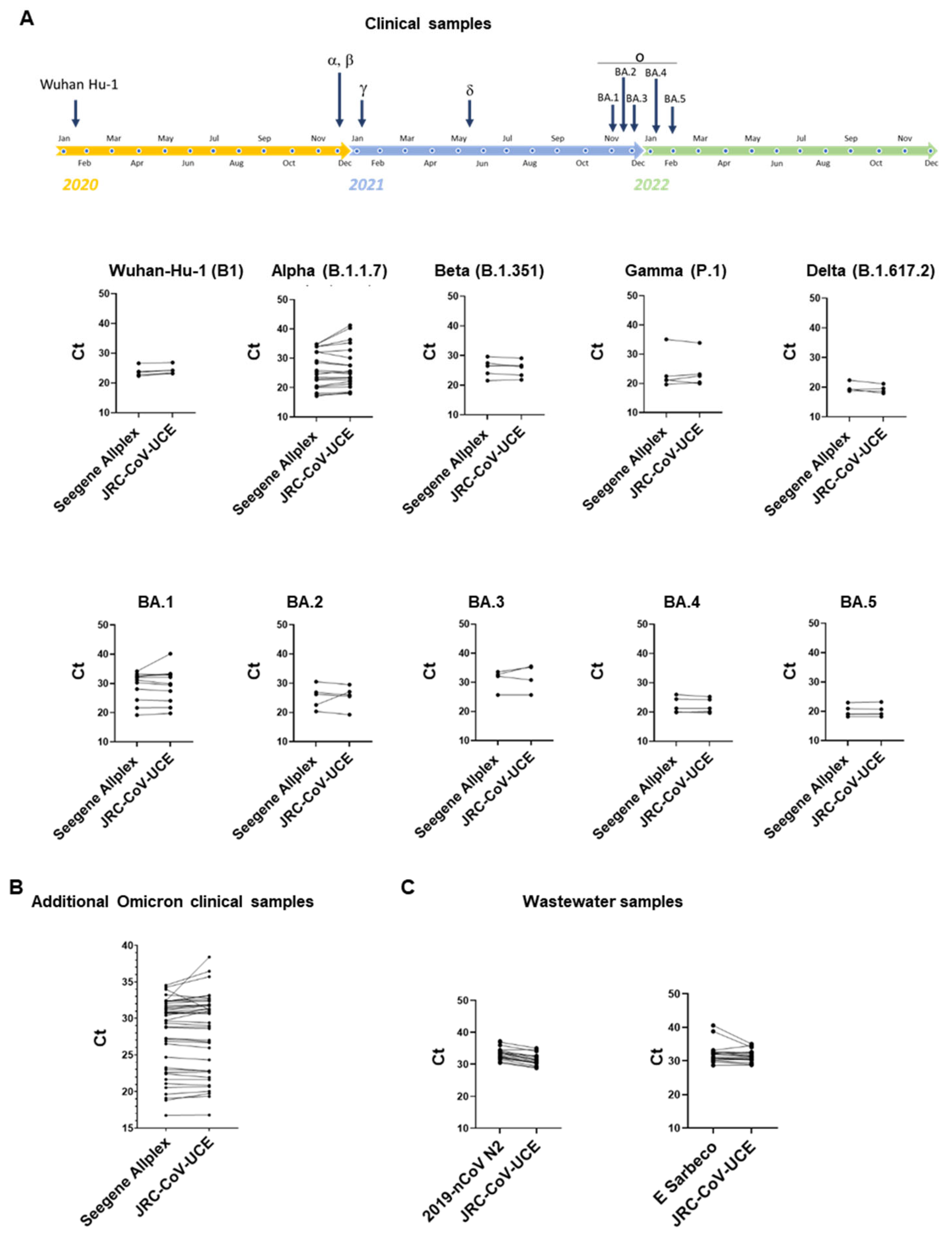
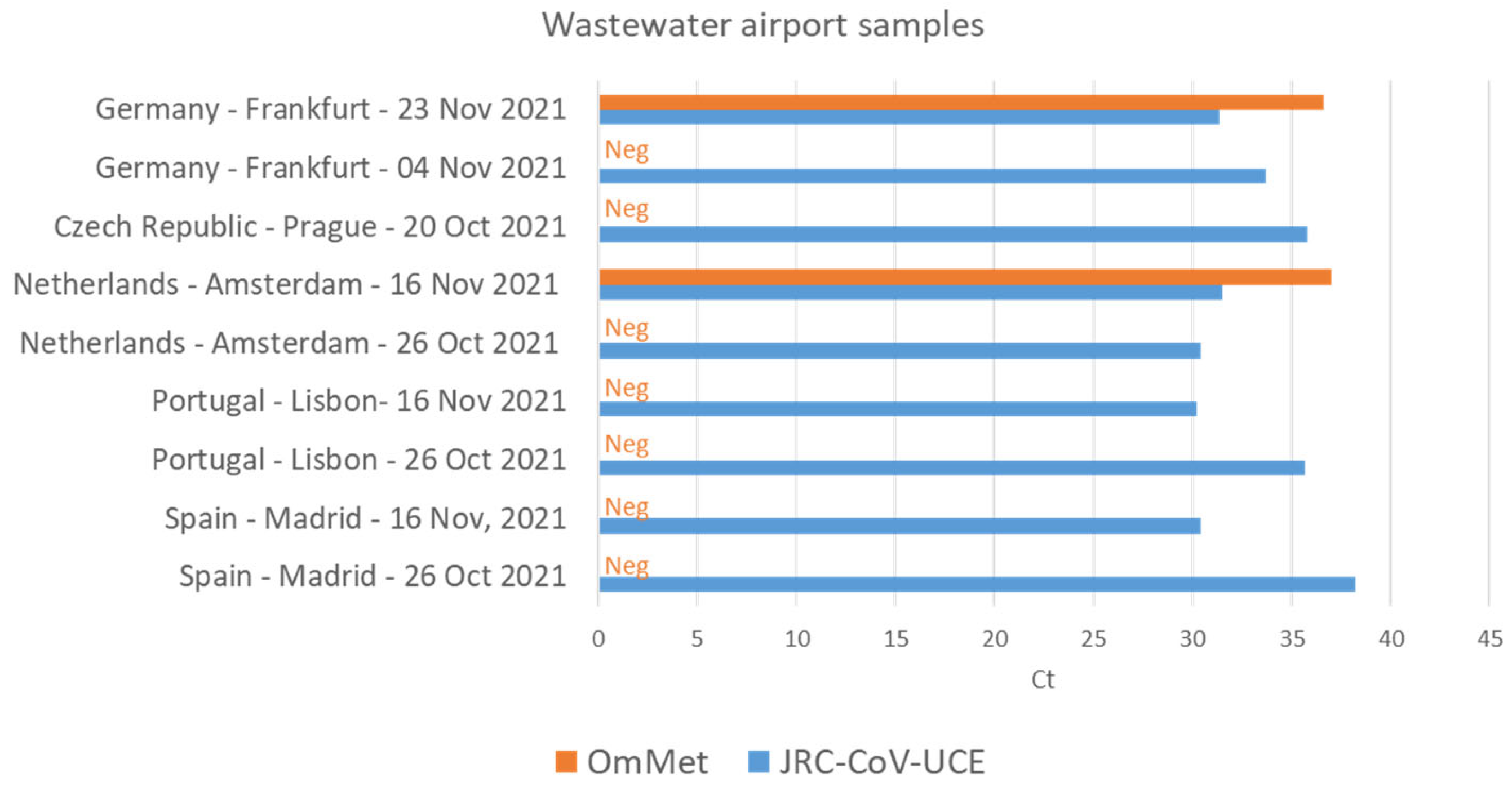
3.3. JRC-CoV-UCE Assay Performance in Clinical and Wastewater Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rambaut, A.; Holmes, E.C.; O’Toole, A.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Emma, B. Hodcroft. CoVariants: SARS-CoV-2 Mutations and Variants of Interest. 2021. Available online: https://covariants.org/ (accessed on 5 October 2022).
- World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. 2021. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 5 October 2022).
- Callaway, E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021, 600, 21. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Lan, W.; Wu, X.; Zhao, T.; Duan, B.; Yang, P.; Ren, Y.; Quan, L.; Zhao, W.; Seto, D.; et al. Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Signal Transduct. Target Ther. 2022, 7, 138. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. [Google Scholar] [CrossRef] [PubMed]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.C.; Ng, K.C.; Ching, R.H.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef]
- Rossler, A.; Riepler, L.; Bante, D.; von Laer, D.; Kimpel, J. SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. N. Engl. J. Med. 2022, 386, 698–700. [Google Scholar] [CrossRef]
- Pulliam, J.R.C.; van Schalkwyk, C.; Govender, N.; von Gottberg, A.; Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science 2022, 376, eabn4947. [Google Scholar] [CrossRef]
- Kuhlmann, C.; Mayer, C.K.; Claassen, M.; Maponga, T.; Burgers, W.A.; Keeton, R.; Riou, C.; Sutherland, A.D.; Suliman, T.; Shaw, M.L.; et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 2022, 399, 625–626. [Google Scholar] [CrossRef]
- Syed, A.M.; Ciling, A.; Taha, T.Y.; Chen, I.P.; Khalid, M.M.; Sreekumar, B.; Chen, P.Y.; Kumar, G.R.; Suryawanshi, R.; Silva, I.; et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. Proc. Natl. Acad. Sci. USA 2022, 119, e2200592119. [Google Scholar] [CrossRef]
- Singh, J.; Pandit, P.; McArthur, A.G.; Banerjee, A.; Mossman, K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 2021, 18, 166. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, N.; Gupta, Y.; Goyal, M.; Khaiboullina, S.F.; Baranwal, M.; Verma, S.C. Mutational Frequencies of SARS-CoV-2 Genome during the Beginning Months of the Outbreak in USA. Pathogens 2020, 9, 565. [Google Scholar] [CrossRef] [PubMed]
- Amoutzias, G.D.; Nikolaidis, M.; Tryfonopoulou, E.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G. The Remarkable Evolutionary Plasticity of Coronaviruses by Mutation and Recombination: Insights for the COVID-19 Pandemic and the Future Evolutionary Paths of SARS-CoV-2. Viruses 2022, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Arena, F.; Pollini, S.; Rossolini, G.M.; Margaglione, M. Summary of the Available Molecular Methods for Detection of SARS-CoV-2 during the Ongoing Pandemic. Int. J. Mol. Sci. 2021, 22, 1298. [Google Scholar] [CrossRef]
- Tahan, S.; Parikh, B.A.; Droit, L.; Wallace, M.A.; Burnham, C.D.; Wang, D. SARS-CoV-2 E Gene Variant Alters Analytical Sensitivity Characteristics of Viral Detection Using a Commercial Reverse Transcription-PCR Assay. J. Clin. Microbiol. 2021, 59, e0007521. [Google Scholar] [CrossRef]
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: A systematic review and meta-analysis. Lancet Infect. Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Mautner, L.; Baillie, C.K.; Herold, H.M.; Volkwein, W.; Guertler, P.; Eberle, U.; Ackermann, N.; Sing, A.; Pavlovic, M.; Goerlich, O.; et al. Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol. J. 2020, 17, 160. [Google Scholar] [CrossRef]
- Wang, X.; Huang, Z.; Song, J.; Xiao, Y.; Wang, H. Analytical sensitivity comparison of 14 conventional and three rapid RT-PCR assays for SARS-CoV-2 detection. J. Virol. Methods 2021, 293, 114144. [Google Scholar] [CrossRef]
- Gdoura, M.; Abouda, I.; Mrad, M.; Ben Dhifallah, I.; Belaiba, Z.; Fares, W.; Chouikha, A.; Khedhiri, M.; Layouni, K.; Touzi, H.; et al. SARS-CoV2 RT-PCR assays: In vitro comparison of 4 WHO approved protocols on clinical specimens and its implications for real laboratory practice through variant emergence. Virol. J. 2022, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Catherine Muenker, M.; Moore, A.J.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Corbisier, P.; Petrillo, M.; Marchini, A.; Querci, M.; Buttinger, G.; Bekliz, M.; Spiess, K.; Polacek, C.; Fomsgaard, A.; Van den Eede, G. A qualitative RT-PCR assay for the specific identification of the SARS-CoV-2 B.1.1.529 (Omicron) Variant of Concern. J. Clin. Virol. 2022, 152, 105191. [Google Scholar] [CrossRef] [PubMed]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef]
- Chen, A.T.; Altschuler, K.; Zhan, S.H.; Chan, Y.A.; Deverman, B.E. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife 2021, 10, e63409. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Gans, J.D.; Wolinsky, M. Improved assay-dependent searching of nucleic acid sequence databases. Nucleic Acids Res. 2008, 36, e74. [Google Scholar] [CrossRef]
- O’Toole, A.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Heijnen, L.; Medema, G. Surveillance of influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in the Netherlands. J. Water Health 2011, 9, 434–442. [Google Scholar] [CrossRef]
- Stinear, T.; Matusan, A.; Hines, K.; Sandery, M. Detection of a single viable Cryptosporidium parvum oocyst in environmental water concentrates by reverse transcription-PCR. Appl. Environ. Microbiol. 1996, 62, 3385–3390. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Coronavirus Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Primers and Probes. 2020. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed on 5 October 2022).
- Centers for Disease Control and Prevention. Cdc 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel: Instructions for Use. 2020. Available online: https://www.fda.gov/media/134922/download (accessed on 5 October 2022).
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Chinese Center for Disease Control and Prevention. Technical Guidelines for COVID-19 Laboratory Testing. China CDC Wkly. 2020, 2, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef]
- Shirato, K.; Nao, N.; Katano, H.; Takayama, I.; Saito, S.; Kato, F.; Katoh, H.; Sakata, M.; Nakatsu, Y.; Mori, Y.; et al. Development of Genetic Diagnostic Methods for Detection for Novel Coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020, 73, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Institut Pasteur, Paris. Protocol: Real-Time RT-PCR Assays for the Detection of SARS-CoV-2. Available online: https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2 (accessed on 5 October 2022).
- Department of Medical Sciences, Ministry of Public Health, Diagnostic Detection of Novel Coronavirus 2019 by Real Time RT-PCR. Available online: https://www.who.int/docs/default-source/coronaviruse/conventional-rt-pcr-followed-by-sequencing-for-detection-of-ncov-rirl-nat-inst-health-t.pdf?sfvrsn=42271c6d_4 (accessed on 5 October 2022).
- Van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef]
- De Smet, D.; Vanhee, M.; Maes, B.; Swaerts, K.; De Jaeger, P.; Maelegheer, K.; Van Hoecke, F.; Martens, G.A. Cycle Threshold Probability Score for Immediate and Sensitive Detection of B.1.351 SARS-CoV-2 Lineage. Am. J. Clin. Pathol. 2022, 157, 731–741. [Google Scholar] [CrossRef]
- Agrawal, S.; Orschler, L.; Tavazzi, S.; Greither, R.; Gawlik, B.M.; Lackner, S. Genome Sequencing of Wastewater Confirms the Arrival of the SARS-CoV-2 Omicron Variant at Frankfurt Airport but Limited Spread in the City of Frankfurt, Germany, in November 2021. Microbiol. Resour. Announc. 2022, 11, e0122921. [Google Scholar] [CrossRef]
- Waterfall, C.M.; Eisenthal, R.; Cobb, B.D. Kinetic characterisation of primer mismatches in allele-specific PCR: A quantitative assessment. Biochem. Biophys. Res. Commun. 2002, 299, 715–722. [Google Scholar] [CrossRef]
- Christopherson, C.; Sninsky, J.; Kwok, S. The effects of internal primer-template mismatches on RT-PCR: HIV-1 model studies. Nucleic Acids Res. 1997, 25, 654–658. [Google Scholar] [CrossRef]
- Kwok, S.; Kellogg, D.E.; McKinney, N.; Spasic, D.; Goda, L.; Levenson, C.; Sninsky, J.J. Effects of primer-template mismatches on the polymerase chain reaction: Human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990, 18, 999–1005. [Google Scholar] [CrossRef]
- Alkhatib, M.; Carioti, L.; D’Anna, S.; Ceccherini-Silberstein, F.; Svicher, V.; Salpini, R. SARS-CoV-2 Mutations and Variants May Muddle the Sensitivity of COVID-19 Diagnostic Assays. Microorganisms 2022, 10, 1559. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control/World Health Organization Regional Office for Europe. Methods for the Detection and Identification of SARS-CoV-2 Variants: Second Update, August 2022; ECDC: Stockholm, Sweden; WHO European Region: Copenhagen, Denmark, 2022. [Google Scholar]
- Amicone, M.; Borges, V.; Alves, M.J.; Isidro, J.; Ze-Ze, L.; Duarte, S.; Vieira, L.; Guiomar, R.; Gomes, J.P.; Gordo, I. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol. Med. Public Health 2022, 10, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C. Watcher in the wastewater. Nat. Biotechnol. 2020, 38, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, A.; Monis, P.; Deere, D.; Ryan, U. Wastewater-based epidemiology-surveillance and early detection of waterborne pathogens with a focus on SARS-CoV-2, Cryptosporidium and Giardia. Parasitol. Res. 2021, 120, 4167–4188. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Orschler, L.; Schubert, S.; Zachmann, K.; Heijnen, L.; Tavazzi, S.; Gawlik, B.M.; de Graaf, M.; Medema, G.; Lackner, S. Prevalence and circulation patterns of SARS-CoV-2 variants in European sewage mirror clinical data of 54 European cities. Water Res. 2022, 214, 118162. [Google Scholar] [CrossRef]
- Whale, A.S.; von der Heide, E.K.; Kohlenberg, M.; Brinckmann, A.; Baedker, S.; Karalay, O.; Fernandez-Gonzalez, A.; Busby, E.J.; Bustin, S.A.; Hauser, H.; et al. Digital PCR can augment the interpretation of RT-qPCR Cq values for SARS-CoV-2 diagnostics. Methods 2022, 201, 5–14. [Google Scholar] [CrossRef]
- Kuypers, J.; Jerome, K.R. Applications of Digital PCR for Clinical Microbiology. J. Clin. Microbiol. 2017, 55, 1621–1628. [Google Scholar] [CrossRef]
- Sun, Y.; Ding, C.; Chen, Q.; Xie, J.; Yu, J.; Shi, Y.; Jiang, C.; Zhang, Z.; He, H.; Ge, Y.; et al. Digital PCR assay for the effective detection of COVID-19 patients with SARS-CoV-2 low viral load. J. Virol. Methods 2021, 295, 114185. [Google Scholar] [CrossRef]
- Vasudevan, H.N.; Xu, P.; Servellita, V.; Miller, S.; Liu, L.; Gopez, A.; Chiu, C.Y.; Abate, A.R. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci. Rep. 2021, 11, 780. [Google Scholar] [CrossRef]
- Heijnen, L.; Elsinga, G.; de Graaf, M.; Molenkamp, R.; Koopmans, M.P.G.; Medema, G. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci. Total Environ. 2021, 799, 149456. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Stange, C.; Suhrborg, R.; Wurzbacher, C.; Drewes, J.E.; Tiehm, A. SARS-CoV-2 wastewater surveillance in Germany: Long-term RT-digital droplet PCR monitoring, suitability of primer/probe combinations and biomarker stability. Water Res. 2022, 210, 117977. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Smith, W.J.M.; Metcalfe, S.; Jackson, G.; Choi, P.M.; Morrison, M.; Field, D.; Gyawali, P.; Bivins, A.; Bibby, K.; et al. Comparison of RT-qPCR and RT-dPCR Platforms for the Trace Detection of SARS-CoV-2 RNA in Wastewater. ACS ES T Water 2022, 2, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- G7 Health Ministers’ Communiqué, 20 May 2022, Berlin. Available online: https://www.bundesregierung.de/blueprint/servlet/resource/blob/974430/2042058/5651daa321517b089cdccfaffd1e37a1/2022-05-20-g7-health-ministers-communique-data.pdf (accessed on 5 October 2022).
- Commission Recommendation (EU) 2021/472 of 17 March 2021 on a Common Approach to Establish a Systematic Surveillance of SARS-CoV-2 and Its Variants in Wastewaters in the EU. C/2021/1925. Available online: http://data.europa.eu/eli/reco/2021/472/oj (accessed on 5 October 2022).
- European Commission, Joint Research Centre; Gawlik, B.; Tavazzi, S.; Mariani, G.; Skejo, H.; Sponar, M.; Higgins, T.; Medema, G.; Wintgens, T. SARS-CoV-2 Surveillance Employing Sewage: Towards a Sentinel System, Publications Office, 2021. Available online: https://data.europa.eu/doi/10.2760/300580 (accessed on 5 October 2022).


| Assay | Reference Primers and Probes | Reference Amplicon (Size, bp) | SARS-CoV-2 Target [Genomic Location] 1 |
|---|---|---|---|
| JRC-CoV-UCE.1 | For: 5′-ACTCTTGTTACAATGCCACTTGG-3′ (900) Rev: 5′-GCTGGCACTTTGAGAGATCTC-3′ (900) P: 5′-VIC-GGCTTAAATTTGGAAGAAGCTGCTCGG-QSY-3′ | ACTCTTGTTACAATGCCACTTGGCTATGTAACACATGGCTTAAATTTGGAAGAGCTGCTCGGTATATGAGATCTCTCAAAGTGCCAGC (89) | Orf1a(NSP3) [4595–4683] |
| JRC-CoV-UCE.2 | For: 5′-AAGGGCCAATTCTGCTGTC-3′ (900) Rev: 5′-TGTAGTACCGGCAGCACAAG-3′ (900) P: 5′-ABY-AGCTTAGTCCTGTTGCACTACGACA-QSY-3′ | AAGGGCCAATTCTGCTGTCAAATTACAGAATAATGAGCTTAGTCCTGTTGCACTACGACAGATGTCTTGTGCTGCCGGTACTACA (85) | Orf1a(NSP9) [12658–12742] |
| Developer | NAAT Name | Reference Primers and Probes (Concentration, nM) | Annealing Temperature (°C) |
|---|---|---|---|
| US CDC, USA | 2019-nCoV N1 | For: 5′-GACCCCAAAATCAGCGAAAT-3′ (500) Rev: 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ (500) P: 5′-FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1-3′ (125) | 55 |
| US CDC, USA | 2019-nCoV-N2 | For: 5′-TTACAAACATTGGCCGCAAA-3′ (500) Rev: 5′-GCGCGACATTCCGAAGAA-3′ (500) P: 5′-FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1-3′ (125) | 55 |
| Charité, DE | E Sarbeco | For: 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′ (400) Rev: 5′-ATATTGCAGCAGTACGCACACA-3′ (400) P: 5′-FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ650-3′(200) | 58 |
| NIH, TH | WH-NIC N | For: 5′-CGTTTGGTGGACCCTCAGAT-3′ (800) Rev: 5′-CCCCACTGCGTTCTCCATT-3′ (800) P: 5′-FAM-CAACTGGCAGTAACCA- BHQ1-3′ (200) | 55 |
| CDC, CN | CDC-N | For: 5′-GGGGAACTTCTCCTGCTAGAAT-3′ (400) Rev: 5′-CAGACATTTTGCTCTCAAGCTG-3′ (400) P: 5′-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3′ (200) | 60 |
| HKU, HK | HKU-N | For: 5′-TAATCAGACAAGGAACTGATTA-3′ (500) Rev: 5′-CGAAGGTGTGACTTCCATG-3′ (500) P: 5′-FAM-GCAAATTGTGCAATTTGCGG-BBQ1-3′ (250) | 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchini, A.; Petrillo, M.; Parrish, A.; Buttinger, G.; Tavazzi, S.; Querci, M.; Betsou, F.; Elsinga, G.; Medema, G.; Abdelrahman, T.; et al. New RT-PCR Assay for the Detection of Current and Future SARS-CoV-2 Variants. Viruses 2023, 15, 206. https://doi.org/10.3390/v15010206
Marchini A, Petrillo M, Parrish A, Buttinger G, Tavazzi S, Querci M, Betsou F, Elsinga G, Medema G, Abdelrahman T, et al. New RT-PCR Assay for the Detection of Current and Future SARS-CoV-2 Variants. Viruses. 2023; 15(1):206. https://doi.org/10.3390/v15010206
Chicago/Turabian StyleMarchini, Antonio, Mauro Petrillo, Amy Parrish, Gerhard Buttinger, Simona Tavazzi, Maddalena Querci, Fay Betsou, Goffe Elsinga, Gertjan Medema, Tamir Abdelrahman, and et al. 2023. "New RT-PCR Assay for the Detection of Current and Future SARS-CoV-2 Variants" Viruses 15, no. 1: 206. https://doi.org/10.3390/v15010206
APA StyleMarchini, A., Petrillo, M., Parrish, A., Buttinger, G., Tavazzi, S., Querci, M., Betsou, F., Elsinga, G., Medema, G., Abdelrahman, T., Gawlik, B., & Corbisier, P. (2023). New RT-PCR Assay for the Detection of Current and Future SARS-CoV-2 Variants. Viruses, 15(1), 206. https://doi.org/10.3390/v15010206








