Complete Genome and Molecular Characterization of a New Cyprinid Herpesvirus 2 (CyHV-2) SH-01 Strain Isolated from Cultured Crucian Carp
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Virus and DNA Extraction
2.2. DNA Sequencing, Genome Assembly, and Annotation
2.3. Analysis of the Genome Structure and Molecular Characterization of CyHV-2 SH-01
2.4. Comparison of Genomic Structure and Evolutionary Relationships among SH-01 and the Other Seven Strains
3. Results
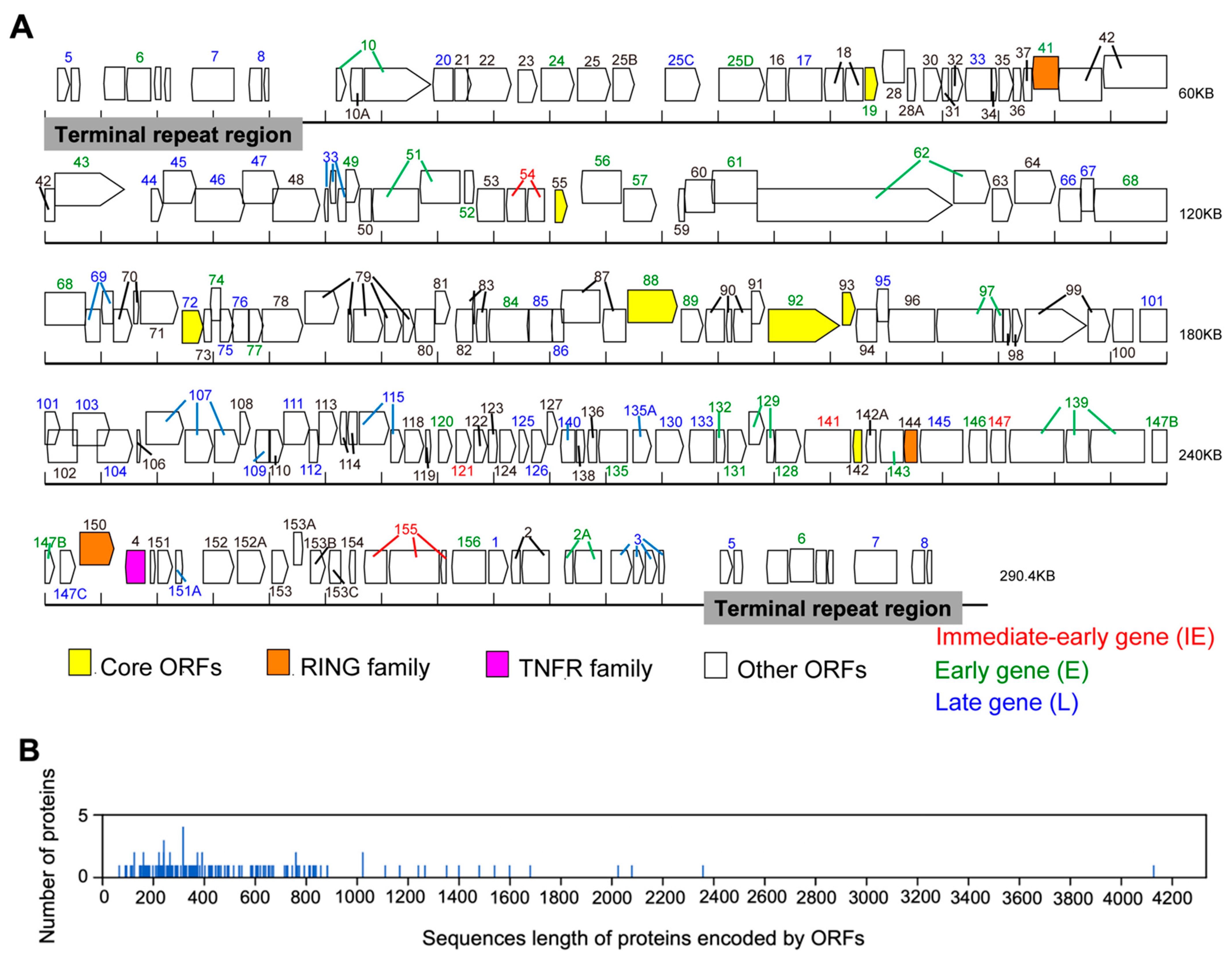
3.1. Genome Structure and Composition
3.2. Chronological Characteristics of Gene Expression
3.3. Features of the Predicted Functional Protein Encoded by SH-01
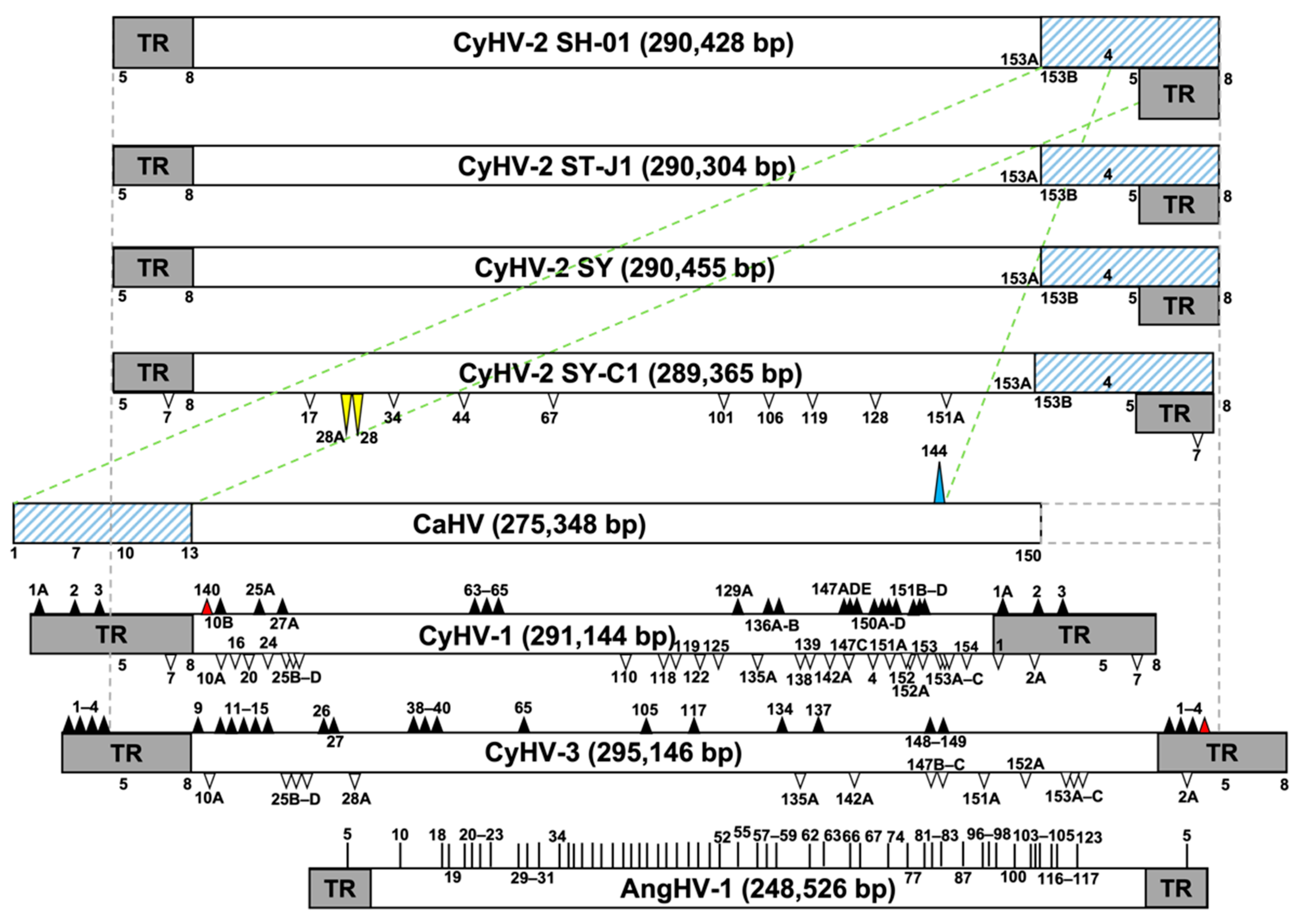
3.4. Comparison of Genome Structure and ORFs Arrangement
3.5. Genomic Evolutionary Relationships among SH-01 and the Other Seven Strains
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jung, S.J.; Miyazaki, T. Herpesviral haematopoietic necrosis of goldfish, Carassius auratus (L.). J. Fish Dis. 1995, 18, 211–220. [Google Scholar] [CrossRef]
- Stephens, F.J.; Raidal, S.R.; Jones, B. Haematopoietic necrosis in a goldfish (Carassius auratus) associated with an agent morphologically similar to herpesvirus. Aust. Vet. J. 2004, 82, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Groff, J.M.; LaPatra, S.E.; Munn, R.J.; Zinkl, J.G. A viral epizootic in cultured populations of juvenile goldfish due to a putative herpesvirus etiology. J. Vet. Diagn. Investig. 1998, 10, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.E.; Khoo, L.; LaPatra, S.E.; Bonar, C.; Key, D.W.; Garner, M.; Lee, M.V.; Hanson, L. Goldfish hematopoietic necrosis herpesvirus (cyprinid herpesvirus 2) in the USA: Molecular confirmation of isolates from diseased fish. J. Aquat. Anim. Health 2006, 18, 11–18. [Google Scholar] [CrossRef]
- Jeffery, K.R.; Bateman, K.; Bayley, A.; Feist, S.W.; Hulland, J.; Longshaw, C.; Stone, D.; Woolford, G.; Way, K. Isolation of a cyprinid herpesvirus 2 from goldfish, Carassius auratus (L.), in the UK. J. Fish Dis. 2007, 30, 649–656. [Google Scholar] [CrossRef]
- Doszpoly, A.; Maria, B.A.; György, C.; Ádám, D.; Mária, L.; Balázs, H. Introduction of the family Alloherpesviridae: The first molecular detection of herpesviruses of cyprinid fish in Hungary. Magy. Allatorv. Lapja 2011, 133, 174–181. [Google Scholar]
- Daněk, T.; Kalous, L.; Vesel, T.; Krásová, E.; Reschová, S.; Rylková, K.; Kulich, P.; Petrtýl, M.; Pokorová, D.; Knytl, M. Massive mortality of Prussian carp Carassius gibelio in the upper Elbe basin associated with herpesviral hematopoietic necrosis (CyHV-2). Dis. Aquat. Org. 2012, 102, 87–95. [Google Scholar] [CrossRef]
- Wang, L.; He, J.; Liang, L.; Zheng, X.; Jia, P.; Shi, X.; Lan, W.; Xie, J.; Liu, H.; Xu, P. Mass mortality caused by Cyprinid Herpesvirus 2 (CyHV-2) in Prussian carp (Carassius gibelio) in China. Bull. Eur. Assoc. Fish Pathol. 2012, 32, 164–173. [Google Scholar]
- Fichi, G.; Cardeti, G.; Cocumelli, C.; Vendramin, N.; Toffan, A.; Eleni, C.; Siemoni, N.; Fischetti, R.; Susini, F. Detection of Cyprinid herpesvirus 2 in association with an Aeromonas sobria infection of Carassius carassius (L.), in Italy. J. Fish Dis. 2013, 36, 823–830. [Google Scholar] [CrossRef]
- Boitard, P.-M.; Baud, M.; Labrut, S.; de Boisséson, C.; Jamin, M.; Bigarré, L. First detection of Cyprinid Herpesvirus 2 (CyHV-2) in goldfish (Carassius auratus) in France. J. Fish Dis. 2016, 39, 673–680. [Google Scholar] [CrossRef]
- Haenen, O.L.M.; Way, K.; Gorgoglione, B.; Ito, T.; Paley, R.; Bigarré, L.; Waltzek, T. Novel viral infections threatening Cyprinid fish. Bull. Eur. Assoc. Fish Pathol. 2016, 36, 11–23. [Google Scholar]
- Sahoo, P.K.; Swaminathan, T.R.; Abraham, T.J.; Kumar, R.; Pattanayak, S.; Mohapatra, A.; Rath, S.S.; Patra, A.; Adikesavalu, H.; Sood, N.; et al. Detection of goldfish haematopoietic necrosis herpes virus (Cyprinid herpesvirus-2) with multi-drug resistant Aeromonas hydrophila infection in goldfish: First evidence of any viral disease outbreak in ornamental freshwater aquaculture farms in India. Acta Trop. 2016, 161, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, S.; Bergmann, S.M.; Keeling, C.; Lany, C.; Schütze, H.; Schmidt-Posthaus, H. Herpesviral Hematopoietic Necrosis in Goldfish in Switzerland: Early Lesions in Clinically Normal Goldfish (Carassius auratus). Vet. Pathol. 2016, 53, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Adamek, M.; Hellmann, J.; Jung-Schroers, V.; Teitge, F.; Steinhagen, D. CyHV-2 transmission in traded goldfish stocks in Germany—A case study. J. Fish Dis. 2018, 41, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Kalayci, G.; Ozkan, B.; Pekmez, K.; Kaplan, M.; Mefut, A.; Anil Cagirgan, A. First detection of Cyprinid herpesvirus-2 (CyHV-2) followed by screening and monitoring studies in Goldfish (Carassius Auratus) in Turkey. Bull. Eur. Assoc. Fish Pathol. 2018, 38, 94–103. [Google Scholar]
- Panicz, R.; Sadowski, J.; Eljasik, P. Detection of Cyprinid herpesvirus 2 (CyHV-2) in symptomatic ornamental types of goldfish (Carassius auratus) and asymptomatic common carp (Cyprinus carpio) reared in warm-water cage culture. Aquaculture 2019, 504, 131–138. [Google Scholar] [CrossRef]
- Adler, B.; Sattler, C.; Adler, H. Herpesviruses and Their Host Cells: A Successful Liaison. Trends Microbiol. 2017, 25, 229–241. [Google Scholar] [CrossRef]
- Collins-McMillen, D.; Buehler, J.; Peppenelli, M.; Goodrum, F. Molecular Determinants and the Regulation of Human Cytomegalovirus Latency and Reactivation. Viruses 2018, 10, 444. [Google Scholar] [CrossRef]
- Eide, K.E.; Miller-Morgan, T.; Heidel, J.R.; Kent, M.L.; Bildfell, R.J.; Lapatra, S.; Watson, G.; Jin, L. Investigation of koi herpesvirus latency in koi. J. Virol. 2011, 85, 4954–4962. [Google Scholar] [CrossRef]
- Wei, C.; Iida, H.; Chuah, Q.; Tanaka, M.; Kato, G.; Sano, M. Persistence of cyprinid herpesvirus 2 in asymptomatic goldfish Carassius auratus (L.) that survived an experimental infection. J. Fish Dis. 2019, 42, 913–921. [Google Scholar] [CrossRef]
- Wei, C.; Kakazu, T.; Chuah, Q.Y.; Tanaka, M.; Kato, G.; Sano, M. Reactivation of cyprinid herpesvirus 2 (CyHV-2) in asymptomatic surviving goldfish Carassius auratus (L.) under immunosuppression. Fish Shellfish Immunol. 2020, 103, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Waltzek, T.B.; Kelley, G.O.; Stone, D.M.; Way, K.; Hanson, L.; Fukuda, H.; Hirono, I.; Aoki, T.; Davison, A.J.; Hedrick, R.P. Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae. J. Gen. Virol. 2005, 86, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Waltzek, T.B.; Kelley, G.O.; Alfaro, M.E.; Kurobe, T.; Davison, A.J.; Hedrick, R.P. Phylogenetic relationships in the family Alloherpesviridae. Dis. Aquat. Organ. 2009, 84, 179–194. [Google Scholar] [CrossRef] [PubMed]
- van Beurden, S.J.; Bossers, A.; Voorbergen-Laarman, M.H.A.; Haenen, O.L.M.; Peters, S.; Abma-Henkens, M.H.C.; Peeters, B.P.H.; Rottier, P.J.M.; Engelsma, M.Y. Complete genome sequence and taxonomic position of anguillid herpesvirus 1. J. Gen. Virol. 2010, 91, 880–887. [Google Scholar] [CrossRef]
- Doszpoly, A.; Kovács, E.R.; Bovo, G.; LaPatra, S.E.; Harrach, B.; Benko, M. Molecular confirmation of a new herpesvirus from catfish (Ameiurus melas) by testing the performance of a novel PCR method, designed to target the DNA polymerase gene of alloherpesviruses. Arch. Virol. 2008, 153, 2123–2127. [Google Scholar] [CrossRef]
- Zeng, X.-T.; Chen, Z.-Y.; Deng, Y.-S.; Gui, J.-F.; Zhang, Q.-Y. Complete genome sequence and architecture of crucian carp Carassius auratus herpesvirus (CaHV). Arch. Virol. 2016, 161, 3577–3581. [Google Scholar] [CrossRef]
- Wen, J.; Xu, Y.; Su, M.; Lu, L.; Wang, H. Susceptibility of Goldfish to Cyprinid Herpesvirus 2 (CyHV-2) SH01 Isolated from Cultured Crucian Carp. Viruses 2021, 13, 1761. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar]
- Davison, A.J.; Kurobe, T.; Gatherer, D.; Cunningham, C.; Korf, I.; Fukuda, H.; Hedrick, R.P.; Waltzek, T.B. Comparative genomics of carp herpesviruses. J. Virol. 2013, 87, 2908–2922. [Google Scholar] [CrossRef]
- Yuan, Y. Identification and characterization of herpesviral immediate-early genes. Methods Mol. Biol. 2005, 292, 231–244. [Google Scholar]
- Gruffat, H.; Marchione, R.; Manet, E. Herpesvirus late gene expression: A viral-specific pre-initiation complex is key. Front. Microbiol. 2016, 7, 869. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Lu, L.; Wang, B.; Yu, J.; Wang, H. Identification of the Immediate-Early Genes of Cyprinid Herpesvirus 2. Viruses 2020, 12, 994. [Google Scholar] [CrossRef]
- Naom, I.S.; Morton, S.J.; Leach, D.R.F.; Lloyd, R.G. Molecular organization of sbcC, a gene that affects genetic recombination and the viability of DNA palindromes in Escherichia coli K-12. Nucleic Acids Res. 1989, 17, 8033–8045. [Google Scholar] [CrossRef]
- Zhang, J.; Ge, W.; Zhan, P.; De Clercq, E.; Liu, X. Retroviral restriction factors TRIM5α: Therapeutic strategy to inhibit HIV-1 replication. Curr. Med. Chem. 2011, 18, 2649–2654. [Google Scholar] [CrossRef]
- Maris, C.; Dominguez, C.; Allain, F.H.-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Bailey-Elkin, B.A.; van Kasteren, P.B.; Snijder, E.J.; Kikkert, M.; Mark, B.L. Viral OTU deubiquitinases: A structural and functional comparison. PLoS Pathog. 2014, 10, e1003894. [Google Scholar] [CrossRef]
- Rajcani, J.; Szenthe, K.; Banati, F.; Szathmary, S. Survey of Epstein Barr virus (EBV) immunogenic proteins and their epitopes: Implications for vaccine preparation. Recent Pat. Anti-Infect. Drug Discov. 2014, 9, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Quinn, L.L.; Williams, L.R.; White, C.; Forrest, C.; Zuo, J.; Rowe, M. The Missing Link in Epstein-Barr Virus Immune Evasion: The BDLF3 Gene Induces Ubiquitination and Downregulation of Major Histocompatibility Complex Class I (MHC-I) and MHC-II. J. Virol. 2015, 90, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Bates, P.; Rivera-Gonzalez, R.; Gu, B.; DeLuca, N.A. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 1993, 67, 4676–4687. [Google Scholar] [CrossRef]
- Li, L.; Luo, Y.; Gao, Z.; Huang, J.; Zheng, X.; Nie, H.; Zhang, J.; Lin, L.; Yuan, J. Molecular characterisation and prevalence of a new genotype of Cyprinid herpesvirus 2 in mainland China. Can. J. Microbiol. 2015, 61, 381–387. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, Y.; Li, K.; Hu, X.; Wang, C.; Cao, G.; Xue, R.; Gong, C. The complete genome of Cyprinid herpesvirus 2, a new strain isolated from Allogynogenetic crucian carp. Virus Res. 2018, 256, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Hirono, I.; Kurokawa, K.; Fukuda, H.; Nahary, R.; Eldar, A.; Davison, A.J.; Waltzek, T.B.; Bercovier, H.; Hedrick, R.P. Genome sequences of three koi herpesvirus isolates representing the expanding distribution of an emerging disease threatening koi and common carp worldwide. J. Virol. 2007, 81, 5058–5065. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Treangen, T.J.; Messeguer, X.; Perna, N.T. Analyzing patterns of microbial evolution using the mauve genome alignment system. Methods Mol. Biol. 2007, 396, 135–152. [Google Scholar]
- McGeoch, D.J. Molecular evolution of large DNA viruses of eukaryotes. Semin. Virol. 1992, 3, 399–409. [Google Scholar]
- Nishiyama, Y. Herpes simplex virus gene products: The accessories reflect her lifestyle well. Rev. Med. Virol. 2004, 14, 33–46. [Google Scholar] [CrossRef]
- Isomura, H.; Stinski, M.F. Coordination of late gene transcription of human cytomegalovirus with viral DNA synthesis: Recombinant viruses as potential therapeutic vaccine candidates. Expert Opin. Ther. Targets 2013, 17, 157–166. [Google Scholar] [CrossRef]
- Boyne, J.R.; Whitehouse, A. γ-2 Herpes virus post-transcriptional gene regulation. Clin. Microbiol. Infect. 2006, 12, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Lumangtad, L.A.; Bell, T.W. The signal peptide as a new target for drug design. Bioorg. Med. Chem. Lett. 2020, 30, 127115. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ning, S. New Look of EBV LMP1 Signaling Landscape. Cancers 2021, 13, 5451. [Google Scholar] [CrossRef] [PubMed]
- Patané, J.S.L.; Martins, J., Jr.; Rangel, L.T.; Belasque, J.; Digiampietri, L.A.; Facincani, A.P.; Ferreira, R.M.; Jaciani, F.J.; Zhang, Y.; Varani, A.M.; et al. Origin and diversification of Xanthomonas citri subsp. citri pathotypes revealed by inclusive phylogenomic, dating, and biogeographic analyses. BMC Genom. 2019, 20, 700. [Google Scholar] [CrossRef]
- Yancopoulos, S.; Attie, O.; Friedberg, R. Efficient sorting of genomic permutations by translocation, inversion and block interchange. Bioinformatics 2005, 21, 3340–3346. [Google Scholar] [CrossRef]
- Lunter, G.; Ponting, C.P.; Hein, J. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput. Biol. 2006, 2, e5. [Google Scholar] [CrossRef]

| Virus | Size (bp) | Nucleotide Composition (%) | No. of ORFs | Identity (%) k | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genome | U g | TR h | G+C | Genome i | Unique j | U g | TR h | |||
| CyHV-2 | SH-01 | 290,428 | 260,586 | 14,921 | 51.60 | 154 | 150 | 146 | 4 | *** |
| ST-J1 a | 290,304 | 260,238 | 15,033 | 51.70 | 154 | 150 | 146 | 4 | 99.60 | |
| SY-C1 b | 289,365 | 259,555 | 14,905 | 51.60 | 143 | 140 | 137 | 3 | 98.53 | |
| SY c | 290,455 | 259,749 | 15,353 | 51.60 | 154 | 150 | 146 | 4 | 98.35 | |
| CyHV-1 a | 291,144 | 224,784 | 33,180 | 51.30 | 143 | 137 | 131 | 6 | 42.72 | |
| CyHV-3 d | 295,146 | 250,208 | 22,469 | 59.20 | 163 | 155 | 147 | 8 | 44.54 | |
| AngHV-1 e | 248,526 | 227,258 | 10,634 | 53.00 | 134 | 129 | 124 | 5 | 36.52 | |
| CaHV f | 275,348 | - | - | 51.73 | 150 | 150 | - | - | 92.63 | |
| SH-01 | CyHV-1 | Change (bp) b | CyHV-3 | Change (bp) b | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LCB | Position | Length (bp) | LCB | Position | Length (bp) | LCB | Position | Length (bp) | ||
| LCB1 | 17,215–20,413 | 3199 | LCB1 | 35,374–38,196 | 2823 | −376 | LCB1 | 25,754–29,079 | 3326 | +127 |
| LCB2 | 20,806–35,267 | 14,462 | LCB4 | 47,773–49,683 | 1911 | −12,551 | LCB3 | 38,157–49,904 | 11,748 | −2714 |
| LCB3 | 39,346–44,586 | 5241 | LCB3 | 41,045–43,535 | 2491 | −2750 | LCB2 | 32,129–37,868 | 5740 | +499 |
| LCB4 | 44,914–195,079 | 150,166 | LCB5 | 51,890–183,835 | 131,946 | −18,220 | LCB4 | 50,081–205,795 | 155,715 | +5549 |
| LCB5 | 195,702–204,934 | 9233 | LCB6 | 185,864–193,284 | 7421 | −1812 | LCB5 | 207,192–218,326 | 11,135 | +1902 |
| LCB6 | 207,694–208,337 | 644 | LCB2 | 38,663–39,134 | 472 | −172 | LCB8 | 242,836–243,294 | 459 | −185 |
| LCB7 | 209,037–220,265 | 11,229 | none a | none a | none a | −11,229 | LCB6 | 221,009–231,789 | 10,781 | −448 |
| LCB8 | 220,500–231,255 | 10,756 | LCB7 | 212,442–222,389 | 9948 | –808 | LCB9 | 243,510–255,637 | 12,128 | +1372 |
| LCB9 | 231,395–238,734 | 7340 | LCB8 | 222,900–224,492 | 1593 | −5747 | LCB7 | 233,309–242,408 | 9100 | +1760 |
| LCB10 | 241,885–264,893 | 23,009 | LCB9 | 233,688–253,332 | 19,645 | −3364 | LCB10 | 258,414–274,200 | 15,787 | −7222 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Wen, J.; Xiao, S.; Wei, C.; Yu, F.; Roengjit, P.; Lu, L.; Wang, H. Complete Genome and Molecular Characterization of a New Cyprinid Herpesvirus 2 (CyHV-2) SH-01 Strain Isolated from Cultured Crucian Carp. Viruses 2022, 14, 2068. https://doi.org/10.3390/v14092068
Yang J, Wen J, Xiao S, Wei C, Yu F, Roengjit P, Lu L, Wang H. Complete Genome and Molecular Characterization of a New Cyprinid Herpesvirus 2 (CyHV-2) SH-01 Strain Isolated from Cultured Crucian Carp. Viruses. 2022; 14(9):2068. https://doi.org/10.3390/v14092068
Chicago/Turabian StyleYang, Jia, Jinxuan Wen, Simin Xiao, Chang Wei, Fei Yu, Patarida Roengjit, Liqun Lu, and Hao Wang. 2022. "Complete Genome and Molecular Characterization of a New Cyprinid Herpesvirus 2 (CyHV-2) SH-01 Strain Isolated from Cultured Crucian Carp" Viruses 14, no. 9: 2068. https://doi.org/10.3390/v14092068
APA StyleYang, J., Wen, J., Xiao, S., Wei, C., Yu, F., Roengjit, P., Lu, L., & Wang, H. (2022). Complete Genome and Molecular Characterization of a New Cyprinid Herpesvirus 2 (CyHV-2) SH-01 Strain Isolated from Cultured Crucian Carp. Viruses, 14(9), 2068. https://doi.org/10.3390/v14092068





